Abstract
HDAC1 and -2 are highly conserved enzymes and often coexist in the same coregulator complexes. Understanding the regulation of histone deacetylase activities is extremely important because these enzymes play key roles in epigenetic regulation in normal and cancer cells. We previously showed that HDAC1 is required for glucocorticoid receptor-mediated transcription activation and that its activity is regulated through acetylation by p300 during the induction cycle. Here, we showed that HDAC2 is also required for glucocorticoid receptor-mediated gene activation. HDAC2, however, is regulated through a different mechanism from that of HDAC1. HDAC2 is not acetylated by p300, although 5 of 6 acetylated lysine residues in HDAC1 are also present in HDAC2. More importantly, the activity of HDAC2 is inhibited by acetylated HDAC1. Additionally, we showed that acetylated HDAC1 can trans-regulate HDAC2 through heterodimerization. Thus, this study uncovered fundamental differences between HDAC1 and HDAC2. It also unveiled a new mechanism of collaborative regulation by HDAC1/2 containing coregulator complexes.
Introduction
Histone deacetylases are responsible for the deacetylation of lysine residues at the N-terminal part of the core histones as well as at non-histone proteins. Histone deacetylation plays an important role in transcriptional regulation, cell cycle progression, and developmental events and is often linked to epigenetic repression (1–4). However, emerging evidence also suggests that histone deacetylase activity may be required for transcriptional activation (5–10) and for preventing the cryptic initiation of transcription (11). HDAC1 and HDAC2 belong to class I histone deacetylases and often coexist in multicomponent protein complexes (12). They are closely related enzymes with an 82% overall sequence identity and can partially compensate for each others' functions (13–17). HDAC1 and -2 can also undergo differential post-translational modification, such as phosphorylation and sumoylation (18–20).
We recently reported that HDAC1 is required for glucocorticoid receptor (GR)2-mediated transcription activation. Furthermore, we showed that HDAC1 is acetylated after its association with the GR, and this acetylation event correlates with a change in promoter activity (8). The acetylated HDAC1 loses its deacetylase activity. The majority of acetylated lysine residues of HDAC1 are present in the C-terminal regulatory domain, which reveals only partial homology to the corresponding region of HDAC2. The C-terminal domain of HDAC1 does not have any catalytic activity. The deletion of this region, however, greatly reduces deacetylase activity (21), indicating that the C-terminal domain plays an indispensable role in the regulation of deacetylase activity. Therefore, it is important to compare the C-terminal regions of HDAC1 and HDAC2 functionally and structurally and to determine how these domains collaborate in the regulation of transcription. In this study, we show that HDAC2 is also important for GR-mediated gene activation; however, in contrast to HDAC1, HDAC2 is not acetylated. However, a chimeric protein, in which the C-terminal tail of HDAC2 was swapped with the corresponding region of HDAC1, can be acetylated. More importantly, we showed that acetylated HDAC1 not only inhibits its own deacetylase activity, but also, through direct interaction with HDAC2, abolishes the deacetylase activity of HDAC2. Furthermore, this study provides further insight into the regulation of the deacetylase activity of HDAC1/2 containing coregulator complexes.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The CherryFP-NF1A1.1 (ChFP-NF1) construct was generated by ligating NF1A1.1 cDNA (kindly provided by R. M. Gronostajski, State University of New York, Buffalo) to a ChFP-C1 vector, which was generated by replacing green fluorescent protein in the pEGFP-C1 (Clontech) vector with Cherry fluorescent protein (22). The GFP-HDAC1 plasmids were described previously (8). The GFP-HDAC2 construct was generated by subcloning mouse HDAC2 into a GFP-C1 vector. HDAC1 K432R, HDAC2 R433K, HDAC1 H141A, and HDAC2 H142A were constructed using site-directed mutagenesis according to the manufacturer's protocol (Agilent Technologies, Santa Clara, CA). HDAC1 and HDAC2 tail swapping mutants were constructed by first creating an EcoRI site at amino acids 419 and 420 of HDAC1 and 420 and 421 of HDAC2 without altering the amino acid sequence. The mutant constructs were then digested with EcoRI and the C-terminal cloning enzyme XhoI to release the C-terminal fragment of HDAC1 or HDAC2, respectively. The purified vector that contained HDAC1-(1–420) was then ligated with a purified HDAC2 C-terminal fragment to create HDAC1–2T. HDAC2–1T was created using the same method. GST HDAC1-(1–67) was constructed by PCR. The integrity of all constructs was confirmed by DNA sequencing.
Cell Culture and Transient Transfections with Small Interfering RNA and Plasmid DNA
The 3134 cell line, as well as its derivative 3617 (expressing GFP-GR), was described previously (23). Cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cell line 6062, an NIH 3T3 derivative, which contains stably integrated copies of the MMTV promoter driving a luciferase reporter, was grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. For HDAC1 and HDAC2 small interfering RNA transfections, 6062 cells were seeded in 12-well plates at 5 × 104 cells per well ∼24 h before transfection. HDAC1 and HDAC2 small interfering RNA were transfected with Oligofectamine according to the manufacturer's protocol (Invitrogen). Cells were incubated for 48 h and harvested for Western blot analysis to evaluate HDAC1 and HDAC2 levels, or the cells were treated with Dex (100 nm) and subjected to analysis for luciferase expression levels.
Stable Knockdown of HDAC2 with shRNA
HDAC2 siRNA sequences were cloned into pSuper retroviral vectors, and the recombinant virus was collected and used to infect 3134 cells. The single cell with a stable incorporation of HDAC2 shRNA was selected to produce a stable cell line.
Live Cell Imaging
Prior to live cell imaging, 3617 cells, transiently transfected by electroporation with green fluorescent protein-tagged HDAC1 or HDAC2 and ChRFP-NF1, were transferred to 35-mm glass bottom dishes (MatTek Corp., Ashland, MA) at a density of 2 × 105 cells/ml in phenol red-free Dulbecco's modified Eagle's medium containing 10% charcoal-stripped fetal bovine serum (Hyclone, Logan, UT) and 5 mg/ml tetracycline to suppress GFP-GR expression. The cells were cultured for at least 16 h and were subsequently treated with Dex (100 nm) for 1 h. All live imaging experiments were carried out on a Zeiss 510 confocal microscope with 100× 1.3-N.A. oil immersion objective, and the cells were kept at 37 °C using an air stream stage incubator (Nevtek, Williamsville, VA).
Protein Purification
Expression and purification of FLAG-tagged HDAC1, HDAC2, and mutants from baculovirus, as well as the in vitro acetylation assays, were performed as described previously (8).
In Vitro Deacetylation Assay
3H-Labeled acetylated histones were isolated from Mel cells as described previously (24, 25). Deacetylation assays were carried out as described previously (8).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation against HDAC1 and HDAC2 was performed in control and knockdown cells as described elsewhere (Upstate Biotechnology, Inc.). Briefly, soluble chromatin was immunoprecipitated with antibodies to HDAC1 and HDAC2. DNA isolated from precipitates at various time points was used as a template for PCR amplification. The following primers were used for amplification of the MMTV promoter: sense, 5′-GCGGTTCCCAAGGCTTAAGTAAGTTT-3′, and antisense, 5′-GGTGAATGTTAGGACTGTTGCAAGTT-3′.
Immunoprecipitation and Western Blot
The transfection was performed with Lipofectamine 2000 (Invitrogen). 24–48 h after transfection, the cells were harvested and lysed in a lysis buffer (20 mm Hepes, pH 7.6, 20% glycerol, 1.5 mm MgCl2, 0.2 mm EDTA, 0.1% Triton X-100, 1 mm dithiothreitol) supplemented with a protease inhibitor mixture (Roche Applied Science). The immunoprecipitation and Western blot assays were performed as described previously (8).
In Vitro GST Pulldown Assay
Bacteria-expressed GST or GST-HDAC1 proteins were immobilized on glutathione-Sepharose 4B beads (Pfizer, New York) and washed. The beads were then incubated with 35S-labeled, in vitro-synthesized HDAC1 and HDAC2 proteins in a GST binding buffer (150 mm NaCl, 50 mm Tris-HCl, pH 8.0, 2 mm EDTA, 0.1% Nonidet P-40, and protease inhibitor mixture). The beads were then washed with the binding buffer, and the bound proteins were separated by SDS-PAGE. The dried gels were then subjected to autoradiography.
RESULTS
HDAC2 Is Required for GR-mediated Transcription Activation
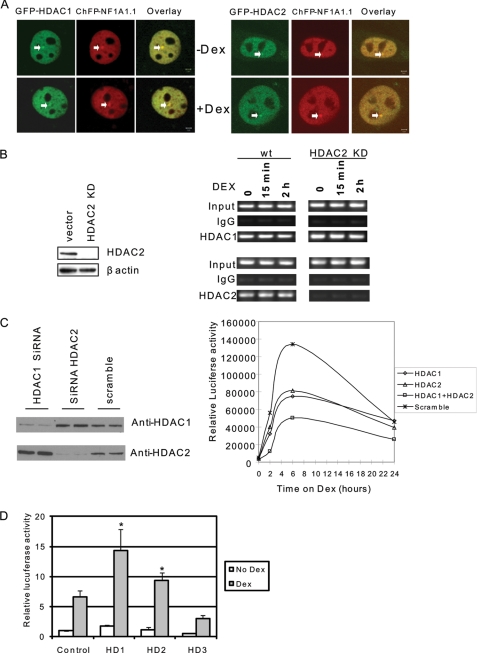
Previously, we found that HDAC1 is required for GR-mediated MMTV gene activation (8). Because HDAC1 and HDAC2 often coexist in the same coregulator complex and HDAC2 can interact with GR (26), we investigated whether HDAC2 is also required for MMTV transcription. First, we determined whether HDAC2 is localized on MMTV chromatin. A GFP-HDAC2 expression construct was cotransfected into mouse mammary tumor cell line 3134 together with a DNA construct expressing ChFP-NF1A1.1, which binds to and identifies MMTV promoter sequences. The 3134 cell line contains ∼200 copies of an MMTV-reporter construct that is stably integrated at one position on chromosome 4 (27). Just like GFP-HDAC1, GFP-HDAC2 clearly localizes to the MMTV array (Fig. 1A). We next examined whether the recruitment of HDAC1 or HDAC2 relies on each other. The stable HDAC2 knockdown cell lines were established by shRNA. The chromatin immunoprecipitation results confirmed that in a vector control cell line, both HDAC1 and HDAC2 bind to the promoter (Fig. 1B). In the HDAC2 knockdown cells, no HDAC2 binding was detected. HDAC1, however, binds to the promoter at a level comparable with the vector control cells, indicating that a lack of HDAC2 in the promoter does not affect the binding of HDAC1 (Fig. 1B). To determine whether HDAC2 regulates MMTV transcription, HDAC2 was transiently knocked down in cell line 6062, an NIH 3T3-based cell line harboring a stably integrated MMTV luciferase reporter construct (8). The efficient knockdown of HDAC2 resulted in a slight increase in HDAC1 expression (Fig. 1C). Importantly, MMTV transcription was reduced by ∼40% by either HDAC1 or HDAC2 knockdown. The combined knockdown of both HDAC1 and HDAC2 further reduced MMTV transcription indicating that HDAC1 and HDAC2 act synergistically in the transcription activation of the MMTV gene promoter (Fig. 1C). These results suggest that HDAC1 and HDAC2 play nonredundant roles in activating the MMTV promoter. To further dissect the role of HDAC2 in MMTV transcription, an HDAC2 expression plasmid was transfected into 6062 cells. The results showed that HDAC2 can activate MMTV transcription just as HDAC1 can. In contrast, HDAC3, which is associated with NcoR/SMRT corepressor complexes, repressed MMTV transcription (Fig. 1D).
FIGURE 1.
Both HDAC1 and HDAC2 are required for MMTV transcription. A, HDAC1 and HDAC2 are localized on MMTV arrays. HDAC1 or HDAC2 were cotransfected with mCherry-NF1A1.1 in cell line 3617, which contains the MMTV array. Images were acquired using green (GFP-HDAC1) and red (ChFP NF1A1.1) fluorescence of the cells before and after 30 min of induction with Dex. NF1A1.1 is present at the array before and after Dex treatment and is used here as a marker for the array. The merged image shows the colocalization of HDAC1 or HDAC2 and NF1A1.1 at the MMTV array. Scale bars in the merged panels indicate 2 μm. B, knockdown of HDAC2 does not prevent the recruitment of HDAC1 on the MMTV promoter. 3134 cells, which contain stably incorporated MMTV promoter arrays, were stably transfected with shRNA of HDAC2. The HDAC2 levels were evaluated by Western blot (left panel) and chromatin immunoprecipitation using antibodies against HDAC1 and HDAC2 in control and knockdown cells (right panel). wt, wild type. C, HDAC1 and HDAC2 are required for MMTV transcription. Mouse NIH 3T3 cells containing stably integrated copies of an MMTV-luciferase reporter (cell line 6062) were transfected with a small interfering RNA pool directed against mouse HDAC1, HDAC2, or a scrambled RNA control. Cells were harvested for Western blot analysis to evaluate HDAC1 and HDAC2 levels (left panel) or treated with Dex and subjected to luciferase analysis to examine MMTV expression levels (right panel). This expression analysis is a representative example from four separate experiments. D, overexpression of HDAC1 or HDAC2 activates MMTV transcription. HDAC1, HDAC2, or HDAC3 in expression vectors were transfected into 6062 cells. Cells were treated with or without Dex for 6 h. The luciferase activity was determined. Each data bar represents the mean ± S.D. from triplicate measurements. Asterisk indicates significant difference (Student's t test, p < 0.01).
HDAC2 Is Not Acetylated by p300
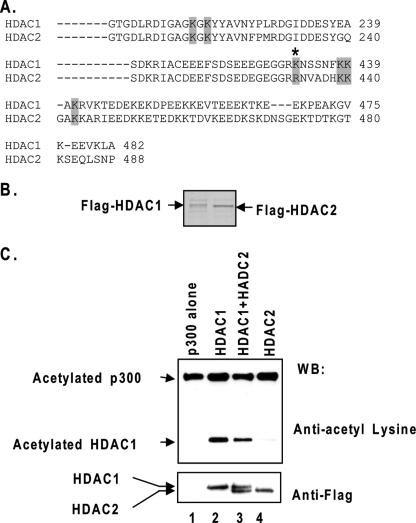
We next investigated whether HDAC2 can also be acetylated. The homology between HDAC1 and -2 is particularly high in the N-terminal catalytic domain and somewhat lower in the C-terminal region. HDAC1 is acetylated at six lysine residues, four of which are located in the C-terminal region (Fig. 2A) (8). Among these four acetylated lysines (lysines 432, 438, 439, and 441), three are present in HDAC2, whereas lysine 432 in HDAC1 corresponds to arginine 433 in HDAC2 (Fig. 2A). FLAG-tagged HDAC1 and HDAC2 were purified from baculovirus-infected Sf9 cells through affinity chromatography (Fig. 2B). HDAC1 and HDAC2 proteins were then incubated with recombinant p300 in the presence of acetyl-CoA and characterized for the acetylation status by Western blot analysis with an anti-acetyl-lysine antibody. As shown in Fig. 2C, the incubation with p300 induced a significant level of acetylation of HDAC1 but not of HDAC2 in the presence or absence of HDAC1.
FIGURE 2.
HDAC2 is not acetylated by p300. A, sequence comparison of human HDAC1 and HDAC2. The six acetylated lysines in HDAC1 are highlighted. The asterisk indicates that the lysine 432 in HDAC1 is not present in HDAC2. B, recombinant HDAC1 and HDAC2 in Coomassie-stained SDS gel. FLAG-tagged HDAC1 and HDAC2 were both expressed from baculovirus vectors in insect Sf9 cells and purified by affinity chromatography. C, HDAC2 is not acetylated by p300 in vitro. FLAG-tagged HDAC1, HDAC2, or both were incubated with purified p300 and acetyl-CoA. The products were then analyzed by SDS-PAGE and Western-blotted (WB) with acetyl-lysine and FLAG-specific antibodies.
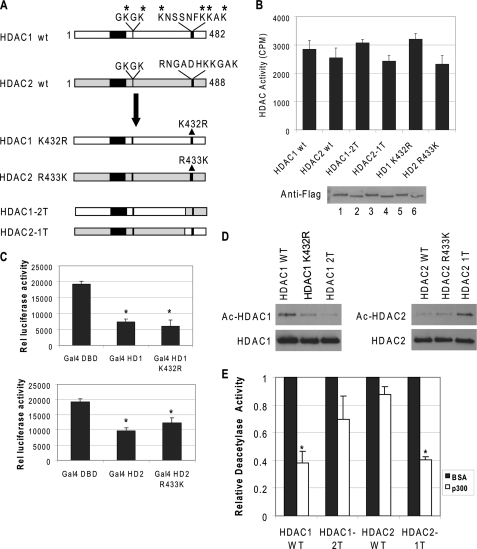
Because acetylated lysine 432 in HDAC1 is not present in HDAC2 (Fig. 2A), it is possible that lysine 432 may be critical for HDAC1 acetylation, and arginine 433 of HDAC2 may be the key residue that prevents HDAC2 acetylation. To test these possibilities, lysine 432 in HDAC1 was mutated to arginine, and arginine 433 in HDAC2 was changed to lysine (Fig. 3A). The mutant proteins were then expressed and purified from baculovirus-infected Sf9 cells. Because the C-terminal regions of HDAC1 and HDAC2 function as regulatory domains (21), it is important to determine that the mutation per se does not affect the enzymatic function. We first showed that the mutation alone does not change deacetylase activity of HDAC1 or HDAC2 in vitro. The histone deacetylase activities of these mutants were determined and compared with their wild type counterparts (Fig. 3B). We also determined that the mutations do not alter the transcription repression effects of HDAC1 and HDAC2 in vivo. HDAC1 K432R and HDAC2 R433K were subcloned into the GAL4 vector and cotransfected with a thymidine kinase minimum promoter driven luciferase reporter containing four GAL4 DNA-binding sites. The repression effect of the mutant proteins did not change when compared with the wild type proteins (Fig. 3C). These results indicate that the lysine and arginine substitution mutations at the HDAC1 and HDAC2 C-terminal regions did not affect their activities and functions. Next, we tested whether these mutations affect acetylation modification. The results showed that the acetylation of HDAC1 K432R by p300 is reduced. In contrast, the HDAC2 R433K mutant protein became acetylated by p300 (Fig. 3D).
FIGURE 3.
C-terminal sequences of HDAC1 are important for acetylation. A, schematic representation of wild type (wt) and mutant HDAC1 and HDAC2 constructs. B, mutations on HDAC1 or HDAC2 do not affect deacetylase activity. Equal amounts of purified proteins were used to determine deacetylase activity and were analyzed by Western blot using FLAG antibody. The results are the average of three independent experiments ± S.D. Bottom panel, the level of recombinant proteins by Coomassie stain. Lane 1, wild type HDAC1; lane 2, wild type (wt) HDAC2; lane 3, HDAC1–2T; lane 4, HDAC2–1T; lane 5, HDAC1 K432R; and lane 6, HDAC2 R433K. C, mutations do not affect HDAC1 or HDAC2 function in vivo. NIH 3T3 cells were transfected with a luciferase reporter driven by a minimal thymidine kinase promoter with four copies of the GAL4 DNA-binding sites. The fusions of the GAL4 DNA binding domain to HDAC1 (top panel), HDAC2 (bottom panel), or various mutants were cotransfected, and the effects on luciferase expression were determined. The results are the average of three independent experiments ± S.D. D, C-terminal sequences of HDAC1 are important for acetylation. FLAG-tagged HDAC1, HDAC2, or mutant proteins were incubated with purified p300 and acetyl-CoA. The products were then analyzed by Western blot with antibodies specific for acetyl-lysine and HDAC1 (left panel) or HDAC2 (right panel). E, acetylated HDAC2–1T lost deacetylase activity. The wild type and mutant proteins were acetylated with p300 or mock-acetylated with bovine serum albumin. The reaction products were then subjected to deacetylase assay. The relative deacetylase activity of HDACs treated with p300 was normalized with the deacetylase activity of the enzyme treated with bovine serum albumin. The results are the average of three independent experiments ± S.D. Asterisk indicates significant difference (Student's t test, p < 0.01).
The observation that HDAC1 K432R only partially lost acetylation modification and HDAC2 R433K was only acetylated at low levels together with the fact that the C terminus is not as highly conserved between HDAC1 and HDAC2 led to the possibility that sequences flanking the acetylation sites are also required for the efficient acetylation of HDAC1. We next exchanged the C-terminal tails of HDAC1 and HDAC2 by ligating the N terminus of HDAC1 with the C-terminal tail of HDAC2 (HDAC1–2T) and vice versa (HDAC2–1T) (Fig. 3A). These mutated proteins are fully functional enzymes (Fig. 3B). However, HDAC1–2T cannot be acetylated by p300, whereas HDAC2–1T was acetylated at levels comparable with that of the wild type HDAC1 counterpart (Fig. 3D). This result indicates that although Lys-432 is an important residue for HDAC1 acetylation and Arg-433 is a critical residue for HDAC2 to prevent HDAC2 acetylation, the C-terminal flanking sequences around these residues are also important for acetylation modifications. To further test whether acetylated HDAC2–1T lost deacetylase activity as did wild type HDAC1, the purified wild type and tail swapping mutants of HDAC1 and HDAC2 proteins were incubated with p300. The reaction products were tested for deacetylase activity using acetylated histones. The deacetylase activity of acetylated HDAC2–1T was significantly inhibited (Fig. 3E). This result indicates that the C-terminal domain of HDAC1 is important for regulating the deacetylase activity of HDAC1. When the C-terminal region of HDAC1 was swapped for HDAC2, HDAC2–1T functions just like HDAC1, whereas HDAC1–2T behaved just like wild type HDAC2.
HDAC1 Acetylation Inhibits HDAC2 Activity
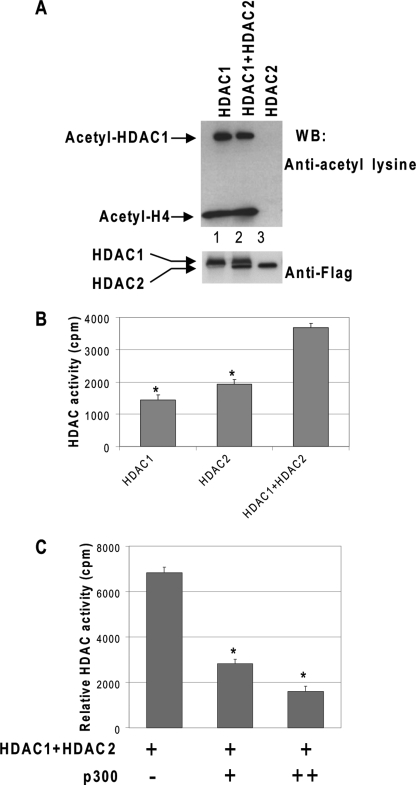
Because HDAC1 and HDAC2 coexist in many corepressor complexes and HDAC2 is not acetylated although HDAC1 is, we investigated if and how HDAC2 activity is regulated. We first examined whether acetylated HDAC1 affects the deacetylase activity of HDAC2. Purified HDAC1 and/or HDAC2 proteins were incubated with p300 proteins in the presence of acetyl-CoA. The acetyl-CoA was then removed by dialysis, and acetylated core histones were subsequently added as substrates. As expected, HDAC1 was acetylated, and its deacetylase activity was inhibited by acetylation (Fig. 4A, lane 1). HDAC2 itself was not acetylated; therefore, its deacetylase activity was not affected by the reaction (Fig. 4A, lane 3). However, in the presence of acetylated HDAC1, HDAC2 was unable to deacetylate core histones (Fig. 4A, lane 2), despite the fact that the wild type nonacetylated HDAC1 and HDAC2 have an additive effect on deacetylase activity (Fig. 4B). The reduction of deacetylase activities from both enzymes correlated with increased concentration of p300 in the reaction, presumably due to the increased HDAC1 acetylation levels (Fig. 4C). This result indicates that acetylated HDAC1 not only inhibits its own deacetylase activity but also the deacetylase activity of HDAC2.
FIGURE 4.
Acetylated HDAC1 inhibits HDAC2 activity through direct interaction. A, acetylated HDAC1 inhibits the deacetylase activity of HDAC2. Recombinant HDAC1, HDAC2, or both were incubated with p300 in the presence of acetyl-CoA. After the acetylation reaction, the acetyl-CoA was removed through dialysis, and acetylated histone was added to examine the overall deacetylase activity. The levels of HDAC1 protein and histone acetylation were examined by Western blot (WB) with anti-acetyl-lysine antibody. HDAC1 or HDAC2 protein in the reactions was also determined by Western blot with FLAG antibody. B, HDAC1 and HDAC2 have an added effect on deacetylase activity. Purified HDAC1, HDAC2, or HDAC1 plus HDAC2 were tested for deacetylase activity by incubating them with tritium-labeled acetylated histone. Each data bar represents the mean ± S.D. from triplicate measurements. C, recombinant HDAC1 and HDAC2 were incubated with an increased amount of p300 for acetylation. Bovine serum albumin was added to normalize the total protein content in the reaction. The reaction mixture was then tested for deacetylase activity by incubating with tritium-labeled acetylated histone. Each data bar represents the mean ± S.D. from triplicate measurements. Asterisk indicates significant difference (Student's t test, p < 0.01).
HDAC1 Regulates HDAC2 Activity through Dimerization
It has been shown that the N-terminal region of HDAC1 can directly interact with HDAC1 and -2 in vitro (28). We further determined the minimum sequences of HDAC1 and -2 interaction domains. The GST pulldown experiments were performed with 35S-labeled HDAC1 or -2 and GST HDAC1 deletion mutants. The results show that HDAC1 amino acids 20–40 are important for HDAC1 and -2 interactions (Fig. 5A). Interestingly, the sequences within this region are identical between HDAC1 and HDAC2. This result indicates that HDAC1 and -2 can form homo- and heterodimers with similar affinities. To test whether the inhibition of HDAC2 activity is mediated by direct interactions with acetylated HDAC1, and not by p300, we tested whether HDAC1 mutants that mimic acetylated HDAC1 (HDAC1 6Q, see Ref. 8) can inhibit the activity of HDAC2 in the absence of p300. Like acetylated HDAC1, HDAC1 6Q had no deacetylase activity. In this experiment, increased concentrations of recombinant HDAC1 6Q were incubated with wild type HDAC2. The HDAC1 6Q/HDAC2 mixture was then tested for deacetylase activity. The result showed that HDAC1 6Q inhibits the deacetylase activity of HDAC2 in a dose-dependent manner (Fig. 5B). This result indicates that acetylated HDAC1 can directly inhibit the deacetylase activity of HDAC2. The acetylated HDAC1 also inhibited the deacetylase activity of nonacetylated HDAC1 in a similar manner (Fig. 5C). These results suggest that acetylated HDAC1 can directly inhibit the deacetylase activity of HDAC2 or HDAC1.
FIGURE 5.
HDAC1 regulates HDAC2 activity through dimerization. A, identification of HDAC1 and -2 interaction domain. Top panel, GST pulldown with 35S-labeled HDAC1 and HDAC2. The amount of GST HD1 protein used for the assay is shown with Coomassie stain. Bottom panel, alignment of HDAC1 and -2 N-termini, showing that HDAC1 and HDAC2 interaction domains, which are located at amino acids 20–40, are identical between HDAC1 and -2. B, mutation mimicking acetylated HDAC1 (HDAC1 6Q) inhibits the deacetylase activity of HDAC2. Recombinant HDAC2 was incubated with an increased amount of HDAC1 6Q first, and then acetylated histone was added to examine the overall deacetylase activity on acetylated histone H3 and acetylated histone H4. The levels of histone loading on each lane were determined by Western blot (WB) using histone H4 antibody. HDAC1 6Q and HDAC2 levels were determined by Western blot with HDAC1- and HDAC2-specific antibodies, respectively. Lane 1, acetylated histone input; lanes 2–4, increased amount of HDAC1 6Q in the reaction; lane 5, HDAC2 alone; lanes 6–8, HDAC2 with an increased amount of HDAC1 6Q in the reaction. C, HDAC1 6Q inhibits deacetylase activity of HDAC1. Lanes 1–3 increased amount of HDAC1 6Q in the reaction; lane 4, HDAC2 alone; lanes 5–7, HDAC2 with an increased amount of HDAC1 6Q in the reaction. D, loss of deacetylase activity on recombinant mutant proteins, HDAC1 H141A and HDAC2 H142A. Equal amounts of recombinant HDAC1, HDAC2, or mutant proteins were tested for deacetylase activity. Each data bar represents the mean ± S.D. from triplicate measurements. WT, wild type. E, HDAC1 H141A inhibits the deacetylase activity of HDAC2. The recombinant HDAC2 was incubated with an increased amount of HDAC1 point mutant H141A, and then acetylated histone was added to examine the overall deacetylase activity. Lane 1, acetylated histone input; lanes 2–4, increased amount of HDAC1 H141A in the reaction; lanes 5–7, HDAC2 with an increased amount of HDAC1 H141A in the reaction; lane 8, HDAC2 alone. F, HDAC2 H142A inhibits the deacetylase activity of HDAC1. The recombinant HDAC1 was incubated with an increased amount of HDAC2 point mutant H142A and then tested for the overall deacetylase activity. Lanes 1–3, increased amount of HDAC2 H142A in the reaction; lane 4, HDAC1 alone; lanes 5–7, HDAC1 with an increased amount of HDAC2 H142A in the reaction. G, interaction of HDAC1 6Q with histones. Recombinant FLAG HDAC1 and FLAG HDAC1 6Q were incubated with acetylated core histones and subsequently immunoprecipitated (IP) with FLAG antibody. The coprecipitated histones were examined by Western blotting with antibodies as indicated. The levels of immunoprecipitated HDAC1 and 6Q were also determined with FLAG antibodies. H, truncated protein that contains only the dimerization domain of HDAC1 can inhibit the deacetylase activity of wild type HDAC1 protein. Left panel, GST and GST HDAC1-(1–67) were purified from E. coli. Right panel, recombinant HDAC1 protein was incubated with GST or GST HDAC1-(1–67), the reaction mix was then tested for deacetylase activity on histone H4.
Next, we addressed the mechanism of how acetylated HDAC1 inhibits HDAC1/2 activity. Although we showed that HDAC1 and HDAC2 can associate with each other via amino acids 20–40, it is unclear whether they form dimers or oligomers (28). Because the crystal structures of HDAC homologous proteins histone deacetylase-like protein, from Aquifex aeolicus, and HDAC8 show that they form dimers (29–31), we speculated that HDAC1 and HDAC2 form dimers as well, either homodimers or heterodimers. There are two scenarios by which acetylated HDAC1 can affect HDAC2 activity. First, the conformation changes in acetylated HDAC1 may affect the conformation and activity of HDAC2; second, the loss of HDAC1 deacetylase activity may directly interfere with the activity of HDAC2. If the loss of deacetylase activity of HDAC1 is the key for acetylated HDAC1 to inhibit HDAC2 deacetylase activity, then a deacetylase-defective HDAC1 should also influence HDAC2 activity. We used an HDAC1 mutant (HDAC1 H141A) that has a point mutation in the catalytic core that results in a loss of HDAC1 deacetylase activity (Fig. 5D) (32). When incubating the purified HDAC1 H141A with wild type HDAC2, HDAC1 H141A inhibits HDAC2 deacetylase activity in a dose-dependent manner just like HDAC1 6Q did (Fig. 5E). This result indicates that HDAC1 regulates HDAC2 activity through its own deacetylase activity. Similarly, the deacetylase-defective HDAC2 mutant protein, HDAC2 H142A (Fig. 5D), can inhibit the deacetylase activity of HDAC1 (Fig. 5F). These results suggest that deacetylase-defective HDAC1 or HDAC2 can influence the activity of their wild type counterpart through dimer formation or oligomerization.
There are two possibilities how the dimerization with defective mutants can inhibit wild type HDAC1 or HDAC2 deacetylase activity. One possibility is that HDAC1 6Q does not bind to histones. Thus, through dimerization with HDAC1 or -2, HDAC1 6Q sequesters wild type enzymes from histone substrates. We tested whether HDAC1 6Q can bind to histones. The FLAG-tagged HDAC1 or HDAC1 6Q was incubated with acetylated core histones. The reaction mixture was then immunoprecipitated with FLAG antibodies and Western-blotted for the presence of acetylated histone H4. The result shows that the wild type HDAC1 and 6Q can pull down similar amounts of acetylated H4 (Fig. 5G), indicating that 6Q does not inhibit the activity of wild type HDAC1 or -2 through sequestering them from the histone substrate. The second possibility is that two functional deacetylase domains are required to form an active HDAC1 homo- or heterodimer. Thus, HDAC1 6Q forms inactive dimers with wild type HDAC1 or -2. If this model is true, any dimer with only one functional deacetylase domain will not be active. We tested this model by examining the activity of dimers containing full-length wild type HDAC1 and truncated HDAC1 that contains only the dimerization domain. The first 67 amino acids of HDAC1 were expressed bacterially as a GST fusion protein. This region of HDAC1 interacts with HDAC1 or HDAC2 and does not contain a deacetylase domain (Fig. 5A) (28). When incubating GST HDAC1-(1–67) with HDAC1, it inhibited the deacetylase activity of HDAC1. In contrast, GST alone did not affect HDAC1 deacetylase activity (Fig. 5H). This result indicates that although GST HDAC1-(1–67) can dimerize with wild type HDAC1, the lack of functional deacetylase domains results in the loss of deacetylase activity from the dimer. This result support our model that active HDAC1/2 dimer requires two functional deacetylase molecules.
Acetylated HDAC1 Inhibits the Overall Deacetylase Activities of Corepressor Complexes
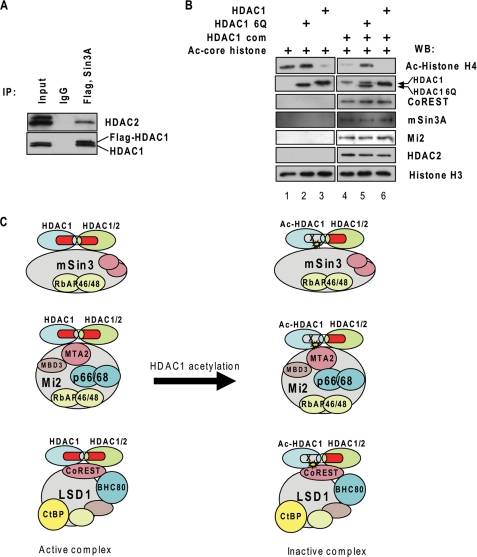
Given that HDAC1 can regulate HDAC2 enzymatic activity by forming heterodimers, we further tested whether HDAC1 could also regulate HDAC1/2-containing corepressor complex activity. First, we determined whether HDAC1 and -2 formed homo- and heterodimer or oligomer in the corepressor complexes. The nuclear extract of the 3134 cell line, which stably expressed FLAG-tagged HDAC1, was immunoprecipitated with FLAG antibody and eluted with FLAG peptide. The precipitant was subsequently immunoprecipitated with Sin3A antibody. The bound fraction was subjected to Western blot analysis. The results show that endogenous HDAC1 and HDAC2 are present in the HDAC1-Sin3 complex (Fig. 6A). This result indicates that FLAG-HDAC1 can associate with endogenous HDAC1 as homodimer or HDAC2 as heterodimer or oligomer in corepressor complexes. Next, we investigated whether HDAC1 6Q can repress the activity of endogenous HDAC1/2-containing corepressor complexes. HDAC1-containing protein complexes were purified through immunoprecipitation with FLAG antibodies from nuclear extracts of 3134 cells that stably expressed FLAG HDAC1. The purified complexes contain components of the mSin3A complex, the CoREST complex, and the NuRD complex (Fig. 6B, lane 4). The isolated protein complexes were incubated with HDAC1 6Q, and the overall deacetylase activity was determined by incubating the complexes with acetylated histones. HDAC1 6Q inhibited the overall deacetylase activity of the protein complexes (Fig. 6B, lanes 4 and 5). In contrast, wild type nonacetylated HDAC1 increased the overall deacetylase activity (Fig. 6A, lanes 4 and 6). This result indicates that HDAC1 6Q can interact with HDAC1 or HDAC2 in the complex to repress the deacetylase activity of HDAC1 and -2. Therefore, the acetylation of HDAC1 results in the repression of the overall deacetylase activity of HDAC1/2-containing corepressor complexes (Fig. 6C).
FIGURE 6.
HDAC1 6Q represses the deacetylase activity of the HDAC1-containing protein complexes. A, HDAC1 can form homo- or heterodimers in the corepressor complex. The nuclear extracts from 3134 cells, which overexpressed FLAG-tagged HDAC1, were prepared and subsequently immunoprecipitated (IP) with FLAG and mSin3A antibodies. The presence of HDAC2 as well as endogenous and FLAG-tagged HDAC1 was examined by Western blot. B, HDAC1 containing protein complexes were purified from 3134 cells overexpressing FLAG-tagged HDAC1. HDAC1 complexes were then incubated with purified HDAC1 or HDAC1 6Q and acetylated histones to examine their overall deacetylase activity. The wild type HDAC1 from the complex and recombinant HDAC1 6Q levels were examined by Western blot (WB) with HDAC1 antibody. The presence of HDAC2, mSin3A, Mi2, or CoREST from the complexes were also examined. Lane 1, acetylated histone input; lane 2, HDAC1 6Q input; lane 3, HDAC1 input; lane 4, HDAC1 complex alone; lane 5, HDAC1 complex with HDAC1 6Q in the reaction; lane 6, HDAC1 complex with wild type HDAC1 in the reaction. C, model of active and inactive HDAC1/2-containing complexes. Acetylation on HDAC1 inhibits the deacetylase activity of both HDAC1 and HDAC2, resulting in the inactivation of corepressor complexes.
DISCUSSION
Here we reported that HDAC1 and HDAC2 are regulated differently because HDAC1 can be acetylated and HDAC2 cannot. This differential regulation of HDAC1 and HDAC2 is due to a sequence difference at the C terminus of the two proteins. A major role for the acetylation of HDAC1 is exerted by lysine 432, which is an arginine at the corresponding position in HDAC2. More importantly, we showed that the acetylation of HDAC1 inhibits the activity of itself and also the activity of HDAC2. Interestingly, deacetylase-defective mutants of HDAC1, such as HDAC1 H141A, can also inhibit HDAC2 activity, suggesting that acetylated HDAC1 inhibits HDAC2 activity due to the loss of the functional deacetylase domain. Thus, our study unveils important fundamental aspects of the structural and functional differences between HDAC1 and HDAC2, documenting that one deacetylase can trans-regulate the activity of another deacetylase.
Crystal structures of HDLP and HDAC8 show that both enzymes form homodimers with the catalytic pockets facing each other (29–31). The crystal structures of HDAC1 and HDAC2 have not yet been solved; however, the N-terminal dimerization domain L1 loop of HDLP shares strong sequence homology to that of HDAC1 and HDAC2 (29). We speculate that HDAC1 and HDAC2 can form homo- or heterodimers with the catalytic domains facing each other as is the case for HDLP and HDAC8. When one enzyme is inactive, it results in the inhibition of function for the other enzyme. To test this model, we will need to determine the crystal structures of HDAC1 and HDAC2. A recent report indicated that the catalytic activity of HDAC6 depends on both intact deacetylase domains, which appears to support our model (33).
Although HDAC1 and HDCA2 often associate with repressor complexes to inhibit transcription, in this study we showed that both HDAC1 and HDAC2 are activators for GR-mediated transactivation. It is unclear what complex they associate with when they function as coactivators. Current effort is directed at identifying the target of deacetylation for MMTV gene activation.
Some HDAC1- or HDAC2-containing complexes have more than one type of histone-modifying enzymes. The CoREST complex contains histone deacetylases and histone demethylase LSD1, and Mi2 complexes contain histone deacetylase and ATPase containing chromatin-remodeling enzymes. It has been reported that deacetylase activity is required for LSD1 demethylase activity on nucleosome substrates, and the histone deacetylase inhibitor, trichostatin A, can inhibit demethylase activity (34). It will be important to investigate whether HDAC1 acetylation is involved in regulating the demethylase activity of LSD1 as well as the remodeling activity of Mi2 within the complex.
HDAC1 can undergo several post-translational modifications, such as phosphorylation, sumoylation, and acetylation (8, 13, 16). All modifications occur at the C-terminal region. It is shown here that most of the mutations that affect the modification of HDAC1 reduce its deacetylase activity. Because all the modification sites are in close proximity, it will be interesting to investigate whether there is a cross-talk among different modifications. There is strong evidence that a cross-talk occurs between different post-translational modifications in histone and non-histone proteins (35–37). The study of cross-regulation among HDAC1 modifications will be an important topic for future studies. The in-depth study of the structures and functions of HDAC1 and HDAC2 may also provide further insight into developing inhibitors that target the unique domains of HDAC1 or HDAC2.
Acknowledgments
We thank Drs. Ed Seto and Stephen Sugrue for advice with the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL090589 and R01 HL091929 (to S. H.) and R01 DK52356 (to J. B.). This work was also supported by grants from the Florida Department of Health (to Y. Q. and S. H.).
- GR
- glucocorticoid receptor
- MMTV
- mouse mammary tumor virus
- shRNA
- short hairpin RNA
- GST
- glutathione S-transferase
- Dex
- dexamethasone.
REFERENCES
- 1.Xu L., Glass C. K., Rosenfeld M. G. (1999) Curr. Opin. Genet. Dev. 9, 140–147 [DOI] [PubMed] [Google Scholar]
- 2.Ng H. H., Bird A. (2000) Trends Biochem. Sci. 25, 121–126 [DOI] [PubMed] [Google Scholar]
- 3.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 4.Tsai C. C., Fondell J. D. (2004) Vitam. Horm. 68, 93–122 [DOI] [PubMed] [Google Scholar]
- 5.Cho Y., Griswold A., Campbell C., Min K. T. (2005) Genomics 86, 606–617 [DOI] [PubMed] [Google Scholar]
- 6.Chang S., Pikaard C. S. (2005) J. Biol. Chem. 280, 796–804 [DOI] [PubMed] [Google Scholar]
- 7.Reid G., Métivier R., Lin C. Y., Denger S., Ibberson D., Ivacevic T., Brand H., Benes V., Liu E. T., Gannon F. (2005) Oncogene 24, 4894–4907 [DOI] [PubMed] [Google Scholar]
- 8.Qiu Y., Zhao Y., Becker M., John S., Parekh B. S., Huang S., Hendarwanto A., Martinez E. D., Chen Y., Lu H., Adkins N. L., Stavreva D. A., Wiench M., Georgel P. T., Schiltz R. L., Hager G. L. (2006) Mol. Cell 22, 669–679 [DOI] [PubMed] [Google Scholar]
- 9.Zupkovitz G., Tischler J., Posch M., Sadzak I., Ramsauer K., Egger G., Grausenburger R., Schweifer N., Chiocca S., Decker T., Seiser C. (2006) Mol. Cell. Biol. 26, 7913–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith C. L. (2008) BioEssays 30, 15–24 [DOI] [PubMed] [Google Scholar]
- 11.Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 12.Yang X. J., Seto E. (2008) Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagger G., O'Carroll D., Rembold M., Khier H., Tischler J., Weitzer G., Schuettengruber B., Hauser C., Brunmeir R., Jenuwein T., Seiser C. (2002) EMBO J. 21, 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery R. L., Davis C. A., Potthoff M. J., Haberland M., Fielitz J., Qi X., Hill J. A., Richardson J. A., Olson E. N. (2007) Genes Dev. 21, 1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trivedi C. M., Luo Y., Yin Z., Zhang M., Zhu W., Wang T., Floss T., Goettlicher M., Noppinger P. R., Wurst W., Ferrari V. A., Abrams C. S., Gruber P. J., Epstein J. A. (2007) Nat. Med. 13, 324–331 [DOI] [PubMed] [Google Scholar]
- 16.Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 17.Barnes P. J. (2005) Expert Opin. Ther. Targets 9, 1111–1121 [DOI] [PubMed] [Google Scholar]
- 18.Pflum M. K., Tong J. K., Lane W. S., Schreiber S. L. (2001) J. Biol. Chem. 276, 47733–47741 [DOI] [PubMed] [Google Scholar]
- 19.Tsai S. C., Seto E. (2002) J. Biol. Chem. 277, 31826–31833 [DOI] [PubMed] [Google Scholar]
- 20.David G., Neptune M. A., DePinho R. A. (2002) J. Biol. Chem. 277, 23658–23663 [DOI] [PubMed] [Google Scholar]
- 21.Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. (1997) Cell 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 22.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 23.Walker D., Htun H., Hager G. L. (1999) Methods 19, 386–393 [DOI] [PubMed] [Google Scholar]
- 24.Carmen A. A., Rundlett S. E., Grunstein M. (1996) J. Biol. Chem. 271, 15837–15844 [DOI] [PubMed] [Google Scholar]
- 25.Huang S., Brandt S. J. (2000) Mol. Cell. Biol. 20, 2248–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito K., Yamamura S., Essilfie-Quaye S., Cosio B., Ito M., Barnes P. J., Adcock I. M. (2006) J. Exp. Med. 203, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer P. R., Fragoso G., Pennie W., Htun H., Hager G. L., Sinden R. R. (1999) J. Biol. Chem. 274, 28590–28597 [DOI] [PubMed] [Google Scholar]
- 28.Taplick J., Kurtev V., Kroboth K., Posch M., Lechner T., Seiser C. (2001) J. Mol. Biol. 308, 27–38 [DOI] [PubMed] [Google Scholar]
- 29.Finnin M. S., Donigian J. R., Cohen A., Richon V. M., Rifkind R. A., Marks P. A., Breslow R., Pavletich N. P. (1999) Nature 401, 188–193 [DOI] [PubMed] [Google Scholar]
- 30.Vannini A., Volpari C., Filocamo G., Casavola E. C., Brunetti M., Renzoni D., Chakravarty P., Paolini C., De Francesco R., Gallinari P., Steinkühler C., Di Marco S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vannini A., Volpari C., Gallinari P., Jones P., Mattu M., Carfí A., De Francesco R., Steinkühler C., Di Marco S. (2007) EMBO Rep. 8, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taunton J., Hassig C. A., Schreiber S. L. (1996) Science 272, 408–411 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Gilquin B., Khochbin S., Matthias P. (2006) J. Biol. Chem. 281, 2401–2404 [DOI] [PubMed] [Google Scholar]
- 34.Lee M. G., Wynder C., Bochar D. A., Hakimi M. A., Cooch N., Shiekhattar R. (2006) Mol. Cell. Biol. 26, 6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latham J. A., Dent S. Y. (2007) Nat. Struct. Mol. Biol. 14, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 36.Yang X. J., Seto E. (2008) Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurash J. K., Lei H., Shen Q., Marston W. L., Granda B. W., Fan H., Wall D., Li E., Gaudet F. (2008) Mol. Cell 29, 392–400 [DOI] [PubMed] [Google Scholar]