Abstract
Nascent mRNAs produced by transcription in the nucleus are subsequently processed and packaged into mRNA ribonucleoprotein particles (messenger ribonucleoproteins (mRNPs)) before export to the cytoplasm. Here, we have used the poly(A)-binding protein Nab2 to isolate mRNPs from yeast under conditions that preserve mRNA integrity. Upon Nab2-tandem affinity purification, several mRNA export factors were co-enriched (Yra1, Mex67, THO-TREX) that were present in mRNPs of different size and mRNA length. High-throughput sequencing of the co-precipitated RNAs indicated that Nab2 is associated with the bulk of yeast transcripts with no specificity for different mRNA classes. Electron microscopy revealed that many of the mRNPs have a characteristic elongated structure. Our data suggest that mRNPs, although associated with different mRNAs, have a unifying core structure.
Introduction
Nucleocytoplasmic transport occurs via nuclear pore complexes, which are embedded into the nuclear membrane. For nuclear mRNA export, a conserved export receptor, Mex67-Mtr2, in yeast (TAP-p15 or NXF1-NXT1 in metazoans) and a number of export adaptor proteins were identified (for review, see Refs. 1 and 2).
The TREX (transcription export) complex belongs to these adaptors and is composed of transcription elongation (THO complex) and export factors (Sub2 and Yra1). These proteins are loaded co-transcriptionally onto the pre-mRNA in yeast (3, 4), before recruitment of Mex67-Mtr2 occurs via an interaction with Yra1. A different assembly (Sac3-Thp1-Cdc31-Sus1 or TREX-2) is thought to facilitate the repositioning of transcribed genes to nuclear pore complexes and, thus, also helps to integrate transcription and export steps (2). Another adapter RNA-binding protein, Npl3, is recruited co-transcriptionally to the mRNA (5) and can interact with Mex67 in the nucleus when dephosphorylated (6).
Finally, Nab2 is a poly(A)+ RNA-binding protein that shuttles between nucleus and cytoplasm and is required for mRNA export (1). Nab2 is mainly localized in the nucleus (7, 8) and interacts with the poly(A)-binding protein (Pab1) to affect poly(A) tail length (9). Moreover, Nab2 interacts with Mlp1, a nuclear pore complex-associated protein at the nuclear basket involved in mRNP quality control (10), with Gfd1 and Gle1, factors involved in mRNA export (11), and with the Sac3-Thp1 complex, which plays a role in coupling transcription with mRNA metabolism (12). When the pools of mRNAs immune-precipitated by Npl3, Nab2, and Nab4 were determined in microarrays and compared, it was suggested that distinct sets of mRNAs are bound to each of these proteins (13). In vitro, the RNA helicase Dbp5 was shown to dissociate Nab2 from the mRNP3 (14) and because of its localization on the cytoplasmic side of the nuclear pore complex, it was suggested that Dbp5 could release Nab2 from mRNPs after the nuclear exit. In addition, the binding of the import receptor Kap104 to Nab2 after export of the Nab2-containing mRNP into the cytoplasm was suggested to trigger release of Nab2 from the mRNA cargo (15). Because both Kap104 and Dbp5 localize to the distal bud tip and the bud neck during cell division, a local release of Nab2 and Nab4 by these two proteins was suggested to generate the translation-competent mRNAs release and increase protein synthesis in the emerging daughter cell (16).
In this study we used Nab2 as bait to isolate yeast mRNPs and characterize them biochemically and structurally. Purified Nab2 is associated with several mRNP biogenesis and/or export factors such as THO/TREX members and Mex67 and with poly(A)+ RNAs of different lengths. High-throughput sequencing indicated that Nab2 is associated with the bulk of yeast transcripts, and electron microscopy revealed a common core structure of the Nab2-containing mRNPs.
EXPERIMENTAL PROCEDURES
Growth Conditions and Cell Lysis
Yeast cells were grown at 30 °C in 2l YPD medium (2% yeast extract, 1% peptone, 2% glucose) to A600 3.5. Cells were harvested by centrifugation, rapidly frozen in liquid nitrogen, and ground in a “mixer mill” (Retsch MM 301) using a 20-mm diameter steel ball. Ground pellets were stored at −80 °C until purification was performed.
Tandem Affinity Purification (TAP)
TAP affinity purifications were performed as described (17, 18). TAP-tagged proteins were purified from 2 ground pellets resuspended in 15 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2, 0,075% Nonidet P-40, 100 mm dithiothreitol) supplemented with phenylmethylsulfonyl fluoride, protease inhibitor FY (Serva), and 4 mm ribonucleoside vanadyl complex (RVC; Sigma) (18). After binding the lysate to IgG-Sepharose, beads were washed with lysis buffer plus 1 mm RVC before two additional washes without RVC. Recombinant RNase inhibitor (Fermentas) was added during TEV cleavage and the subsequent purification on calmodulin beads (250 units/ml). When split-tag affinity purification was applied, we further purified the TEV eluate on anti-FLAG antibody beads (according to the manufacturer's instructions; Sigma) instead of the calmodulin beads purification. A flow scheme of the purification with and without preservation for RNA stability is shown in supplemental Fig. S1.
Sucrose Gradient Centrifugation to Separate mRNPs
EGTA eluates of Nab2-TAP affinity purifications (derived from 4 pellets; see above) were loaded on a 10–30% linear sucrose gradient (SW40 tubes). Where indicated, micrococcal RNase (1 units/μl) was added to Nab2-TAP eluate before sucrose gradient centrifugation (27,000 rpm in SW40 rotor (Beckman Coulter) for 16 h). The sucrose gradient was fractionated with a continuous monitoring of A254 nm. Fractions of 800 μl were collected and analyzed for protein and RNA content.
High-Throughput RNA Sequencing (GS-FLX-454)
1 μg of purified RNAs extracted from gradient fractions 4–6 (see Fig. 2A) was used for high-throughput sequencing by the GS-FLX-454 Technology (performed by the company Eurofins MWG Operon) following standard protocols (19). Systematic BLAST analysis was performed as described under supplemental “Methods.” Genome ontology (GO) annotation (20) was carried out using SlimMapper. For comparison with the complete yeast transcriptome, all yeast genes were downloaded from the Saccharomyces Genome Database. Transcript abundance data from the yeast ORF transcriptome (21) were taken from the Young laboratory of the Whitehead Institute, Cambridge, MA.
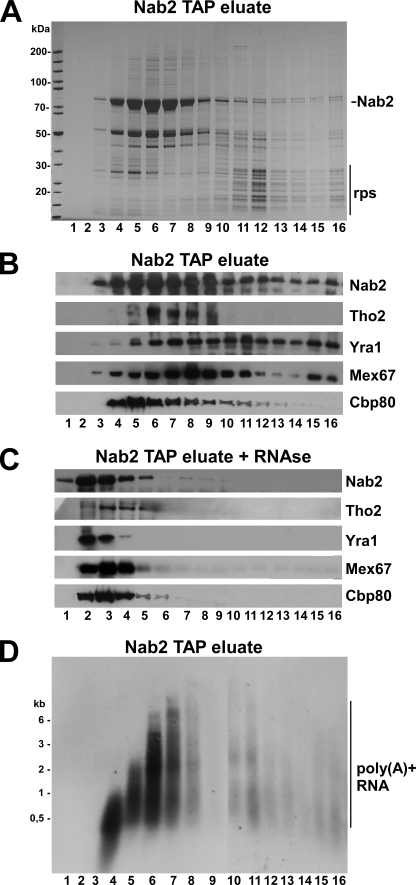
FIGURE 2.
Sucrose gradient centrifugation of Nab2-precipitated mRNPs. A, affinity-purified Nab2-TAP under conditions of mRNA protection was separated on a 10–30% sucrose gradient. Fractions 1 (top) to 15 (bottom) were analyzed by SDS-PAGE and Coomassie staining. Nab2 and ribosomal proteins (rps) were indicated. B, shown is Western blot analysis of the sucrose gradient fractions shown in A. Antibodies against the indicated proteins were used to show their distribution on the sucrose gradient. C, shown is Western blot analysis of sucrose gradient fractions using the indicated antibodies, but the Nab2-TAP eluate was treated with RNase before sucrose gradient centrifugation. D, Northern analysis of the gradient fractions (10–30% sucrose) of the Nab2-TAP eluate using an oligo(dT) probe is shown.
EM Analysis by the GraFix Method
We combined the GraFix procedure (22) with sucrose gradient centrifugation. 25% glutaraldehyde (Roth) was added to a final concentration of 0.17% into the 30% sucrose solution before casting the gradient. This results in a 0.07–0.15% linear glutaraldehyde gradient across the sucrose 10–30% gradient. Gradient fractions were loaded on freshly glow-discharged, carbon-coated copper grids and stained in 2% uranyl acetate. Particles were imaged on a Philips Morgagni 268 FEI electron microscope (100 kV) with a 1K format (1024 × 1024 pixels) side-mounted camera (Soft Imaging Software), at a nominal magnification of 71,000×. Sizing of 250/300 single particles for each fraction were carried out using measureIT software (Olympus) with a calibrated pixel size of 0.87 Å.
RESULTS
Isolation of Yeast mRNPs by Nab2-TAP
To isolate mRNPs from yeast, we affinity-purified TAP-tagged Nab2 that was genomically integrated and functional. To preserve the integrity of mRNAs, we lysed yeast cells by grinding in liquid nitrogen similarly to previous reports (23, 24) in the presence of the potent RNase inhibitor RVC. These precautions improved mRNP purification as revealed by SDS-PAGE (Fig. 1A) and Northern analysis (Fig. 1B). Nab2-TAP was co-enriched in Tho2 (THO/TREX subunit) and Yra1 (RNA-binding protein) but largely devoid of the karyopherin Kap104 (Fig. 1C). The absence of Kap104 is a further indication that Nab2 is RNA-associated, as free Nab2 forms a complex with its import receptor Kap104 (Fig. 1A (15)). The co-precipitation of ribosomal proteins by Nab2-TAP could indicate that mRNAs associated with Nab2 may have also pulled down polysomes. On the other hand, ribosomal proteins are known to be frequent contaminants in TAP purifications due to unspecific binding to IgG-Sepharose beads. Hence, it is difficult to conclude whether the ribosomes in the Nab2 preparation are of physiological origin or contaminants.
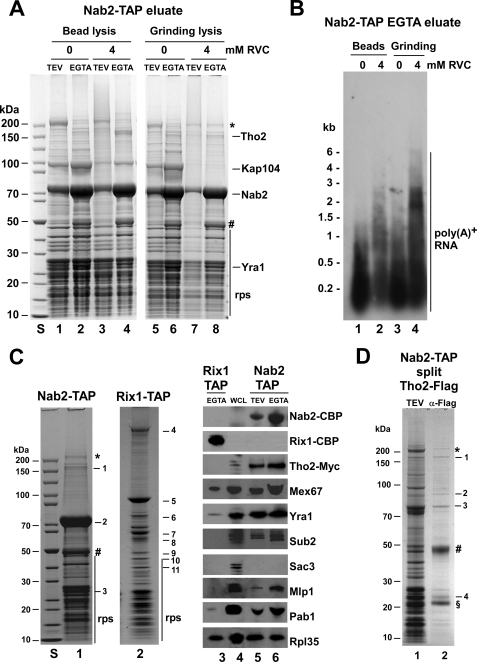
FIGURE 1.
Affinity purification of Nab2-TAP under different mRNA protection conditions. A and B, shown is bead lysis (lanes 1–4) versus N2 grinding (lanes 5–8) with or without RVC. TEV and EGTA eluates of Nab2-TAP were analyzed by SDS-PAGE/Coomassie staining (A) and Northern to reveal poly(A)+ RNA (B). The indicated bands were identified by mass spectrometry. rps, ribosomal proteins; *, fatty acid synthetase; #, RNase inhibitor. S, protein standard. C, shown is a comparison of Nab2-TAP (lane 1) and Rix1-TAP purifications (lane 2) using mRNPs protection conditions by SDS-PAGE and Coomassie staining. The labeled bands 1–11 were identified by mass spectrometry. S, protein standard. 1, Tho2; 2, Nab2; 3, Yra1; 4, Rea1; 5, Rix1-CBP; 6, Nog1; 7, Ipi3; 8, Nug2; 9, Rpl3; 10, Rpl4; 11, Ipi1. Lanes 3–6, shown is Western blot analysis of the Rix1-TAP EGTA eluate (lane 3) and Nab2-TAP TEV (lane 5) and EGTA (lane 6) eluates and RS453 whole cell lysate (lane 4). D, Nab2-TAP/Tho2-FLAG split affinity purification is shown. Nab2-TAP TEV eluate and second eluate after Tho2-FLAG immunoprecipitation (anti-FLAG) is shown. 1, Tho2-FLAG; 2, Hpr1; 3, Nab2-CBP; 4, Yra1; §, flag peptide.
Additional proteins co-precipitated by Nab2-TAP and detected by Western included Sub2, Mex67, Pab1, and Mlp1, which are all factors with a role in mRNP biogenesis and/or export, but the TREX-2 factor Sac3 was not co-enriched (Fig. 1C, lane 4–6). Rix1-TAP served as a negative control, which co-enriched pre-60 S factors and rRNA but not mRNA biogenesis/export factors apart from Mex67 (Fig. 1C, lanes 2 and 3). However, Mex67 is also involved in the export of 60 S subunits (25).
Next, we performed “split tag affinity purifications” to test which of the co-enriched factors was present on the same mRNP. Hence, yeast strains coexpressing either Nab2-TAP and Tho2-FLAG or Nab2-TAP and Yra1-FLAG were constructed and subjected to split affinity purifications (see “Experimental Procedures”). As shown in Fig. 1D, the Nab2-TAP/Tho2-FLAG split affinity purification yielded a preparation that contained Nab2, Tho2, Hpr1 (another THO/TREX subunit), and Yra1, whereas ribosomal proteins were largely absent, suggesting the co-enrichment of predominantly nuclear mRNPs. Using the Nab2-TAP/Yra1-FLAG pair for split affinity purification, Tho2 and Hpr1 were also co-enriched, but ribosomal proteins were still present (data not shown). Altogether, these results indicate that mRNPs isolated via Nab2-TAP contain predominantly mRNP biogenesis/export factors.
mRNAs of Different Lengths Are Co-enriched with Nab2
To show that Nab2 and its co-precipitated proteins are part of distinct mRNA-protein particles, we performed sucrose gradient centrifugation. This analysis showed that Nab2 sedimented over a broad range of the sucrose gradient (Fig. 2, A and B). A similar but non-identical distribution was seen for Tho2, Yra1, Mex67, and Cbp80. However, the co-precipitated ribosomal proteins migrated deeper into the sucrose gradient and, hence, could be separated from the major pool of Nab2-containing particles (Fig. 2A). Treatment of the Nab2-TAP eluate with micrococcal nuclease before sucrose gradient centrifugation caused Nab2 and its associated partners to shift toward the top fractions of the gradient, which contain the soluble proteins (Fig. 2C).
To estimate the length of the mRNAs associated with the Nab2-precipitated mRNPs, a Northern blot using an oligo-dT probe was performed. This analysis showed that poly(A)+ RNAs were predominantly present in the upper half of the sucrose gradient that contains most of Nab2 and its co-purifying partners (Fig. 2D). The length of these mRNAs steadily increased from fraction 4 to 8. Thus, Nab2 and its co-precipitated proteins are present in mRNPs with different mRNA length that can be disintegrated by degrading the RNA.
A Large Pool of mRNAs Is Associated with Nab2
To determine which types of mRNAs were co-precipitated with Nab2, we arbitrarily selected eight transcripts that belong to two major transcripts classes, four encoding transcription factors (two high and two low abundant) and four encoding metabolic enzymes (two high and two low abundant). Among these selected transcripts, five were previously reported to be associated with Nab2, whereas such association was not observed for the remaining three transcripts (13). By performing RT-PCR, we found all eight transcripts co-precipitated by Nab2-TAP with no apparent preference for abundance or transcript family (supplemental Fig. S2). Rix1-TAP, which was devoid of these mRNAs, was used as a negative control.
Next, we performed high-throughput sequencing (GS-FLX-454) of the co-precipitated RNAs (derived from fraction 4–6; see Fig. 2, A and D) to obtain a more comprehensive overview on the gene ontology of Nab2-associated mRNAs (see “Experimental Procedures”). This yielded a total number of 79,526 sequence reads (average length 187 bases) with 3,595 contigs after assembly and an average length of 300 bases (supplemental Tables S2 and S3).
Annotation of these contigs identified 2666 individual transcripts (of ∼6600 in total) co-precipitated with Nab2 (supplemental Table S2). Apparently, sequenced transcripts were devoid of intron (confirmed by RT-PCR, data not shown), and poly(A) tails were present at the expected sites, suggesting that they have passed the splicing and polyadenylation steps. These mRNAs were categorized according to functional classes based on three different GO annotation criteria (i) “function,” (ii) “component,” or (iii) “process” (20) (supplemental Table S2). This GO annotation revealed that the pool of Nab2-associated mRNAs largely follows the distribution of all yeast transcripts (Fig. 3A and supplemental Fig. S3). In the few cases of slightly underrepresented subclasses in the Nab2 precipitate, these transcripts belong often to a GO category, which is not expressed constitutively in yeast (e.g. pseudohyphal growth).
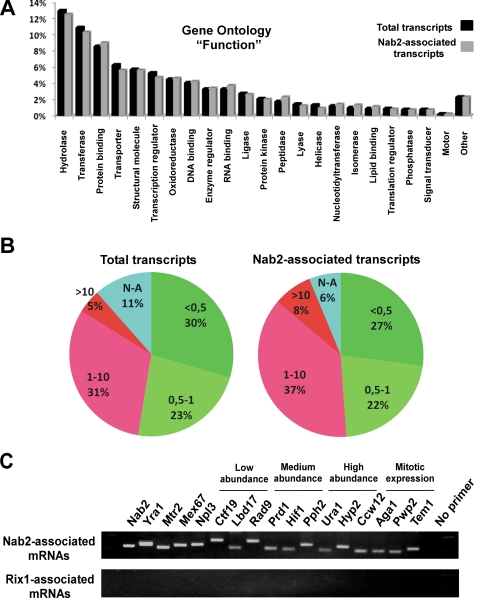
FIGURE 3.
Analysis of Nab2-TAP associated transcripts. A, shown is a comparison of total yeast transcript distribution (black; GO annotation criteria function; for component and process, see supplemental Fig. S3) with Nab2-associated mRNAs identified by high-throughput sequencing (gray). B, shown is a comparison of total yeast transcript distribution (left) according to abundance versus Nab2-associated mRNAs (right). Transcripts were sorted according to abundance and divided in 5 classes: <0.5 t/c, 0.5–1 t/c, 1–10 t/c, >10 t/c, and unknown. C, shown is RT-PCR analysis of the indicated transcripts associated with Nab2-TAP (upper panel) or Rix1-TAP (lower panel).
Next, we examined whether low abundant transcripts tended to be absent from the Nab2-TAP eluate. Transcripts were sorted according to abundance (supplemental Table S2) and divided in 5 classes: <0.5 transcripts/cell (t/c), 0.5–1 t/c, 1–10 t/c, >10 t/c, unknown (Fig. 3B, total transcripts). When all the 2666 transcripts that co-enriched with Nab2-TAP were categorized according to their known abundance, no significant bias for the absence of low abundant transcripts from the Nab2-TAP eluate was observed (Fig. 3B, Nab2-associated transcripts). These data suggest that Nab2 is associated with a broad range of yeast transcripts independent of transcript abundance or gene ontology.
High-throughput sequencing may not have detected all transcripts co-precipitated by Nab2-TAP. Hence, we tested whether nondetected transcripts were present in the Nab2-TAP eluate but escaped due to the intrinsic limitations of the 454 sequencing methods used in this analysis. Among the transcripts not detected were those for Mex67 and Nab2. On the other hand, Mtr2, the small subunit of the Mex67-Mtr2 heterodimer, and Yra1, which associates with the Mex67-Mtr2 export receptor, were detected by high-throughput sequencing (supplemental Table S2). As shown by RT-PCR, all four mRNAs encoding Mex67, Mtr2, Yra1, and Nab2 were significantly co-precipitated by Nab2-TAP (Fig. 3C). In addition, we randomly picked several other transcripts not found by the 454 technology and analyzed them by RT-PCR. All the chosen transcripts, Ctf19, Lbd17, Rad9 (low abundance), Prd1, Hif1, Pph2 (medium abundance), Ura1, Ccw12, Hyp2 (high abundance), and Aga1, Pwp2, Tem1 (expressed mainly during mitosis), were clearly detected by RT-PCR to be present in the Nab2-TAP containing gradient fractions but were not found in the Rix1-TAP eluate (Fig. 3C). We conclude that Nab2 is likely to be associated with the bulk of yeast mRNAs.
EM Analysis of Nab2 mRNPs Reveals an Elongated Core Structure
We sought then to visualize the mRNPs affinity-purified by NAB2-TAP via EM. To date, very little is known about the structure of mRNPs except for the huge worm-like Balbiani ring particles, viewed by electron tomography (26). To stabilize the mRNPs for negative stain EM, we adopted a sample preparation procedure for single-particle termed GraFix (22) using glutaraldehyde to cross-link the mRNPs for better structural preservation (see “Experimental Procedures”). A254 nm measurements of the sucrose gradient fractions without/with glutaraldehyde indicated that mRNPs and ribosomal peaks were shifted by 1–2 fractions toward higher sucrose density when glutaraldehyde was present.4
For the subsequent EM analysis, we focused on the fractions of the sucrose gradient containing mRNPs of different mRNA length but devoid of ribosomal proteins (see Fig. 2A). When a typical mRNP-containing fraction from the GraFix gradient (e.g. fraction 8, which approximately corresponds to fractions 6–7 from the sucrose gradient without glutaraldehyde; see Fig. 2D) was stained with uranyl acetate and inspected by EM, most particles had a similar size and elongated shape (Fig. 4A, overview), whereas few other particles showed a more globular shape. The elongated particles with a ribbon-like structure and lateral constrictions exhibited a length of 20–30 nm and a thickness of 5–7 nm (see also Fig. 4B, gallery).
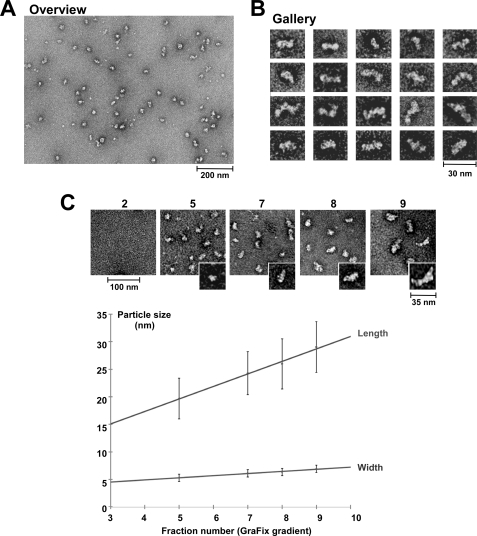
FIGURE 4.
Electron microscopic analysis of mRNPs isolated via Nab2-TAP. Shown is an electron micrograph overview (A) and a gallery (B) of single particles showing mRNPs affinity-purified by Nab2-TAP (fraction 8 from the GraFix sucrose gradient). Scale bars, 200 nm (A) and 30 nm (B). C, shown is an increase in particle size of Nab2-purified mRNPs with increasing gradient fraction number. Electron micrographs of mRNPs present in the GraFix sucrose gradient fractions 2, 5, 7, 8, and 9 (Overview: scale bar, 100 nm; inset: characteristic single mRNP enlarged; scale bar, 35 nm). Below the gallery is a graph with measured mRNP particle size (in nm) plotted against GraFix sucrose gradient fraction number. The upper line indicates the average length, and the lower line indicates the average width of the mRNP particles. 250–300 single particles were selected to measure length and width of the mRNPs, and the S.D. is given. Note that in the GraFix sucrose gradient fractions are shifted by ∼1–2 fractions to higher density when compared with the sucrose gradient without glutaraldehyde (see Fig. 2A).
When neighboring fractions of the sucrose gradient were analyzed by EM, they also contained elongated particles of similar shape but were longer in heavier fractions (e.g. in fraction 9; 29 ± 5 nm) or shorter in lighter fractions (e.g. in fraction 5; 20 ± 4 nm) than the particles in fraction 8 (25 ± 5 nm) (Fig. 4C). As anticipated, no particles were detected in the two first gradient fractions that are devoid of Nab2. When the particle length (in nm; upper line) or particle width (in nm; lower line) was plotted against the sucrose gradient fraction number, it became evident that mRNP particles had a rather constant width, but their length increased steadily the deeper they sedimented into the sucrose gradient (Fig. 4C, lower panel). This finding suggests a correlation between mRNA length and length of the mRNP (see also Fig. 2D).
DISCUSSION
In this study we describe the isolation of mRNPs from yeast using the RNA-binding protein Nab2 as bait. Our method is an advancement to previous purifications of other mRNA binding proteins (see Refs. 23 and 24) as we have used, besides a “soft” grinding lysis method and the addition of the potent RNase inhibitor (RVC), sucrose gradient centrifugation to further purify the mRNPs. This additional step allowed the separation of mRNPs according to their different size and removal of contaminants (e.g. ribosomal proteins, which are frequent impurities in TAP purifications). Thus, our final mRNP material was pure enough to be used for electron microscopic analyses.
The purified mRNPs separated on the sucrose gradient contained transcripts that were fractionated according to their size, the largest mRNPs containing the longest mRNAs. The detection of both poly(A) tail and 5′ cap structure in the purified Nab2-TAP suggested that our purification yielded intact mRNPs. Moreover, the co-enrichment of Cbp80 in the Nab2-TAP eluate indicated that the purified mRNPs, at least a pool of it, may not have undergone a pioneer round of translation in which Cbp80 is known to be released from the mRNA cap and exchanged with eIF4E (27). Thus, the mRNPs isolated in this study largely represent mRNPs that are in the process of assembly and/or export. However, the fact that the mRNAs associated with Nab2 did not contain introns suggested that they had already passed the splicing step.
High-throughput sequencing of mRNAs extracted from Nab2-TAP allowed us to detect a large pool of the yeast transcripts in association with Nab2. Missing transcripts not detected by 454 sequencing could be shown to be co-enriched with Nab2-TAP by RT-PCR. Limitations of the applied 454 sequencing method could be the quality of the filter used for contigs assembly and, thus, lead to under-representation of short mRNAs that could be also lost during the enrichment of DNA capture beads (28). Nevertheless, if we consider the results from both high-throughput sequencing and RT-PCR, we conclude that Nab2 is associated with the bulk of yeast mRNAs. This suggestion is consistent with the finding that Nab2 is a poly(A)-binding protein (29). However, our data do not support a model in which Nab2 is associated with specific subclasses of mRNAs (13). Thus, Nab2 could function in global mRNA export together with the general mRNA export receptor Mex67-Mtr2 and its adaptor protein Yra1, with which it is also associated.
The structural organization of the mRNPs isolated by Nab2-TAP was determined by electron microscopy. Accordingly, Nab2 mRNP particles exhibited an elongated shape (like a ribbon) with lateral constrictions and a length that increased proportionally with the mRNA length (see Fig. 4C). As suggested by this correlation, it will be interesting to find out whether the factors found to co-enrich with Nab2-TAP (i.e. TREX complex, Mex67-Mtr2, Yra1) could serve as a structural unit for RNA binding and RNP organization. To date, very little is known about the structure of mRNPs. Most of our knowledge of mRNP structure stems from studies using Balbiani ring particles, which due to their huge size and tremendous length of the associated mRNA can be well studied by electron tomography (for review, see Ref. 26). Notably, Balbiani ring particles emerging from the chromosome exhibit an elongated structure with lateral constrictions (RNP ribbon) that was also observed for the Nab2-purified mRNPs. However, when the Balbiani ring particles are released from giant chromosomes, the RNP ribbon is bent into a ring-like configuration (50-nm granule). When the 50-nm granule is finally translocated through the nuclear pore complex, it unfolds and regains its elongated ribbon-like conformation. On the cytoplasmic side, the ribbon is unpacked, and an elementary fiber is formed that extends into cytoplasm and binds to ribosomes (30). Isolated Nab2 mRNPs, which also form a ribbon-like structure, are not circular, but the length of the transcripts associated with Nab2 is by far much shorter than the huge Balbiani ring mRNA. However, the elongated linear yeast mRNPs of different length could be exported through the nuclear pore complexes in a similar way as suggested for the elongated ribbon-like Balbiani ring mRNP with the 5′ cap ahead (31). Finally, the ribbon-like structure of yeast mRNPs as described in this study is also consistent with the EM structures of other studied RNPs, which are an influenza virus mRNP (32) and in vitro reconstituted mRNPs (33, 34).
Although our EM characterization of yeast mRNPs is only a start for a more comprehensive study, the shape and size of the mRNPs could suggest that the mRNA within the particle is compacted and condensed (note that a 1-kilobase-long linear RNA has a length of ∼340 nm, whereas our mRNPs carrying a ∼1-kilobase-long mRNA are 25–30 nm long). However, we do not expect that mRNAs due to their heterogeneity in length and sequence will compactly fold into a mRNA core, as is the case for rRNA in ribosomal subunits due to an extensive folding into secondary and tertiary RNA structures (35–37). Although we cannot exclude that naked and linear mRNA exists in the mRNP that is not contrasted by negative staining, we speculate that the mRNA could be wrapped around a protein core consisting of Nab2 and other partners (like DNA in the nucleosome). Consistent with this speculation, we calculated that about 12 Nab2 molecules could be associated with 1 mRNA molecule of 1 kilobase average length, corresponding to 83 RNA bases per 1 Nab2 protein (for this rough calculation we estimated that fraction 6 from the sucrose gradient (see Fig. 2A) contains ∼5 μg or 85 pmol of Nab2 and 2.4 μg or 7 pmol of poly(A)+ RNA average length 1 kilobase; see Fig. 2D). These obtained numbers would be consistent with a model in which a Nab2 protein core structure organizes and compacts the mRNA.
It is not known whether in vivo Nab2 is restricted to the poly(A) tail, which so far is only an in vitro data (7, 29). We speculate that in vivo, Nab2 and its binding partners (e.g. Yra1) could bind at several sites along the mRNA. Indeed, it has shown Yra1 is associated with the nascent mRNA transcript from the 5′ to the 3′ part (38), and the mammalian Yra1 homologue Aly/Ref and members of the human TREX complex were found to associate with the 5′ part of the mRNA involving the cap binding complex (39, 40).
In conclusion, we have purified and characterized mRNPs from yeast that were affinity-purified via the poly(A)+ RNA-binding protein Nab2. Our data show that Nab2 is preferentially associated with THO/TREX complex members and with many if not all poly(A)+ mRNAs. This supports a model in which Nab2 is involved in general mRNP assembly and export. The fact that THO/TREX members are significantly associated with the Nab2 mRNPs suggests that they were assembled co-transcriptionally. Because Nab2 mRNPs are also associated with Mex67, a pool of it may represent transport intermediates on the way to the cytoplasm. Our results, thus, provide novel insights into the structure of mRNPs and how they can be transported to the cytoplasm.
Acknowledgments
We thank Dr. Tamas Fischer, who helped in the beginning of this project. We are grateful to Dr. S. Glinka (454 sequencing), the Lechner laboratory (mass spectrometry), and Dr. A. KÖhler for critical reading of the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2 and Tables S1 (454 sequencing, computational analysis) and S2 (454 sequencing, raw sequences).
J. Batisse, unpublished results.
- mRNP
- messenger ribonucleoprotein
- RVC
- ribonucleoside vanadyl complex
- GO
- genome ontology
- t/c
- transcripts/cell
- EM
- electron microscopy
- TAP
- tandem affinity purification
- contig
- group of overlapping clones
- RT
- reverse transcription
- TEV
- tobacco etch virus protease.
REFERENCES
- 1.Kelly S. M., Corbett A. H. (2009) Traffic 10, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler A., Hurt E. (2007) Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 3.Aguilera A. (2005) Curr. Opin. Cell Biol. 17, 242–250 [DOI] [PubMed] [Google Scholar]
- 4.Vinciguerra P., Stutz F. (2004) Curr Opin. Cell Biol. 16, 285–292 [DOI] [PubMed] [Google Scholar]
- 5.Lei E. P., Krebber H., Silver P. A. (2001) Genes Dev. 15, 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert W., Guthrie C. (2004) Mol. Cell 13, 201–212 [DOI] [PubMed] [Google Scholar]
- 7.Anderson J. T., Wilson S. M., Datar K. V., Swanson M. S. (1993) Mol. Cell. Biol. 13, 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Truant R., Fridell R. A., Benson R. E., Bogerd H., Cullen B. R. (1998) Mol. Cell. Biol. 18, 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hector R. E., Nykamp K. R., Dheur S., Anderson J. T., Non P. J., Urbinati C. R., Wilson S. M., Minvielle-Sebastia L., Swanson M. S. (2002) EMBO J. 21, 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasken M. B., Stewart M., Corbett A. H. (2008) J. Biol. Chem. 283, 27130–27143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suntharalingam M., Alcázar-Román A. R., Wente S. R. (2004) J. Biol. Chem. 279, 35384–35391 [DOI] [PubMed] [Google Scholar]
- 12.Gallardo M., Luna R., Erdjument-Bromage H., Tempst P., Aguilera A. (2003) J. Biol. Chem. 278, 24225–24232 [DOI] [PubMed] [Google Scholar]
- 13.Kim Guisbert K., Duncan K., Li H., Guthrie C. (2005) Rna 11, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007) Mol. Cell 28, 850–859 [DOI] [PubMed] [Google Scholar]
- 15.Lee D. C., Aitchison J. D. (1999) J. Biol. Chem. 274, 29031–29037 [DOI] [PubMed] [Google Scholar]
- 16.van den Bogaart G., Meinema A. C., Krasnikov V., Veenhoff L. M., Poolman B. (2009) Nat. Cell Biol. 11, 350–356 [DOI] [PubMed] [Google Scholar]
- 17.Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. (2002) EMBO J. 21, 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 19.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L., Jarvie T. P., Jirage K. B., Kim J. B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., Rothberg J. M. (2005) Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Cell 95, 717–728 [DOI] [PubMed] [Google Scholar]
- 22.Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Lührmann R., Stark H. (2008) Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 23.López de Heredia M., Jansen R. P. (2004) BMC Biochem. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. (2007) Nat. Methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 25.Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., Hurt E. (2007) Mol. Cell 26, 51–62 [DOI] [PubMed] [Google Scholar]
- 26.Daneholt B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortes P., Inada T., Preiss T., Hentze M. W., Mattaj I. W., Sachs A. B. (2000) Mol. Cell 6, 191–196 [PubMed] [Google Scholar]
- 28.Torres T. T., Metta M., Ottenwälder B., Schlötterer C. (2008) Genome Res. 18, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly S. M., Pabit S. A., Kitchen C. M., Guo P., Marfatia K. A., Murphy T. J., Corbett A. H., Berland K. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12306–12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehlin H., Skoglund U., Daneholt B. (1991) Exp. Cell Res. 193, 72–77 [DOI] [PubMed] [Google Scholar]
- 31.Mehlin H., Daneholt B., Skoglund U. (1992) Cell 69, 605–613 [DOI] [PubMed] [Google Scholar]
- 32.Wu W. W., Weaver L. L., Pante N. (2009) J. Vis. Exp. 24, 1105–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto K., Tanaka K. J., Aoki K., Sameshima M., Tsujimoto M. (2003) Biochem. Biophys. Res. Commun. 306, 53–58 [DOI] [PubMed] [Google Scholar]
- 34.Skabkin M. A., Kiselyova O. I., Chernov K. G., Sorokin A. V., Dubrovin E. V., Yaminsky I. V., Vasiliev V. D., Ovchinnikov L. P. (2004) Nucleic Acids Res. 32, 5621–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 36.Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Nature 407, 327–339 [DOI] [PubMed] [Google Scholar]
- 37.Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. (2001) Science 292, 883–896 [DOI] [PubMed] [Google Scholar]
- 38.Zenklusen D., Vinciguerra P., Wyss J. C., Stutz F. (2002) Mol. Cell. Biol. 22, 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H., Dufu K., Lee C. S., Hsu J. L., Dias A., Reed R. (2006) Cell 127, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 40.Nojima T., Hirose T., Kimura H., Hagiwara M. (2007) J. Biol. Chem. 282, 15645–15651 [DOI] [PubMed] [Google Scholar]