Abstract
15-Acetyldeoxynivalenol (15-AcDON) is a low molecular weight sesquiterpenoid trichothecene mycotoxin associated with Fusarium ear rot of maize and Fusarium head blight of small grain cereals. The accumulation of mycotoxins such as deoxynivalenol (DON) and 15-AcDON within harvested grain is subject to stringent regulation as both toxins pose dietary health risks to humans and animals. These toxins inhibit peptidyltransferase activity, which in turn limits eukaryotic protein synthesis. To assess the ability of intracellular antibodies (intrabodies) to modulate mycotoxin-specific cytotoxocity, a gene encoding a camelid single domain antibody fragment (VHH) with specificity and affinity for 15-AcDON was expressed in the methylotropic yeast Pichia pastoris. Cytotoxicity and VHH immunomodulation were assessed by continuous measurement of cellular growth. At equivalent doses, 15-AcDON was significantly more toxic to wild-type P. pastoris than was DON. In turn, DON was orders of magnitude more toxic than 3-acetyldeoxynivalenol. Intracellular expression of a mycotoxin-specific VHH within P. pastoris conveyed significant (p = 0.01) resistance to 15-AcDON cytotoxicity at doses ranging from 20 to 100 μg·ml−1. We also documented a biochemical transformation of DON to 15-AcDON to account for the attenuation of DON cytotoxicity at 100 and 200 μg·ml−1. The proof of concept established within this eukaryotic system suggests that in planta VHH expression may lead to enhanced tolerance to mycotoxins and thereby limit Fusarium infection of commercial agricultural crops.
Introduction
Fusarium head blight of cereals and Fusarium ear rot of maize are caused by morphologically similar species (Fusarium graminearum and Fusarium culmorum, etc.) common throughout global agricultural regions. With few exceptions, Fusarium epidemics are characterized by cyclical and highly aggressive infection of commercial crops with economic impacts on food and feed industries that are immediate and far reaching. For example, losses associated with the most recent Fusarium outbreak in North America in the 1990s were estimated to range from 1.3 to 3.0 billion United States dollars (1).
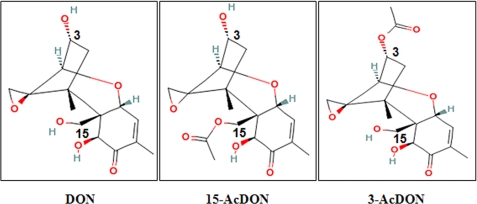
A toxin class commonly found within agricultural commodities infected by Fusarium are trichothecene mycotoxins. Trichothecenes represent a highly diverse group of over 180 sesquiterpenoid low molecular weight (typically 200–500 Da) mycotoxins characterized by a tricyclic ring structure containing a double bond at C-9,10 and an epoxide group at C-13 (2). Regardless of size and structural composition, trichothecenes are potent inhibitors of eukaryotic protein synthesis with specific activity on ribosomal protein L3 within the 60 S subunit resulting in inhibition of peptidyltransferase activity (3, 4). Although the capacity to inhibit protein synthesis is regarded as central to trichothecene cytotoxicity (5, 6), adverse effects on eukaryotic cells may actually be attributed to dysregulation of cellular signaling and alterations in downstream gene expression (7). As a result, trichothecenes such as deoxynivalenol (DON),2 15-acetyldeoxynivalenol (15- AcDON), and 3-acetyldeoxynivalenol (3-AcDON) (Fig. 1) are considered to be inherently hazardous feed- and foodborne contaminants (2, 8).
FIGURE 1.
Structure, composition, and molecular weight of trichothecene mycotoxins used within this study (2).
Numerous studies have demonstrated a correlation between in planta DON accumulation and Fusarium virulence in susceptible cultivars of wheat (9) and maize (10). Based on these findings, mechanisms that convey innate and acquired host plant resistance to DON and other trichothecene toxins have received considerable attention. To date, in planta trichothecene resistance has been achieved through mechanisms that alter targeted proteins within host cell ribosomes (11, 12), promote metabolic transformation to less toxic forms, e.g. DON-glucosyl conjugate (13) or to 3-AcDON (14), and/or reduce intracellular concentrations to effectively limit mycotoxin exposure to sensitive cellular targets. Collectively, such research can be applied to impart novel mechanisms of trichothecene resistance in higher order plants.
Yeast is well suited as a eukaryotic model organism to identify and validate mechanisms involved in host plant resistance to mycotoxins (12, 13, 15). Test systems based on yeast offer cost-effective convenience and flexibility as one can validate a wide range of novel detoxification mechanisms within a short period of time at a minimal cost using common/nonspecialized laboratory equipment. Assessment of mycotoxin resistance mechanisms is likewise straightforward as reproducible treatment-specific effects can be precisely determined based on simple measurements of cellular growth and function over time.
Single domain heavy chain antibody fragments (i.e. VHH) from the camelidae heavy chain IgG subfamily are among the smallest functional recombinant antibody (Ab) fragments at 14–15 kDa. VHH fragments exhibit the same exquisite specificity as larger immunoglobulins, with added biochemical advantages of high solubility, stability, and robust expression in various recombinant systems (16, 17). VHH fragments have been generated against low molecular weight ligands (haptens) and toxins (18). Based on favorable physicochemical properties and efficacy against a wide range of antigens, single domain recombinant Ab fragments have been developed and tested as immunotherapeutic reagents with applications ranging from pharmacology (16, 19) to plant science (20).
This study demonstrates that intrabody expression of a VHH fragment isolated from a hyper-immunized phagemid library with affinity for 15-AcDON (18) can impart real time immunomodulation of mycotoxin-specific cytotoxicity within a model eukaryotic system. Pichia pastoris was selected as the host organism based on an expected high level expression of functional VHH intrabody fragments and anticipated sensitivity to 15-AcDON. This system was established as a “proof of concept” to demonstrate that intrabody expression of recombinant VHH fragments could impart a novel means of mycotoxin-specific resistance.
EXPERIMENTAL PROCEDURES
VHH Genes
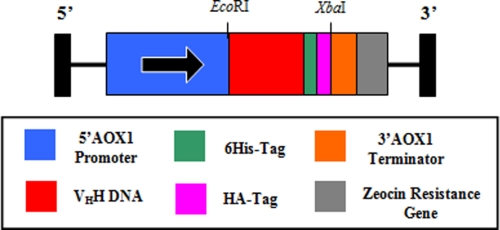
NAT-267 VHH DNA sequence (GenBankTM EU676170.1) (18) was used to design a P. pastoris codon optimized version of the gene to which was added 3′ HA and His6 epitope tags, as well as EcoRI- and XbaI-cloning sites at the respective 5′ and 3′ ends (GeneArtTM, Toronto, Ontario, Canada). A nonspecific VHH gene (B-24) isolated from a hyper-immunized phagemid llama library with confirmed nonspecificity for 15-AcDON, or any other trichothecene mycotoxin, was used as a VHH intrabody control. Both VHH fragments were of the same immunoglobulin family and had similar molecular weights and isoelectric points. Like NAT-267, B-24 VHH DNA was PCR-amplified to include 3′ HA and His6 epitope tags and restriction cloning sites EcoRI and XbaI with the following gene-specific primers: VHH-B24For (5′-GGAATTCCATGCAGGTAAAGCTGGAGGAG-5′) and VHH-B24Rev (5′-GGGTCTAGACCCTATGCTCGGCCGGAACCGTAG-3′). A homology chart of nucleotide and peptide sequences of NAT-267 and B-24 VHHs is provided to show respective framework and complementarity-determining regions, endonuclease sites, and epitope tags (supplemental Fig. S1). VHH DNA inserts (∼0.46 kb) were ligated into the pPICZB vector (Invitrogen). Constructs were sequenced by StemCore Labs (Ottawa, Ontario, Canada) using the pPICZB-specific primers 5′ AOX1For (5′-GACTGGTTCCAATTGACAAGC-3′) and 3′ AOX1Rev (5′-GCAAATGGCATTCTGACATCC-3′).
Transformation
Expression constructs (Fig. 2) were linearized with SacI and electroporated into P. pastoris strain KM71H (Invitrogen). Transformants were plated onto yeast extract, peptone, dextrose, and sorbitol (YPDS) agar containing 100 μg·ml−1 Zeocin and incubated for 3 days at 30 °C until colonies formed. Ten colonies from each transformant were re-streaked on yeast extract, peptone, and dextrose (YPD) agar plates with 100 μg ml−1 Zeocin to ensure pure clonal isolates.
FIGURE 2.
Diagram of pPICZB expression vector (3745 bp) with VHH gene (NAT-267 = 387 bp, B-24 = 390 bp). Legend in the box shows schematic representation for each component of transgenic DNA for expression in P. pastoris.
Induction of VHH Expression
Single colonies of VHH and pPICZB control transformants were used to inoculate 5 ml of YPD media. Cultures were grown overnight (30 °C, 300 rpm). One ml of each culture was used to inoculate 100 ml of minimal glycerol medium with histidine (MGYH) and cultured in 1-liter baffled flasks (30 °C, 250–300 rpm) for 1 day. Cells were harvested by centrifugation (3000 × g for 5 min) at room temperature. To induce VHH expression, Pichia cells were resuspended in 20 ml of minimal methanol medium with histidine (MMH), transferred to 125-ml baffled flasks, and incubated at 30 °C with shaking at 300 rpm. Methanol (100%) was added to a final concentration of 0.5% (% v/v) every 24 h. Western blot analysis of 10 VHH (NAT-267 and B-24) transformants from 0, 24, 48, 72, 96, and 120 h post-induction was used to select clones and induction time points corresponding to the highest overall protein expression.
Preparation of Soluble VHH Extracts and Western Blot Analysis
Cell pellets were thawed on ice and resuspended in 100 μl of lysis buffer (50 mm Tris, pH 7.5, 1 mm EDTA, 0.5% Triton X-100, 1 mm dithiothreitol, 1× Complete Protease Inhibitor Mixture (Roche Applied Science)). Equal volumes of acid-washed 0.5-mm glass beads were added to each resuspended pellet, followed by successive 30-s cycles of vortexing and incubation on ice for a total of 10 cycles. Cell lysate samples were clarified by centrifugation (14,000 rpm for 10 min) at 4 °C. One hundred microliters of 2× SDS loading buffer (Bio-Rad) was added to the supernatant followed by boiling (95 °C for 5 min) and electrophoresis on 12% SDS-polyacrylamide gels. Samples were transferred to polyvinylidene difluoride membrane (Bio-Rad) and blocked overnight in 1% blocking reagent (Roche Applied Science) in TBS at 4 °C. VHH intrabody fragments were detected by probing for 1 h with rabbit anti-HA IgG primary antibody (Sigma) diluted 1:1000 in 0.5% blocking buffer followed by two washes with TBS + 0.1% Tween (TBST) and two washes with TBS + 0.5% blocking reagent. Secondary antibody (mouse anti-rabbit IgG horseradish peroxidase conjugate; Jackson ImmunoResearch, West Grove, PA), diluted 1:50,000 in 0.5% blocking buffer, was used to probe the membrane for 1 h, followed by four consecutive 15-min washes with TBST. VHH proteins were visualized with ECL Plus Western blotting detection system (GE Healthcare). Transformants with the highest VHH intrabody expression levels were used in subsequent in vivo cytotoxicity assays.
Cytotoxicity Assays
Cells were induced as described previously. Forty eight hours post-induction, P. pastoris cells were diluted to an A600 of 0.25 in YPD and incubated for 4 h (30 °C, 300 rpm). After this “recovery period,” cells were diluted to an A600 of 0.1 in YPD and immediately transferred (250 μl·well−1) to an F96 microwell assay plate (Nalge Nunc Inc., Naperville, IL). Pre-calibrated concentrations of the four ribotoxin treatments (15-AcDON, DON, 3-AcDON, or cycloheximide) prepared in DMSO and a DMSO-only control were added to P. pastoris cells contained in the wells of assay plates. All treatments, respective controls, and cell-free wells containing only YPD media (i.e. blank wells) were established in triplicate on each plate. Plates were sealed with Progene pressure-sensitive optical sealing film (Ultident, St.-Laurent, Quebec, Canada) prior to initiation of cytotoxicity assays. Cellular growth (30 °C, 300 rpm) was measured in real time based on measurement of A620 values at 25-min intervals, following 5 min of shaking (600 rpm) in a Polarstar Optima Microplate Reader (BMG Labtech, Gmbh, Offenberg, Germany). Assay results were considered valid if similar and reproducible effects were observed in three separate experiments, conducted in triplicate under the same test parameters.
Data Analysis
Results were presented as P. pastoris growth curves over a 24-h time period. Mean (n = 3) A620 values for VHH and pPICZB (empty vector) transformants used in ribotoxin and control treatments, minus A 620 values of YPD media (blank) wells, were plotted against time (i.e. 25-min intervals). Standard error values and paired t tests (p = 0.05 and 0.01) were used to assess statistically significant differences at the end of each 25-min time interval.
The time to doubling of initial P. pastoris A620 cellular growth values was calculated from cytotoxicity assay data to assess relative toxicity of each ribotoxin treatment. A comparative ranking of various ribotoxin treatments on P. pastoris growth was calculated based on time required to double A620 values of t = 0.0 h pPICZB empty vector transformants.
Differential area under curve (ΔAUC) values were calculated to quantify relative differences in P. pastoris growth over the full time course of each cytotoxicity assay. Relative differences in cellular growth were established by subtracting mean A620 values of pPICZB (empty vector) wells from corresponding values for VHH transformants. ΔAUC values for each toxophore treatment were calculated based on addition of differences in cellular growth between VHH and control transformants at each 25-min time interval across the full time course of each assay.
Quantitative Western Blot Analysis
20-ml cultures were centrifuged (1500 × g for 5 min at 4 °C) 48 h after methanol induction. Culture pellets were lysed and prepared for SDS-PAGE as described above. VHH lysate samples (500 μl) were diluted 1:5; 1:25; and 1:50 in nonreducing SDS sample buffer (Bio-Rad). pPICZB vector only (control) samples were not diluted. A reference VHH recombinant Ab fragment of a precisely defined concentration, and of similar size to NAT-267 and B-24 VHH with C-terminal HA and His6 epitope tags, was used to establish a standard dilution series at final protein concentrations of 300.0, 150, 75, 37.5, and 18.75 ng·lane−1. To decrease the bias of Western blot band intensities, VHH standard dilution series was prepared in P. pastoris cell lysate from pPICZB empty vector sample.
Samples were loaded onto 12.5% SDS-polyacrylamide gels and electrophoresed followed by transfer to a polyvinylidene difluoride membrane (Millipore, Billerica, MA) followed by a 1-h blocking with 3% nonfat skimmed milk in PBST. Mouse anti-His6 IgG primary antibody (GE Healthcare) diluted 1:2500 in 3% milk was added to membranes for 1 h followed by 1 h of blocking and three consecutive washes with PBST. Alkaline phosphatase-labeled goat anti-mouse IgG conjugate (Cedarlane Laboratories, Burlington, Ontario, Canada), diluted 1:3000 in blocking buffer, was used to probe the membrane for 1 h followed by three consecutive washes with PBST. VHH proteins were visualized using an alkaline phosphatase conjugate substrate kit (Bio-Rad).
Quantitative Western blots were dried and photographed with an AlphaImager 3400 (Alpha Innotech Corp., San Leandro, CA). Analysis was performed using the AlphaEaseFc software package (version 7.0.1, Alpha Innotech Corp.). Densitometric values for a standard five data point VHH dilution series (18.75–300 ng of VHH·lane−1) were used to produce regression equations. Densitometric values measured for 1:5, 1:25, and 1:50 dilutions of NAT-267 or B-24 VHH samples were expressed as VHH mass per μl of P. pastoris cell pellet (ng·μl−1) based on the following: VHH quantity (nanogram) (from standard regression equation) divided by cell pellet lysis volume loaded into each well (microliter). Cellular densities for each cytotoxicity assay were determined by multiplication of A600 of 1.0 ml of culture by 5 × 107 cells per ml of culture (constant). VHH concentration (fg·cell−1) was calculated as VHH mass per μl of cell pellets (ng·μl−1) (above) divided by cell density (number of cells·μl−1 culture). VHH intrabody concentration expressed as attomoles of VHH per cell (amol·cell−1) was calculated based on the following: VHH mass per cell (fg·cell−1) divided by molecular weight of NAT-267 or B-24 VHH proteins (16,637 and 16,976 Da, respectively). Finally, intracellular VHH molarity (μmol of VHH ·liter−1) was calculated as VHH concentration (amol·cell−1) divided by cell volume. P. pastoris cell volume was estimated to be 29 fl per cell (fl·cell−1) based on the average of independent measurements of generic yeast cell volume as follows: 42, 37, 70, and 83 fl (21–24) multiplied by 0.5 to account for an estimate that the cytosol represents 50% of total yeast cell volume (25).
Mean and standard error values of intracellular VHH expression values were calculated based on five independent quantitative Western blot assays. Additional details are summarized in supplemental Table S1.
VHH Immunolocalization
Cultures of P. pastoris cells expressing either NAT-267 VHH or pPICZB (empty vector) were induced with methanol (as described). Cells (20 A600 units) were pelleted and washed three times with PBS and resuspended in 500 μl of PBS. Fifty-μl aliquots of P. pastoris cells were incubated in 500 μl of fixative solution (1:1 acetone:methanol) for 20 min at −20 °C. Cells were pelleted (1000 × g for 1 min), washed three times with PBS, and resuspended in 500 μl of PBS supplemented with Lyticase (5 units·μl−1). After incubation (30 °C, 15 min.), cells were pelleted (as described) and incubated in 500 μl of permeabilization solution (3% bovine serum albumin, 0.5% Triton X-100 in PBS) for 30 min at room temperature. After washing with incubation solution (1% bovine serum albumin, 0.5% Tween 20), cells were probed with a 1:100 dilution of monoclonal anti-HA-fluorescein isothiocyanate antibody (Sigma) for 1–2 h at room temperature. Cells were mounted in PBS supplemented with 4′,6-diamidino-2-phenylindole dihydrochloride stain (Sigma) at a final concentration of 1.3 μg/ml. Cells were observed after 5 min using a Carl Zeiss LSM 510 Meta confocal microscope equipped with diode 405-nm and argon/2 line 488-nm lasers for excitation. Emission images were taken at 420–480 and 505–565 nm for 4′,6-diamidino-2-phenylindole dihydrochloride and fluorescein isothiocyanate, respectively.
Mycotoxin Biotransformation Assays
After 48 h of induction (see above), cultures were diluted to an A600 of 0.25 in YPD and incubated for 4 h (30 °C, 300 rpm). Cultures were diluted to A600 0.1 in YPD, and 2.5-ml aliquots of each culture were transferred to 50-ml tubes. Highly purified DON was added to respective samples to a final concentration of 100 μg·ml−1 followed by incubation (30 °C, 300 rpm). Culture samples (0.5 ml) were taken at intervals of 30 min and 16 and 24 h after mycotoxin addition and were used to measure A600 and culture pH. Samples were pelleted (3500 × g for 5 min). Supernatant was removed and transferred to 2.0-ml tubes. Samples were frozen in liquid nitrogen and stored at −80 °C. Lysate samples were prepared from cell pellets (as described above). All samples were split to enable concurrent anti-HA Western blot analysis (as described above) and mycotoxin biotransformation assays.
Supernatant, cell lysate, and pellet of selected samples were washed with double distilled H2O and passed through a 0.8-μm filter (Millipore). Filtrates were passed through Chromosep C18 columns (C18 Sep-Park cartridge, Waters) and pre-washed with 10 ml of 100% (w/v) HPLC grade methanol and 10 ml of double distilled H2O. After washing with double distilled water (10 ml), mycotoxin fractions were eluted with 10 ml of 100% (v/v) HPLC grade methanol and dried under a stream of N2. Selected culture samples (as described in supplemental Table S2) were diluted in 0.5 ml of HPLC grade methanol (100%) and 50.0-μl subsamples were used for HPLC analysis.
Trichothecenes were separated on a Shimadzu HPLC system equipped with a 150 × 4.6-mm C18 column (5 μm packing size) and a Hichrome UV flow cell. Standards and samples were eluted at a flow rate of 1.0 ml·min−1 with a 20-min linear gradient of 95% solvent A (degassed double distilled H2O), 5% solvent B (acetonitrile) to 40% solvent A, 60% solvent B. The column was washed from 20 to 25 min by ramping the gradient to 100% solvent B, holding at 100% solvent B for 5 min, and then returning to the starting conditions over 2 min. Assessment of DON biotransformation was based on assessment of retention times of sample peaks relative to trichothecene standard peaks.
Gas chromatography/mass spectrometry analysis, based on a standard Canadian Food Inspection Agency approved method (26), was used to further characterize and validate HPLC samples for structural composition of trichothecene analytes. Biotransformation samples from HPLC analysis were derivatized with 1.5 mg·ml−1 dimethylaminopyridine in toluene:acetonitrile (95:5) solution and trifluoroacetic anhydride. This solution was heated at 60 °C for 30 min and neutralized twice with 5% KH2PO4. Derivatized samples were analyzed on a Saturn 2000 GC/ion-trap mass spectrometer equipped for chemical ionization using acetonitrile.
1H NMR spectra of pooled P. pastoris lysate samples and subsequent HPLC fractions of those samples were obtained on a Bruker AM 500 NMR spectrometer in CDCl3. Chemical shifts are referenced to residual CHCl3 at 7.24 ppm for 1H spectra and reported (δ) relative to tetramethylsilane.
RESULTS
Sensitivity to Ribotoxin Treatments
The structure, composition, and molecular weight of the mycotoxins used in these experiments are shown in Fig. 1. The sensitivity of wild-type P. pastoris to trichothecene and cycloheximide (control) ribotoxin treatments, was governed by the dose and chemical structure tested. Treatments based on 15-AcDON resulted in the most immediate and largest overall reduction of cellular growth (Table 1). P. pastoris was comparatively less sensitive to equivalent doses of DON and cycloheximide. It was not possible to establish a dose-specific sensitivity to 3-AcDON relative to toxin-free DMSO media as cellular growth was not inhibited at the highest concentration (200 μg·ml−1) (Table 1).
TABLE 1.
Sensitivity of P. pastoris pPICZB empty vector transformants to ribotoxin treatments tested relative to toxin-free DMSO media
Least significant differences are based on a significant analysis of variance (p = 0.05). Standard deviation (σ) for each mean value is shown in parentheses.
| Dose (μg·ml−1) | Time to doubling of initial A620 cellular growth (min) |
||||
|---|---|---|---|---|---|
| 15-AcDON | DON | 3-AcDON | Cycloheximide | L.S.D.a | |
| 0b | 205 (2.2) | 221 (14.4) | 219 (21.7) | 221 (14.4) | 14.5 |
| 20 | 322 (4.4) | NDc | ND | ND | ND |
| 30 | 384 (4.4) | ND | ND | ND | ND |
| 40 | 439 (6.6) | ND | ND | ND | ND |
| 50 | 494 (8.8) | 308 (7.2) | 190 (124) | 283 (7.2) | 71.7 |
| 100 | 642 (138) | 417 (47.3) | 194 (119) | 310 (20.1) | 99.3 |
| 200 | 996 (312) | 590 (58.1) | 217 (13.0) | 341 (7.2) | 166.6 |
| L.S.D. | 117.2 | 40.2 | 91.3 | 14.0 | |
a L.S.D. means least significant difference (p = 0.05).
b Ribotoxin-free DMSO media were used.
c ND means not determined.
Transformation and VHH Intrabody Expression
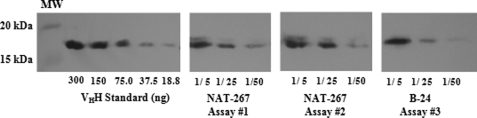
P. pastoris KM71H cells were successfully transformed with linearized NAT-267 (treatment) or B-24 (control) VHH DNA ligated into expression vector pPICZB (Fig. 2). Lysate fractions of methanol-induced transformants were assessed by anti-HA epitope Western blot analysis, and robust intracellular VHH expression was confirmed by the presence of a 17-kDa band at 48 and 72 h post-methanol induction (data not shown).
Quantitative Western Blot Assay
Mean levels of soluble intracellular VHH expression (t = 0 h) within transformants tested within cytotoxicity assays were determined by quantitative Western blot analysis (Fig. 3). The initial concentration of NAT-267 VHH in P. pastoris cells within cytotoxicity assays of 15-AcDON (Fig. 4) was 4.01 ± 0.31 amol of VHH·cell−1 (Table 2). Accounting for an estimated mean P. pastoris cytosol volume of 29 fl·cell−1 (as previously described), initial expression of NAT-267 VHH was equivalent to an intracellular concentration of 138 ± 10.8 μmol of VHH·liter−1. Likewise, mean NAT-267 intrabody concentration within cells used in cytotoxicity assays of DON, 3-AcDON, and cycloheximide (Fig. 5) was 3.18 ± 0.42 amol·cell−1 or 110 ± 14.4 μmol·liter−1 (Table 2). The average initial concentration of B-24 control VHH intrabody in cytotoxicity assays of 15-AcDON and cycloheximide (Fig. 6) was 1.93 ± 0.14 amol·cell−1 or 66.6 ± 4.3 μmol·liter−1 (Table 2).
FIGURE 3.
Quantitative Western blot analysis of VHH intrabody expression within cytotoxicity assays. 1st blot is VHH standard dilution series from 300.0 to 18.75 ng·well−1. 2nd blot is NAT-267 VHH expression for cytotoxicity assay 1 (Fig. 4). 3rd blot is NAT-267 VHH expression for cytotoxicity assay 2 (Fig. 5). 4th blot is B-24 (control) VHH expression for cytotoxicity assay 3 (Fig. 6). Bio-Rad molecular weight marker (MW) is indicated on the left. Protein dilutions are shown along the bottom of 2nd to 4th blots.
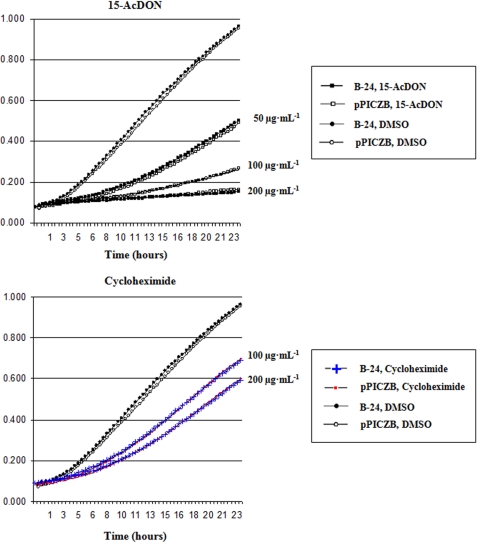
FIGURE 4.
Cytotoxicity assays showing the effect of various concentrations (20–200 μg·ml−1) of 15-AcDON on the growth of P. pastoris pPICZB and NAT-267 VHH transformants as measured by absorbance at 620 nm (OD620). ΔAUC values for NAT-267 VHH minus pPICZB empty vector are shown on each graph. At all concentrations of 15-AcDON (except 200 μg·ml−1) where blue versus red symbols are used, there was a significant difference (p = 0.01) at each time between NAT-267 and pPICZB transformants.
TABLE 2.
Mean VHH intrabody concentration (t = 0 h) within P. pastoris transformants used in cytotoxicity assays (Figs. 4–6)
Values were derived from data generated from quantitative Western blot assays (Fig. 3) as summarized in supplemental Table S1. VHH concentration values are expressed in femtograms and attomoles of VHH per cell, as well as micromoles·liter−1 VHH equivalent within P. pastoris cytosol. Standard errors for all means (n = 9) are shown in parentheses.
| VHH gene | Cytotoxicity assay | VHH intrabody concentration | ||
|---|---|---|---|---|
| fg·cell−1 | amol·cell−1 | μmol·liter−1 | ||
| NAT-267 | 15-AcDON (Fig. 4) | 66.8 (±5.2) | 4.01 (±0.31) | 138 (±10.8) |
| NAT-267 | DON, 3-AcDON, and cycloheximide (Fig. 5) | 53.0 (±7.0) | 3.18 (±0.42) | 110 (±14.4) |
| B-24 | 15-AcDON and cycloheximide (Fig. 6) | 32.8 (±2.3) | 1.93 (±0.14) | 66.6 (±4.3) |
FIGURE 5.
Cytotoxicity assays showing the effect of various concentrations (50–200 μg·ml−1) of DON and cycloheximide on the growth of P. pastoris pPICZB and NAT-267 VHH transformants as measured by absorbance at 620 nm (OD620). ΔAUC values for NAT-267 VHH minus pPICZB empty vector are shown for each concentration of DON. ΔAUC values for cycloheximide treatments were nil. At all concentrations of DON where green versus red symbols are used, there was a significant difference (p = 0.01) at each time between the NAT-267 and pPICZB transformants; this was not the case for all cycloheximide treatments.
FIGURE 6.
Cytotoxicity assays showing the effect of various concentrations (50–200 μg·ml−1) of 15-AcDON and cycloheximide on the growth of P. pastoris pPICZB and nonspecific B-24 VHH transformants as measured by absorbance at 620 nm (OD620). ΔAUC values for B-24 VHH minus pPICZB empty vector for both 15-AcDON and cycloheximide are nil. At all concentrations of DON and cycloheximide, there was no significant difference at each time between the B-24 and pPICZB transformants.
Cytotoxicity Assays
Intrabody VHH expression did not influence the cellular growth of transformed P. pastoris cultures in mycotoxin-free DMSO media, as A620 values and overall growth rate of NAT-267 and B-24 VHH transformants were similar to pPICZB empty vector control cells (Figs. 4–6).
15-AcDON
Significant differences (p = 0.01) in cellular growth were observed between NAT-267 VHH and pPICZB control transformants grown in media spiked with 20–100 μg·ml−1 15-AcDON (Fig. 4). The impact of NAT-267 VHH intrabody expression on cellular growth was dependent on the concentration of 15-AcDON used. At the lowest dose (20 μg·ml−1), growth of NAT-267 VHH-expressing cells was similar to cell lines grown in DMSO (control) media.
Time to significantly improved (p = 0.01) growth of NAT-267 VHH intrabody transformants was dose-dependent. Significant (p = 0.01) differences in A620 values were observed after 7.5 h of growth for cells cultured in medium supplemented with 15-AcDON at both 20 and 50 μg·ml−1 (Fig. 4). Assays spiked with 30, 40, and 100 μg·ml−1 15-AcDON had significant differences in A620 values after 4.6, 2.9, and 11.3 h of growth, respectively. No differences in A620 values were observed between NAT-267 VHH and control transformants at 200 μg·ml−1 15-AcDON (Fig. 4).
ΔAUC analysis was used to quantify relative differences in cellular growth between NAT-267 and B-24 (control) VHH transformants. The largest ΔAUC value (5.25) corresponded to NAT-267 VHH-expressing P. pastoris grown with 15-AcDON at 30 μg·ml−1. Cytotoxicity assays based on 15-AcDON at 20, 40, and 50 μg·ml−1 had similar ΔAUC values (4.48, 4.50, and 4.45, respectively) (Fig. 4). Accordingly, assays at 100 μg·ml−1 produced a substantially lower ΔAUC value (1.20), whereas NAT-267 VHH intrabody expression conferred no immunomodulation when spiked with 200 μg·ml−1 15-AcDON (Fig. 4).
DON, 3-AcDON, and Cycloheximide
NAT-267 VHH intrabody expression resulted in a dose-dependent response to DON. Although no beneficial impact on cellular growth was observed in cultures supplemented with 50 μg·ml−1 DON, NAT-267 VHH expression resulted in significantly improved cellular growth at 100 and 200 μg·ml−1 DON relative to pPICZB control transformants (Fig. 5A). Comparative differences in growth were greater for yeast spiked with 100 relative to 200 μg·ml−1 DON (ΔAUC values = 2.11 and 1.68, respectively). In addition, time to significant differences in A620 values was several hours later for the higher dose (Fig. 5A).
Differences in cellular growth between NAT-267 VHH and pPICZB control P. pastoris cells could not be established for cytotoxicity assays spiked with 3-AcDON because this trichothecene had little or no adverse effect on P. pastoris growth (Table 1). Assays based on cycloheximide (control ribotoxin) exhibited a dose-dependent effect on cellular growth; however, no differences were observed between P. pastoris NAT-267 VHH and pPICZB transformants spiked with doses of 50, 100, and 200 μg·ml−1 (Fig. 5B).
B-24 VHH Control
Expression of B-24 control VHH intrabody had no effect on cellular growth rate relative to pPICZB control transformants in DMSO or in media supplemented with either 15-AcDON or cycloheximide (Fig. 6). These results suggest that trichothecene-specific immunomodulation imparted by NAT-267 VHH intrabody expression was trichothecene-specific, as Pichia cells expressing a nonspecific VHH intrabody were equally as sensitive as pPICZB control transformants.
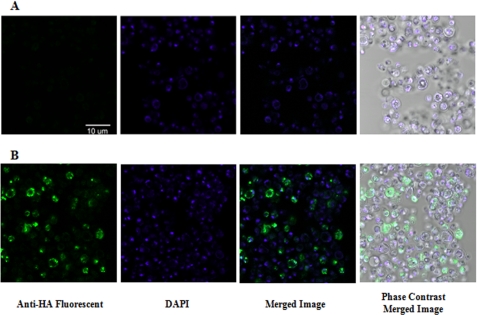
VHH Immunolocalization
NAT-267 VHH intrabody expression and distribution within P. pastoris transformants were further validated with fluorescent confocal microscopy (Fig. 7). Detection of the HA tag on NAT-267 VHH intrabody fragments within transformants grown in YPD media to A600 = 20 further confirmed robust intrabody expression within our test system.
FIGURE 7.
Representative confocal microscopy photographs of P. pastoris (KM71H) cells isolated, washed, and immunoprobed with anti-HA epitope monoclonal antibody-fluorescein isothiocyanate conjugate and 4′,6-diamidino-2-phenylindole (DAPI) stain after growing in YPD media to A600 = 20 (OD600). A, control, pPICZB empty vector. B, P. pastoris transformants expressing NAT-267 VHH intrabody shown by fluorescent green cells.
Mycotoxin Biotransformation Assays
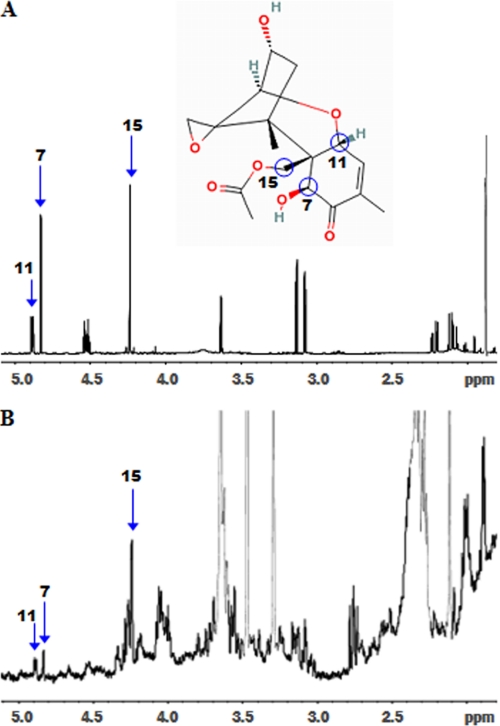
Given that NAT-267 VHH was previously confirmed to have binding affinity limited to 15-AcDON (18), we were surprised to observe a dose-dependent amelioration to DON cytotoxicity within NAT-267 VHH transformants (Fig. 5A). To validate this observation and re-confirm in vivo antigen specificity of NAT-267 VHH intrabody expression, we assessed the potential biotransformation of DON within our P. pastoris test system by spiking with DON. HPLC analysis of 16- and 24-h cultures grown in DON-supplemented YPD media consistently yielded another peak in addition to the DON peak at ∼5.85 min (supplemental Table S2). This peak appeared at ∼9.5 min and matched the retention time of 15-AcDON (supplemental Table S2). HPLC of cell lysate and supernatant samples of pPICZB and NAT-267 VHH transformants provided clear evidence of a metabolic conversion of DON to 15-AcDON. No trichothecenes were found within cell pellet fractions (supplemental Table S2). HPLC analysis revealed no evidence of DON-glucosyl metabolites or any structurally similar compounds within the samples tested. Gas chromatography/mass spectrometry analysis confirmed the presence of 15-AcDON within pooled cell lysate samples (data not shown). The GC retention time and mass spectrum of the peak confirmed the presence of 15-AcDON and absence of 3-AcDON within the samples tested (data not shown). Finally, the 1H NMR spectra of the total filtrate extract was dominated by DON and other impurities from the medium. However, the presence of 15-AcDON was confirmed in the NMR spectrum of the HPLC fraction isolated at ∼9.5 min from the pooled lysate samples (Fig. 8).
FIGURE 8.
A, representative 500-mHz 1H NMR spectrum of 15-AcDON standard. B, HPLC fraction of P. pastoris cell lysate sample (24 h). Unique chemical shifts of 15-AcDON are labeled due to H-11 (4.87 ppm), H-7 (4.81 ppm), and H-15 (4.21 ppm) protons, which confirm the presence of 15-AcDON within the cell lysate.
DISCUSSION
Various strategies have been used to develop plants with enhanced resistance to trichothecene cytotoxicity and associated Fusarium pathogenesis. These approaches tend to be based on mechanisms that alter host cellular targets (11, 12), reduce mycotoxin cytotoxicity (13, 14), or enhance innate host resistance through crop breeding techniques (27). Our goal was to evaluate the ability of a mycotoxin-specific VHH intrabody to immunomodulate, or attenuate, the cytotoxic effects of 15-AcDON using a yeast model.
Aside from application as a bioassay-indicator organism (28, 29), several species of yeast have been used to evaluate the in situ function of various trichothecene-specific genes (13, 30) or to assess the efficacy of mycotoxin-specific transgenes prior to in planta application (12, 13, 15). The methylotropic yeast P. pastoris (strain KM71H) was selected as our eukaryotic model based on expected sensitivity to target ribotoxins and known capacity to express high levels of functional heterologous proteins (31). We demonstrated that it was possible to transform, assess, and validate P. pastoris cells within a matter of weeks, thus creating an ideal platform for screening mycotoxin-specific constructs before in planta evaluation.
Trichothecene cytotoxicity is determined by the C-12,13 epoxide group common to this class of mycotoxins (3, 4, 6). However, the number and position of hydroxyl and acetyl ester groups on the trichothecene structure can also influence the mechanism of protein synthesis inhibition and relative toxicities within eukaryotic cells (32–34). In this regard, we found that the position of an acetyl ester group on either carbon 3 or 15 of DON (Fig. 1) had a profound effect on their cytotoxicity to P. pastoris (Table 1). Our findings align with results established in other yeast species where 15-AcDON was significantly more toxic than DON (12, 13), which was more toxic than 3-AcDON (35). These results also support the findings of structure-activity studies established in planta that demonstrated 15-AcDON is more toxic than DON (13, 34) and 3-AcDON (2, 14). Our observation of no measurable cytotoxicity of 3-AcDON is in agreement with previous studies that demonstrated that addition of an acetyl group at C-3 (Fig. 1) serves to eliminate cytotoxicity as part of a metabolic defense mechanism during trichothecene synthesis (36, 37).
Our results also quantify relative differences in trichothecene cytotoxicity over time, not at a specific assay end point. Our P. pastoris model system enables real time evaluation of cellular effects during the initial phases of ribotoxin exposure. We propose that this biological system is very sensitive (i.e. compared with whole plant systems) and shows clear effects on cellular growth within the first hours of exposure thereby indicating the potency of trichothecene-mediated cytotoxicity.
A sequential two-step process was required before commencement of each cytotoxicity assay. First, transformants were induced with methanol for 48 h. After induction of VHH intrabody expression, P. pastoris cells were transferred to YPD media for a 4-h recovery period to re-stimulate cellular growth. Mycotoxin or cycloheximide (control) treatments were then added (at t = 0 h) to initiate each cytotoxicity assay. Given the well established negative impact on eukaryotic protein synthesis, it was an obvious necessity to induce VHH expression prior to addition of ribotoxin treatments. The 4-h recovery in YPD media was required because induction conditions did not support an optimal cellular division and growth over time. We also noted that, by virtue of its design, our test system ensured a fixed ratio of VHH antibody to ribotoxin antigen throughout the course of each cytotoxicity assay. We recognized the potential disadvantage of culturing induced cells in YPD as leading to a continuous dilution of VHH concentration within the cytosol of rapidly dividing cells. However, we sought to maintain a fixed ratio of Ab:antigen within each microtiter well to ensure unbiased measurements of overall cellular growth.
Soluble NAT-267 and B-24 control VHH intrabody fragments were well expressed within P. pastoris. Average protein expression levels (Table 2) were found to be much higher than previously reported in P. pastoris (38–40). Furthermore, images generated by confocal immunomicroscopy confirmed that NAT-267 VHH fragments were well distributed within the cytosol of P. pastoris transformants even after growth to a very high A600 value (Fig. 7).
Our principal hypothesis was that the expression of mycotoxin-specific VHH intrabody fragments would effectively reduce (i.e. immunomodulate) bioavailable 15-AcDON concentrations within the cell and thereby limit cellular toxicity, thereby resulting in enhanced growth of P. pastoris. VHH-mediated mycotoxin binding was assessed by time-to-significant immunomodulation and ΔAUC values between VHH transformants and pPICZB (control) cell lines for each ribotoxin treatment tested (Figs. 4–6). The initial doses of 15-AcDON tested are generally equivalent to those found during Fusarium infection of agricultural crops. For example, intracellular levels of DON have been reported at concentrations ranging from 13.3 to 88.7 μg·g−1 within F. graminearum-inoculated cereal tissue (41, 42).
Attenuation of 15-AcDON activity clearly demonstrates that NAT-267 VHH intrabody expression conveys significant (p = 0.01) resistance to mycotoxin-specific cytotoxicity (Fig. 4) by regulating the availability of free and VHH-bound 15-AcDON. Consequently, results were dose-dependent as time to immunomodulation was optimal for 30 and 40 μg·ml−1 15-AcDON, when compared with the lower (20 μg·ml−1) and higher concentrations (i.e. 50 and 100 μg·ml−1) of the toxophore. At 15-AcDON concentrations of 100 μg·ml−1, immunomodulation was substantially reduced and delayed, although at 200 μg·ml−1, there was no benefit associated with VHH expression (Fig. 4).
NAT-267 VHH intrabody immunomodulation was trichothecene-specific as no attenuation of 15-AcDON toxicity was observed in assays supplemented with control ribotoxin (cycloheximide) (Fig. 5). It was not possible to assess the effect of NAT-267 VHH on the cytotoxicity of 3-AcDON because all doses tested were not toxic to P. pastoris cells (Table 1). Further confirmation of NAT-267 VHH specificity was shown with cytotoxicity assays using P. pastoris transformants expressing a control VHH intrabody, i.e. B-24. Furthermore, there was no immunomodulation of either 15-AcDON- or cycloheximide-specific cytotoxicity when pPICZB control cells were used (Fig. 6).
The attenuation of DON cytotoxicity at 100 and 200 μg·ml−1 (Fig. 5) was an unexpected result because we previously reported that NAT-267 VHH has no affinity for DON (18). In vivo immunomodulation of DON was explained by confirmation of biotransformation of DON to 15-AcDON in P. pastoris cell lysate and culture supernatant samples of NAT-267 and pPICZB transformants (supplemental Table S2 and Fig. 8). Data mining of the genome of NRRL Y-11430 P. pastoris (the parent strain of KM71H) by Integrated Genomics Inc. (Chicago) revealed no similarities or related homologs to a previously characterized fungal gene responsible for acetylation of DON to 15-AcDON (43). We therefore attributed this biochemical conversion to a previously uncharacterized acetyltransferase gene within the P. pastoris genome.
To predict the impact of NAT-267 VHH intrabody binding to reduce effective 15-AcDON toxin concentrations within the cytosol, we adopted a previously described (44) mass-balance model based on Equation 1.
We assumed a linear, dose-independent binding of VHH to 15-AcDON within an aqueous environment, where 1 g = 1 ml. The dissociation constant (KD) of NAT-267 VHH was taken as 1.24 μm (18) with 1:1 stoichiometry of Ab:antigen binding. NAT-267 ([VHH]) concentration within the P. pastoris cytosol was estimated to be 138 μmol·liter−1 (supplemental Table 2). We assumed optimal disulfide bond formation and post-translational VHH folding within the endoplasmic reticulum and cytosol (45) with no NAT-267 VHH intrabody leakage from the P. pastoris cell membrane. The concentration of 15-AcDON within the cytosol was taken as equivalent to culture media, and we also assumed that there were no trichothecene targets outside of the cells.
Based on total 15-AcDON concentration of 50 μg·ml−1 (or 148 μm) (Fig. 4), this model predicts (138 × 148 μm) ÷ (138 + 1.24 μm) = 147 μm 15-AcDON (or >99.9%) toxin was bound, and ∼1.3 μm (or <0.1%) of 15-AcDON was free. This simple model indicates that 15-AcDON cytotoxicity at 50 μg·ml−1 should be completely eliminated by expression of NAT-267 VHH; however, although the effects of 15-AcDON were significantly ameliorated by VHH expression at this concentration, they were not completely eliminated (Fig. 4).
A more complete assessment of NAT-267 VHH immunomodulation should also account for in situ competition effects and binding affinity between 15-AcDON and its major cellular target, i.e. ribosome binding. Binding affinity is also an important consideration because NAT-267 VHH has a relatively low affinity (KD = 1.24 μm) for 15-AcDON that is mediated by weak, noncovalent interactions (e.g. van der Waals forces, hydrogen bonding, etc.) (46). Due to the rapid dissociation rate constant for NAT-267, we hypothesize that 15-AcDON is subject to continuous turnover in terms of mycotoxin binding to the VHH. In other words, the efficiency of NAT-267 VHH in terms of limiting the cytotoxicity of 15-AcDON is limited by the short “residence time” of hapten binding to the VHH (47). Further evidence for this hypothesis resides in the fact that even when the VHH intrabody is expressed in excess (138 μmol·liter−1; Table 2) compared with 15-AcDON, i.e. at 89 μm (30 μg ml−1), NAT-267 cannot fully immunomodulate the cytotoxic effects of 15-AcDON when compared with controls (Fig. 4). Thus, at higher doses of ≥30 μg·ml−1 15-AcDON, we postulate that the VHH simply cannot compete with a stronger and quite possibly longer residence time of 15-AcDON on the 60 S ribosomal protein subunit L3 (Rpl3) of P. pastoris.
We also assert that, at very high doses, 15-AcDON may cause other potentially irreversible effects such as membrane disruption, inhibited RNA and DNA synthesis, and various other apoptotic effects (reviewed in Refs. 4–6), which may also severely limit cellular growth. Thus, to accurately determine the efficacy of NAT-267 VHH intrabody in P. pastoris cells, one must account for the dynamic nature of mycotoxin-mediated cytotoxicity and in vivo binding kinetics of 15-AcDON for other cellular binding targets. Our assertion that attenuation of trichothecene-specific cytotoxicity is not a simple process is in agreement with previous work that demonstrated only partial remediation of DON cytotoxicity based on in planta expression of an altered Rpl3 target protein (11, 12) or the expression of Tri101 (14, 36).
Most phytopathogenic Fusarium species are believed to produce 15-AcDON or 3-AcDON as acetylated precursors to DON (15). Production of each acetylated compound has been described as geography-dependent; 3-AcDON chemotypes dominate in Asia and Europe, although Fusarium species, which produce 15-AcDON, are more prevalent in North America (48, 49). Therefore, the accumulation of DON within plant tissue is believed to be a metabolic process conferred by fungal and plant carboxylesterases that continuously deacetylate either 15-AcDON or 3-AcDON within plant tissue (15). Confirmation of NAT-267 efficacy observed in this work is very significant because 15-AcDON was the most cytotoxic compound tested. If 15-AcDON is as toxic to plants as it is to yeast, then expressing NAT-267 VHH within the cytosol before trichothecene accumulation may help limit in vivo pathogenesis and metabolism to DON during Fusarium infection of plants such as corn and wheat.
Future experiments will focus on the development of an anti-15-AcDON VHH with an improved dissociation constant (KD) to ensure a longer association between the VHH and target ligand for improved in vivo efficacy (47). It would also be of great interest to develop and test novel VHH fragments with affinity for various other trichothecenes (e.g. neosolaniol, diacetoxyscripenol, T-2 toxin, etc.) and various other mycotoxin classes (e.g. fumonisins, aflatoxins, etc.) using this test system. A logical subsequent application would be constitutive in planta expression of optimized mycotoxin-specific VHH fragments, possibly with catalytic activity, to bind and deactivate/degrade mycotoxins during critical initial periods of plant pathogenesis.
Acknowledgments
We thank Dr. Barbara Blackwell at Agriculture and Agri-Food Canada (Ottawa) for assistance, insights, and expertise in HPLC, gas chromatography/mass spectrometry, and NMR assays to characterize mycotoxin biotransformation samples. We are also indebted to colleagues at the Canadian Food Inspection Agency for gas chromatography/mass spectrometry analysis of biotransformation samples.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs Program, and the Ontario Ministry of Agriculture, Food, and Rural Affairs (to J. C. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
- DON
- deoxynivalenol
- 15-AcDON
- 15-acetyldeoxynivalenol
- 3-AcDON
- 3-acetyldeoxynivalenol
- ΔAUC
- differential area under curve
- HA
- hemagglutinin
- HPLC
- high pressure liquid chromatography
- Ab
- antibody
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Demcey Johnson D., Flaskerud G. K., Taylor R. D., Satyanarayana V. (2003) Fusarium Head Blight of Wheat and Barley (Leonard K. J., Bushnell W. R. eds) pp. 461–483, American Phytopathological Society, St. Paul, MN [Google Scholar]
- 2.Desjardins A. E. (2006) Fusarium Mycotoxins Chemistry, Genetics, and Biology, pp. 13–64, American Phytopathological Society, St. Paul, MN [Google Scholar]
- 3.Carter C. J., Cannon M. (1977) Biochem. J. 166, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha O., Ansari K., Doohan F. M. (2005) Food Addit. Contam. 22, 369–378 [DOI] [PubMed] [Google Scholar]
- 5.Ueno Y. (1984) Fundam. Appl. Toxicol. 4, S124–S132 [DOI] [PubMed] [Google Scholar]
- 6.Rotter B. A., Prelusky D. B., Pestka J. J. (1996) J. Tox. Env. Health 48, 1–34 [DOI] [PubMed] [Google Scholar]
- 7.Pestka J. J. (2008) Food Addit. Contam. 25, 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Mello J. P. F., Placinta C. M., MacDonald A. M. C. (1999) Anim. Feed Sci. Tech. 80, 183–205 [Google Scholar]
- 9.Mesterházy Á. (2002) Eur. J. Plant Pathol. 108, 675–684 [Google Scholar]
- 10.Harris L. J., Desjardins A. E., Plattner R. D., Nicholson P., Butler G., Young J. C., Weston G., Proctor R. H., Hohn T. M. (1999) Plant Dis. 83, 954–960 [DOI] [PubMed] [Google Scholar]
- 11.Harris L. J., Gleddie S. C. (2001) Physiol. Mol. Plant Pathol. 58, 173–181 [Google Scholar]
- 12.Mitterbauer R., Poppenberger B., Raditsching A., Lucyshyn D., Lemmens M., Glössl J., Adam G. (2004) Plant Biotech. J. 2, 329–340 [DOI] [PubMed] [Google Scholar]
- 13.Poppenberger B., Berthiller F., Lucyshyn D., Sieberer T., Schuhmacher R., Krska R., Kuchler K., Glössl J., Luschnig C., Adam G. (2003) J. Biol. Chem. 278, 47905–47914 [DOI] [PubMed] [Google Scholar]
- 14.Alexander N. J. (2008) World Mycotoxin J. 1, 31–37 [Google Scholar]
- 15.Mitterbauer R., Adam G. (2002) Eur. J. Plant Pathol. 108, 699–703 [Google Scholar]
- 16.Holliger P., Hudson P. J. (2005) Nat. Biotechnol. 23, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 17.Muyldermans S. (2001) J. Biotechnol. 74, 277–302 [DOI] [PubMed] [Google Scholar]
- 18.Doyle P. J., Arbabi-Ghahroudi M., Gaudette N., Furzer G., Savard M. E., Gleddie S., McLean M. D., Mackenzie C. R., Hall J. C. (2008) Mol. Immunol. 45, 3703–3713 [DOI] [PubMed] [Google Scholar]
- 19.Saerens D., Ghassabeh G. H., Muyldermans S. (2008) Curr. Opin. Pharmacol. 8, 1–9 [DOI] [PubMed] [Google Scholar]
- 20.Jobling S. A., Jarman C., Teh M. M., Holmberg N., Blake C., Verhoeyen M. E. (2003) Nat. Biotechnol. 21, 77–80 [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M. (2002) Science 297, 395–400 [DOI] [PubMed] [Google Scholar]
- 22.Tyson C. B., Lord P. G., Wheals A. E. (1979) J. Bacteriol. 138, 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman F. (2000) Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 24.Tamaki H., Yun C. W., Mizutani T., Tsuzuki T., Takagi Y., Shinozaki M., Kodama Y., Shirahige K., Kumagai H. (2005) Genes Cells 10, 193–206 [DOI] [PubMed] [Google Scholar]
- 25.Biswas S. K., Yamaguchi M., Naoe N., Takashima T., Takeo K. (2003) J. Electron Microsc. 52, 133–143 [DOI] [PubMed] [Google Scholar]
- 26.Schwadorf K., Müller H. M. (1991) Chromatographia 32, 137–142 [Google Scholar]
- 27.Snijders C. H. A. (2004) Toxicol. Lett. 153, 37–46 [DOI] [PubMed] [Google Scholar]
- 28.Abolmaali S., Mitterbauer R., Spadiut O., Peruci M., Weindorfer H., Lucyshyn D., Ellersdorfer G., Lemmens M., Moll W. D., Adam G. (2008) J. Microbiol. Methods 72, 306–312 [DOI] [PubMed] [Google Scholar]
- 29.Binder J. (1999) Nat. Toxins 7, 401–406 [DOI] [PubMed] [Google Scholar]
- 30.Alexander N. J., McCormick S. P., Hohn T. M. (1999) Mol. Gen. Genet. 261, 977–984 [DOI] [PubMed] [Google Scholar]
- 31.Cereghino J. L., Cregg J. M. (2000) FEMS Microbiol. Rev. 24, 45–66 [DOI] [PubMed] [Google Scholar]
- 32.Betina V. (1989) Chem. Biol. Interact. 71, 105–146 [DOI] [PubMed] [Google Scholar]
- 33.Ehrlich K. C., Daigle K. W. (1987) Biochim. Biophys. Acta 923, 206–213 [DOI] [PubMed] [Google Scholar]
- 34.Desjardins A. E., McCormick S. P., Appell M. (2007) J. Agric. Food Chem. 55, 6487–6492 [DOI] [PubMed] [Google Scholar]
- 35.Binder J., Horvath E. M., Schatzmayr G., Ellend N., Danner H., Krska R., Braun R. (1997) Cereal Res. Commun. 25, 343–346 [Google Scholar]
- 36.Kimura M., Kaneko I., Komiyama M., Takatsuki A., Koshino H., Yoneyama K., Yamaguchi I. (1998) J. Biol. Chem. 273, 1654–1661 [DOI] [PubMed] [Google Scholar]
- 37.Kimura M., Takahashi-Ando N., Nishiuchi T., Ohsato S., Tokai T., Ochiai N., Fujimura M., Kudo T., Hamamoto H., Yamaguchi I. (2006) Pest. Biochem. Physiol. 86, 117–123 [Google Scholar]
- 38.Gurkan C., Symeonides S. N., Ellar D. J. (2004) Biotechnol. Appl. Biochem. 39, 115–122 [DOI] [PubMed] [Google Scholar]
- 39.Omidfar K., Rasaee M. J., Kashanian S., Paknejad M., Bathaie Z. (2007) Biotechnol. Appl. Biochem. 46, 41–49 [DOI] [PubMed] [Google Scholar]
- 40.Rahbarizadeh F., Rasaee M. J., Forouzandeh M., Allameh A. A. (2006) Mol. Immunol. 43, 426–435 [DOI] [PubMed] [Google Scholar]
- 41.Del Ponte E. M., Fernandes J. M. C., Bergstrom G. C. (2007) J. Phytopath. 155, 577–581 [Google Scholar]
- 42.Mudge A. M., Dill-Macky R., Dong Y., Gardiner D. M., White R. G., Manners J. M. (2006) Physiol. Mol. Plant Pathol. 69, 73–85 [Google Scholar]
- 43.McCormick S. P., Hohn T. M., Desjardins A. E. (1996) Appl. Environ. Microbiol. 62, 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almquist K. C., Niu Y., McLean M. D., Mena F. L., Yau K. Y. F., Brown K., Brandle J. E., Hall J. C. (2004) Plant Biotech. J. 2, 189–197 [DOI] [PubMed] [Google Scholar]
- 45.Ellgaard L. (2004) Biochem. Soc. Trans. 32, 663–667 [DOI] [PubMed] [Google Scholar]
- 46.Maynard J., Georgiou G. (2000) Annu. Rev. Biomed. Eng. 2, 339–376 [DOI] [PubMed] [Google Scholar]
- 47.Copeland R. A., Pompliano D. L., Meek T. D. (2006) Nature 5, 730–739 [DOI] [PubMed] [Google Scholar]
- 48.Goswami R. S., Kistler H. C. (2004) Mol. Plant Pathol. 5, 515–525 [DOI] [PubMed] [Google Scholar]
- 49.Moss M. O., Thrane U. (2004) Toxicol. Lett. 153, 23–28 [DOI] [PubMed] [Google Scholar]