Abstract
Proteins containing defined recognition modules mediate readout and translation of histone modifications. These factors are thought to initiate downstream signaling events regulating chromatin structure and function. We identified CDYL1 as an interaction partner of histone H3 trimethylated on lysine 9 (H3K9me3). CDYL1 belongs to a family of chromodomain factors found in vertebrates. We show that three different splicing variants of CDYL1, a, b, and c, are differentially expressed in various tissues with CDYL1b being the most abundant variant. Although all three splicing variants share a common C-terminal enoyl-CoA hydratase-like domain, only CDYL1b contains a functional chromodomain implicated in H3K9me3 binding. A splicing event introducing an N-terminal extension right at the beginning of the chromodomain of CDYL1a inactivates its chromodomain. CDYL1c does not contain a chromodomain at all. Although CDYL1b displays binding affinity to methyl-lysine residues in different sequence context similar to chromodomains in other chromatin factors, we demonstrate that the CDYL1b chromodomain/H3K9me3 interaction is necessary but not sufficient for association of the factor with heterochromatin. Indeed, multimerization of the protein via the enoyl-CoA hydratase-like domain is essential for H3K9me3 chromatin binding in vitro and heterochromatin localization in vivo. In agreement, overexpression of CDYL1c that can multimerize, but does not interact with H3K9me3 can displace CDYL1b from heterochromatin. Our results imply that multimeric binding to H3K9me3 by CDYL1b homomeric complexes is essential for efficient chromatin targeting. We suggest that similar multivalent binding stably anchors other histone modification binding factors on their target chromatin regions.
Introduction
For packaging the chromosomes of an eukaryotic cell into the nucleus the negatively charged DNA is wrapped around a positively charged octamer of histone proteins consisting of two H2A–H2B dimers and one (H3–H4)2 tetramer. 147 bp of DNA are wound around one histone octamer forming the fundamental repeating unit of chromatin, the nucleosome. N- and C-terminal tails of the histones are protruding out of this structural entity.
Histones are subject to a plethora of post-translational modifications (1–3). Among these methylation of lysine residues plays a special role. Specific sites of lysine methylation as well as distinct stages of lysine methylation are associated with different nuclear processes (4, 5). For example, trimethylation of H3 lysine 4 (H3K4me3) is found in euchromatic structures, which are open for transcription and are early replicating during S-phase. In contrast trimethylation of lysine 9 of histone 3 (H3K9me3) is accumulated at heterochromatic regions. These are densely packed, mostly transcriptional silent and late replicating in S-phase (6, 7).
Whereas histone modifications might directly affect chromatin structure (8, 9), a number of protein domains have been identified that specifically bind certain histone modifications (10, 11). These factors are thought to read-out and translate the effects of individual histone modifications or combinations thereof. Different proteins containing chromodomains have been implicated in binding methylated histone lysine residues preferentially when in higher (tri- and dimethylated) states. For example, heterochromatin protein 1 (HP1)4 was shown to recognize H3K9me, whereas Polycomb binds H3K27me (12). Structural analysis of a number of binding domains has identified aromatic cages of at least three residues as central elements in histone methyl-lysine binding (10). Although the interaction of different protein domains with histone modifications has been well studied in vitro using isolated histone tail peptides, the exact parameters by which these proteins are recruited to their target sites on chromatin have not been worked out.
Recently, a new chromodomain containing group of proteins was described (13). The cdy (chromodomain on the Y) family represents a set of related genes in higher eukaryotes. In humans the CDY family comprises two autosomal genes, namely CDYL1 and CDYL2 as well as multiple gene copies of CDY on the Y chromosome (14). Evolutionary, a common ancestor of the cdyl gene appeared first in deuteriostomia. This predecessor was later duplicated to yield the cdyl1 and cdyl2 genes. A very recent multiplication of the cdyl1 gene in the primate lineage lead to multiple cdy copies on the Y chromosome (14–16).
CDY family proteins have a N-terminal chromodomain, a central hinge region, and a C-terminal enoyl-CoA hydratase-like (ECH) domain in common (see Fig. 1A). Recently, we could show that the chromodomains of CDY and CDYL2, which have high sequence similarities with HP1 and Polycomb chromodomains, can recognize di- or trimethylated histone and non-histone lysine residues. The strongest interaction was found for the H3K9me3 modification (17). In contrast, CDYL1 was not able to recognize H3K9me3 because of subtle sequence differences at the very beginning of the chromodomain affecting the first of three aromatic cage residues. Mutagenesis of a few residues at the N terminus of the CDYL1 chromodomain lead to restored binding activity to H3K9me3 peptides in vitro and partial relocalization to heterochromatic regions in vivo. Other studies indicated that CDYL1 binding to H3K9me3 might be increased after methylation by the histone methyltransferase G9a (18). Interestingly, biochemical pull-down experiments identified CDYL1 as the binding protein of automethylated G9a (19). The exact reasons for the different methyl-lysine binding behavior of CDYL1 are not understood.
FIGURE 1.
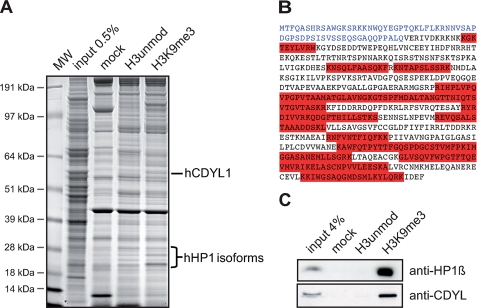
CDYL1 is an H3K9me3 binding factor. A, the indicated histone H3 peptides were used in pulldown experiments of HeLa S3 cell nuclear extract. Beads without coupled peptides were used as control (mock). Specifically recovered proteins were run on SDS-PAGE and stained with Coomassie Blue. Proteins were identified by MS/MS analysis. The positions of CDYL1 and HP1 proteins are indicated on the right. Molecular weight (MW) markers are indicated on the left. B, peptide sequence of CDYL1. The highlighted (red) peptides were identified by MS/MS analysis. The N terminus of CDYL1 is shown in blue. C, Western blot analysis of a histone H3 peptide pulldown experiment of HeLa S3 cell nuclear extract as in A using the indicated antibodies.
Biochemically, CDYL1 was found in the CoREST complex where it bridges the repressor REST and the histone methyltransferase G9a (20–22). The ECH-like domain of CDYL1 was shown to interact with histone deacetylases HDAC1 and HDAC2 likely via CoREST association thereby acting as corepressor during transcriptional repression (23). Conflicting results have suggested that the ECH-like domain of CDY and CDYL1 might constitute a histone acetyltransferase activity in elongating spermatids during hyperacetylation and replacement of histones (24). In peroxisomes and mitochondria, trimeric enoyl-CoA hydratases accomplish the hydration of the double bond of fatty acids during β-oxidation (25, 26). The functional impact of putative multimerization of ECH-like domains onto the CDY family protein function has not yet been investigated.
In this study we present results that clarify the different binding abilities reported for CDYL1 to methylated lysine residues. CDYL1 exists in three different splicing variants, a, b, and c. The b form is not only the most abundant splicing variant but is also exclusively able to recognize H3K9me3 in vitro and in vivo. We further demonstrate that the chromodomain of CDYL1 alone is not sufficient to stably bind to H3K9me3 chromatin in vitro and to mediate localization to DNA-dense heterochromatic regions in vivo. Besides a functional chromodomain multimerization of the ECH-like domain is necessary to bring CDYL1b to heterochromatin.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids for expression of hCDYL1a were described elsewhere (17). cDNAs corresponding to the open reading frame of hCDYL1b or hCDYL1c were amplified from an EST clone (IMAGE: 6140263) using PCR and cloned into a derivative pcDNA3.1 vector (Invitrogen) generating C-terminal fusion to a 2× FLAG-2× HA epitope tag. The GenBankTM accession numbers of the cDNAs used are as follows: hCDYL1a, AF081259; hCDYL1b, BC108725; hCDYL1c, BC119682. For expression in Escherichia coli cDNAs corresponding to the open reading frame of hCDYL1a, hCDYL1b, and hCDYL1c were subcloned into pET11a (Novagen). The hCDYL1b chromodomain (1–78 aa) was cloned into pET16b (Novagen) for expression with a His10 tag. hCDYL1c (309 aa) and hCDYL1cΔCterm (1–236 aa) were cloned into pcDNA3.1myc-His (Invitrogen). cDNAs corresponding to the CDYL1b fragments described in Fig. 5C were PCR amplified from Xenopus laevis EST clone XL213m10 and cloned into the pCS2+ vector (RZPD) generating fusion to a C-terminal 1× FLAG epitope: chromo, 1–64 aa; chromohingeΔNLS, 1- 233 aa; chromohinge, 1–295 aa; hingeECH, 64–541 aa; CDYL1cΔC-term, 233–541 aa; CDYL1cΔNLS, 295–541 aa; ECH, 295–463 aa; chromoCDYL1c, 1–64 aa fused to 233–541 aa; and chromoCDYL1cΔC-term, 1–64 fused to 233–463 aa.
FIGURE 5.
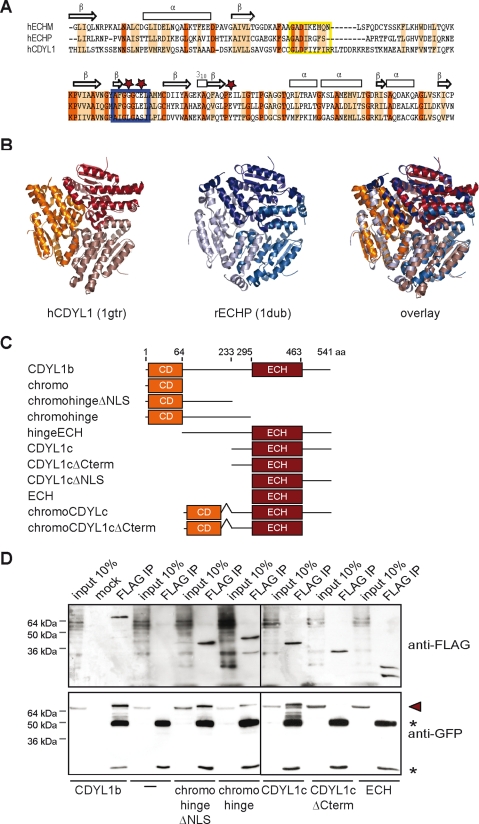
CDYL1b multimerizes via its ECH-like domain. A, sequence alignment of the human CDYL1 ECH-like domain with mitochondrial (ECHM) and peroxisomal enoyl-CoA hydratases (ECHP). Identical residues are highlighted in red; homolog residues are highlighted in orange. Secondary structure elements of ECH enzymes are indicated on the top. Stars mark residues directly involved in ECH catalysis. Blue box, catalytic center; yellow box, adenine binding site. B, tertiary and quaternary structures of human CDYL1 (1gtr, red) and rat ECHP (1dub, blue) as determined by x-ray crystallography are shown. The image on the right represents an overlay of hCDYL1 and rECHP structures. Different color shading indicates different polypeptides. C, schematic representation of XlCDYL1 constructs used for immunoprecipitation and immunofluorescence experiments (see D, and Figs. 6 and 7A). Domain boundaries of the mutant proteins are as indicated by the aa positions. D, the FLAG-tagged CDYL1 proteins indicated on the bottom were produced by in vitro translation together with full-length YFP-CDYL1b. Expressed proteins (input) as well as material bound to beads (mock) or anti-FLAG antibodies (FLAG IP) were analyzed by Western blotting using the indicated antibodies. Arrowhead marks position of YFP-CDYL1b; asterisks mark antibody heavy and light chain.
Real Time PCR
Total RNA of human tissues was a gift from Dr. Thomas Giger (Max Planck Institute for Evolutionary Anthropology, Leipzig). For real time PCR total RNA was isolated according to the TRIzol protocol (Invitrogen). DNA was digested with a DNA free kit (Ambion). cDNA was made using random hexamers and the First Strand Synthesis kit (Invitrogen). cDNA was used for real time PCR using IQ SYBR Green Supermix on a MJ Research DNA engine Opticon (Bio-Rad). The following primers were used for real time PCR detection: hCDYL1a forward, 5′-GGTCAGCCTGGGGAAAAAGC-3′; hCDYL1a reverse, 5′-CGGGAGGCTGCTGTGCC-3′; hCDYL1b forward, 5′-CTTCCGAGGAGCTGTACGAGGTTG-3′; hCDYL1b reverse, 5′-TCTCCGTGTGGCGTCTGTTGAA-3′; hCDYL1c forward, 5′-GCTTCCGAGGAGCTGTACGAGTACATCTC-3′; hCDYL1c reverse, 5′-CAAAAGGCTGGTCTCTTCTGTCGTCAAT-3′.
Western Blotting
For Western blot analysis primary antibodies were used as follows: anti-CDYL (Abcam), 1:1,000; anti-H3, 1:40,000 (Abcam); anti-HP1α and anti-HP1β (Upstate), 1:2,000; anti-FLAG (Sigma), 1:1,000; anti-H3 (Upstate), 1:10,000; anti-green fluorescent protein (Roche), 1:10,000.
Recombinant Chromatin
Expression and purification of WT X. laevis histones was performed as described (27). H3K9me3 was generated by native protein ligation (28). In short, the coding sequence for X. laevis H3Δ1–20,C21A was amplified by PCR and cloned into the pET3d expression vector. The truncated H3 protein was expressed and purified like the WT histones. The H3 N-terminal peptide containing residues 1–20 and trimethylated lysine 9 was synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl)-based solid-phase synthesis and activated at the C terminus by thioesterification. Ligation of the activated H3 peptide to the truncated H3 histone and purification of the ligation product was performed as described (28). Identity and purity of histones was verified by SDS-PAGE as well as by mass spectrometry (supplemental Fig. S2).
Assembly of histone octamers containing H3unmod and H3K9me3 as well as reconstitution of recombinant oligonucleosomes was performed by salt dialysis as described using the 12 × 200 × 601 template (27, 29). Briefly, octamers were reconstituted using H3unmod or H3K9me3 and purified by gel filtration on Superdex 200 (GE Healthcare). Scavenger DNA corresponding to a 148-bp length fragment PCR amplified from pUC18 was used in all reconstitutions. Assembly reactions were titrated at different octamer:DNA ratios. Reproducibly, an octamer:DNA ratio of 1.1:1 resulted in saturated nucleosomal arrays. Assembly reactions were analyzed by native gel electrophoresis and analytical ultracentrifugation. To further control the saturation level of the oligonucleosomal templates, assembled material was analyzed after digestion with the restriction enzyme HhaI, which cuts within every 601 repeat that is not protected by histone octamers in the 12-mer template. Also, material was analyzed by scanning force microscopy after mild fixation with glutaraldehyde and fixation on mica support (see supplemental Fig. S3) (30). Last, responsiveness of reconstituted material to higher order compaction by Mg2+ titration for chromatin templates containing H3unmod and H3K9me3 was compared (supplemental “Materials and Methods” and Fig. S4). Reconstituted material was used for all measurements without further purification after extensive dialysis against 10 mm triethanolamine, 0.1 mm EDTA, pH 7.5.
Peptide and Chromatin Pulldowns
Peptides used for pulldown studies carried a biotinylated lysine residue at the C terminus: H3unmodified, ARTKQTARKSTGGKAPRKQLK-biotin; H3K9me3, ARTKQTARK(me3)STGGKAPRKQLK-biotin. For peptide pulldown 10 μg of biotinylated histone peptide was bound to 40 μl of prewashed strepatvidin-coated magnetic beads (Pierce) for 3 h at room temperature. 1 ml of precleared HeLa S3 nuclear extract (5 mg/ml) prepared as described was incubated with the peptide-bound magnetic beads overnight at 4 °C. Beads were washed six times with 1 ml of PD150 (20 mm HEPES, pH 7.9, 150 mm KCl, 0.2% (v/v) Triton X-100, 20% (v/v) glycerol) supplemented with Protease Inhibitor Complete EDTA-free (Roche Applied Science). After boiling the beads in loading buffer recovered proteins were separated on 4–20% gradient SDS-polyacrylamide gels (Invitrogen). Proteins were either detected by Western blot procedures or analyzed by mass spectrometry (MS). Chromatin pulldowns were carried out accordingly using 2 μg of recombinant chromatin assembled on biotinylated DNA templates.
Chromatin Precipitation
Proteins were translated in vitro using the T7 TnT kit (Promega). 1.75 μg of chromatin and 45 μl of the TnT reaction were mixed in 500 μl of end volume of CP buffer (20 mm Tris-HCl, 0.2% (v/v) Triton X-100, 150 mm NaCl, 1 mm dithiothreitol, protease inhibitor EDTA free) and incubated 2 h at 4 °C. MgCl2 was added to 5 mm final concentration and the reactions were incubated for 15 min at 4 °C. Precipitated chromatin was collected at 13,000 × g for 30 min at 4 °C. Chromatin pellets were washed once with CP buffer supplemented with 5 mm MgCl2. The supernatant was removed and the pellet dissolved by boiling (5 min) in SDS gel loading buffer.
Mass Spectrometry
SDS-PAGE gels were stained with Coomassie Blue, and the entire gel lanes were cut into 23 slices of equal size. Proteins within the slices were digested according to Shevchenko et al. (31). Peptides were extracted and analyzed by LC-coupled tandem MS on an Orbitrap Xl mass spectrometer (Thermo Fisher Scientific). CID fragment spectra were searched against the NCBInr data base using the MASCOT search engine.
Fluorescence Polarization Measurements
Peptides used for fluorescence polarization measurements were as described (17). Fluorescence polarization assays were essentially carried out and analyzed as described (32). A titration series of 10-μl volumes in 384-well plates were read multiple times on a Plate Chameleon II plate reader (HIDEX Oy). Multiple readings and the independent titration series were averaged after data normalization.
Cell Transfection and Immunofluorescence
Mouse embryonic fibroblast (MEF, gift of Dr. Thomas Jenuwein, IMP, Vienna) cells were grown at 37 °C in a humidified atmosphere, 5% CO2 using Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, and 1× penicillin/streptomycin (Invitrogen). Transfection was carried out using JetPei (PolyPlusTransfections). For immunofluorescence, cells were grown and transfected on glass coverslips. 24 h post-transfection cells were fixed with 3% paraformaldehyde in PBS for 15 min at 37 °C. After three washing steps with PBST (1× PBS, 0.2% Triton X-100) and a 10-min permeabilization (PBST, 0.2% Nonidet P-40) at room temperature, cells were blocked for 1 h at room temperature (1× PBS, 5% goat serum, 2% bovine serum albumin, 0.2% Triton X-100). Coverslips were incubated with the indicated primary antibodies diluted in blocking buffer for 1 h at room temperature. Dilutions of primary antibodies were as follows: anti-H3K9me3 (Upstate), 1:1,000; anti-FLAG and anti-HA (Sigma), 1:1,000; anti-myc (Millipore), 1:1,000. Coverslips were washed three times with PBST and then incubated with the appropriate secondary antibodies (anti-mouse Alexa555, 1:2,000 or anti-rabbit Alexa488, 1:2,000; Molecular Probes) for 1 h at room temperature. After washing (3 × 5 min, 1× PBST) cells were mounted with MOWIOL (Calbiochem) containing 50 μg/ml of 4′,6-diamidino-2-phenylindole. Pictures were captured on a Leica SP5 confocal microscope (×60 objective).
Multimerization Assay
15 μg of anti-FLAG antibody per 40 μl of goat anti-mouse IgG magnetic beads (Invitrogen) were incubated 2 h in 1 ml of PBS at 4 °C under constant rotation. YFP-CDYL1b and FLAG-tagged CDYL1b constructs were expressed using the SP6 TnT kit (Promega). 50 μl of the reactions were incubated with the anti-FLAG-coated beads in 1 ml of PD150 overnight at 4 °C under constant rotation. Beads were washed six times in PD150 and boiled in SDS gel loading buffer (5 min).
RESULTS
Identification of CDYL1 as H3K9me3-binding Protein
To gain insight into translation of the H3K9me3 histone modification for regulating heterochromatin formation and maintenance we used H3 peptides trimethylated on lysine 9 for pulldown experiments out of HeLa S3 nuclear extracts. Compared with a mock (beads only) and the unmodified H3 peptide control we identified 25 protein factors specifically bound to the H3K9me3 peptide as analyzed by SDS-PAGE and following MS analysis (Fig. 1A). Specific proteins were assigned with an arbitrary cut-off of at least three sequenced peptides per protein where at least two peptides had to be unique in sequence (not shown). According to gene ontology analysis, out of the 25 factors 18 have functions in nucleobase, nucleoside, nucleotide, and nucleic acid metabolic processes. Among these 18 proteins 10 are involved in chromatin maintenance and architecture. The identified chromatin-related factors include the three mammalian HP1 isoforms (α, β, γ), CAF1, and surprisingly CDYL1. A total of 12 peptides covering 45.6% of the amino acid sequence of CDYL1 could be identified (Fig. 1B). None of the sequenced peptides could be assigned to the very N-terminal region of the CDYL1 polypeptide (originally described by Lahn et al.) (blue sequence in Fig. 1B) (15, 24). Also, no precursor could be detected in the MS analysis of this particular molecular weight region of the SDS-PAGE gels that matched any tryptic peptide derived from the very N-terminal region. Recent studies have reported a new exon acquisition in the mammalian cdyl1 gene (33). Also, we reported that the chromodomain of the originally described CDYL1 polypeptide sequence does not bind to H3K9me3 (17). These observations lead us to hypothesize that alternative splicing might generate different CDYL1 protein species with different functionality.
To independently confirm the results of the MS analysis, we repeated the H3 peptide pulldowns from HeLa S3 nuclear extracts. Using antibodies specific for CDYL1 we could confirm specific enrichment of CDYL1 exclusively on the H3K9me3 matrix compared with the unmodified control peptide and the mock control in Western blots (Fig. 1C). Similarly, HP1β was found highly enriched in the H3K9me3-associated protein fraction. From these results we conclude that CDYL1, in contrast to former results, can associate with the heterochromatic H3K9me3 modification.
CDYL1 Has Three Splicing Variants and CDYL1b Is the Most Abundant Splicing Variant
To clarify whether differential splicing could indeed explain the distinctive H3K9me3 binding behavior of CDYL1 in different experimental settings, we performed data base searches. In silico analysis defined three putatively alternative spliced mRNAs (a, b, and c) transcribed from the CDYL1 locus on chromosome 6 (supplemental Fig. S1). The CDYL1 gene locus contains 10 exons. The previously known CDYL1 mRNA is generated by splicing the first three to the last six exons. From here on, we will refer to this mRNA as CDYL1a as also suggested by Li et al. (33). The second splice variant CDYL1b mRNA emerges from exons 4–10 and a third variant originates from splicing of exon 4 to exons 6–10 of the CDYL gene locus (CDYL1c).
Sequence analysis and domain comparison indicate that the polypeptides corresponding to CDYL1 variants a and b both connect a N-terminal chromodomain via a 238-aa long linker region to a C-terminal ECH-like domain (Fig. 2A). Due to the described splicing events the CDYL1a protein contains a prolonged N-terminal part. Sequence comparison of the chromodomain of CDYL1 splicing variants a and b with the canonical chromodomain of HP1β reveals high sequence homology (Fig. 2D). However, splicing eliminates the first of three aromatic cage residues from the CDYL1a sequence, as the splicing site is located to the very N terminus of the CDYL1 chromodomain (Fig. 2D). Mutagenesis of any of the three HP1 aromatic cage residues in the Drosophila HP1α protein results in loss of H3K9me interaction (34). Mutation of the CDYL1a residues at this position to the corresponding residues in CDY restores H3K9me3 interaction (17). We therefore conclude that alternative splicing of CDYL1 generates two proteins containing a chromodomain signature with differential H3K9me3 interaction potential. In contrast, CDYL1c lacks the chromodomain region and a 175-aa long part of the linker region altogether.
FIGURE 2.
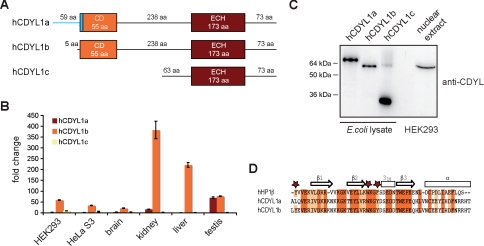
CDYL1b is the major splicing variant. A, schematic representation of the protein domain structure of CDYL1 splicing variants a, b, and c. CD, chromodomain. B, mRNA levels of CDYL1 splicing variants a, b, and c in human cell lines HEK293 and HeLa S3 as well as in the indicated human tissues were analyzed using real time PCR. Relative expression levels were normalized to the lowest expression (hCDYL1a in HEK293), which was set arbitrarily to 1. Error bars represent standard deviation of three independent measurements. C, Western blot analysis of E. coli cells expressing recombinant CDYL1a, -b, or -c and nuclear extracts of HEK293 cells using anti-CDYL antibodies. D, sequence alignment of chromodomains of CDYL1a and -b with the chromodomain of HP1β. Identical residues are highlighted in red; homolog residues are highlighted in orange. Secondary structure elements of HP1β are indicated on the top. Stars mark aromatic amino acids forming an aromatic cage for methyl-lysine recognition.
Real time PCR analysis of cDNAs of two human cell lines (HeLa S3 and HEK293) and four human tissues (brain, liver, kidney, and testis) revealed that CDYL1b is the major splicing variant of the CDYL1 gene (Fig. 2B). In agreement with former results CDYL1a mRNA levels in testis were found elevated and reaching CDYL1b expression suggesting a specific role for this splicing variant in this tissue (14). Also, Western blot analysis of HEK293 human cell nuclear extracts using an antibody specific for CDYL1 showed the highest abundance of CDYL1b (Fig. 2C). Our observations indicate that transcription of the CDYL1 locus results in three splicing variants with different expression levels and different capabilities to bind to H3K9me3.
Only CDYL1b Recognizes Methyl-lysines
To find out whether CDYL1b is indeed a CDYL1 splicing variant that recognizes H3K9me3 we in vitro transcribed and translated FLAG-tagged CDYL1 variants a, b, and c. We then tested binding to H3K9me3 in peptide pulldown experiments. HP1β was used as positive control. As Fig. 3A shows, CDYL1a and -c displayed no preferential binding to the H3 tail trimethylated on lysine 9. In contrast, CDYL1b was clearly enriched in the H3K9me3 peptide-bound fraction.
FIGURE 3.
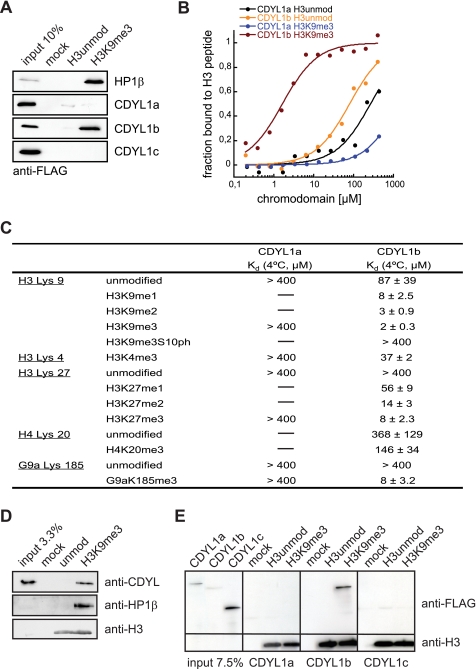
CDYL1b specifically recognizes H3K9me3 in the context of chromatin. A, the indicated FLAG-tagged proteins were produced by in vitro translation and tested for interaction with differentially modified H3 peptides using pulldown assays. Beads without coupled peptides were used as control (mock). Western blot analysis of the input and bound fractions using anti-FLAG antibodies is shown. B, fluorescence polarization binding experiments using the indicated H3 peptides and the recombinant chromodomains of CDYL1a and CDYL1b. The average bound fraction of H3 peptides from three independent experiments is blotted. C, summary of Kd values of interaction of recombinant CDYL1a and CDYL1b chromodomains with the indicated peptides as measured by fluorescence polarization binding assays. The average of at least three independent experiments including standard deviation is given. D, recombinant 12-mer oligonucleosomal arrays reconstituted with the indicated H3 species were immobilized on agarose beads and incubated with HEK293 cell nuclear extract. Beads without coupled chromatin were used as control (mock). Western blot analysis of the input and bound fractions using the indicated antibodies is shown. E, CDYL1a, -b, and -c were produced by in vitro translation and incubated with recombinant 12-mer oligonucleosomal arrays reconstituted with the indicated H3 species. Recombinant chromatin was precipitated by addition of Mg2+ as described under “Experimental Procedures.” Western blot analysis of input recombinant proteins and material recovered after centrifugation using the indicated antibodies is shown. Mock, precipitation in the absence of added recombinant chromatin.
Fluorescence polarization measurements were used to quantify interaction of the CDYL1a and CDYL1b chromodomains to differentially modified histone and non-histone peptides. Fig. 3B shows the binding curves to H3 peptides with or without trimethylation on lysine 9. Although the CDYL1a chromodomain had very little affinity for the H3K9me3 peptide over the unmodified counterpart, the CDYL1b chromodomain displayed significant affinity to H3K9me3 but discriminated against the unmodified H3 peptide. In agreement with the results from the pulldown assays, the chromodomain of CDYL1b was found to interact the strongest with the H3K9me3 peptide (see Fig. 3C for a summary of all binding data). The measured Kd of 2 μm was highly similar to that reported for the HP1/H3K9me3 interaction (12, 34). Similar to HP1, weaker interaction with the dimethylated (1.5-fold) and monomethylated (4-fold) forms of this site as well as with the H3K27 methylation marks was seen. The chromodomain of CDYL1b recognized H3K4me3 and H4K20me3 peptides with much lower affinity.
A phosphate on serine 10 (H3S10) next to H3K9me3 abolished the affinity of the CDYL1b chromodomain for this modification. H3S10 is phosphorylated at the onset of mitosis by the Aurora B kinase (35). We and others (36, 37) have shown previously that this phosphorylation event delocalizes HP1 from heterochromatin during M-phase. In agreement with a putative regulation of CDYL1b chromatin association by a “methyl-phos switch” (38), immunofluorescence analysis of CDYL1b during mitosis showed the absence of the protein from condensed chromatin during M-phase (data not shown).
Previous studies had identified CDYL1 as the binding partner of automethylation of the G9a histone methyltransferase on lysine 185. However, in vitro interaction with the available CDYL1 protein could not be verified (19). In fluorescence polarization assays we found the CDYL1b chromodomain to bind a peptide corresponding to G9aK185me3 with 8 μm affinity. In contrast, the chromodomain of CDYL1a showed no interaction (Fig. 3C). Altogether, our analysis demonstrates that only the chromodomain of CDYL1b but not that of CDYL1a can bind differentially methylated lysine residues in distinct histone and non-histone sequence context.
CDYL1b Recognizes H3K9me3 in Chromatin Context
Reduced binding of recombinant HP1 to chromatin templates compared with isolated histone peptides has been reported (39). Because 15–20-aa long peptides do not represent the targeting and binding situation of CDYL1 in the cell, we wanted to analyze H3K9me3 binding of CDYL1b in the context of chromatin. Therefore, we established an in vitro chromatin reconstitution system that makes use of uniformly K9me3-modified H3 obtained by native protein ligation (supplemental Fig. S2) (28, 40). Oligonucleosomal arrays on a 12 × 200 × 601 sequence were assembled under template saturating conditions and contained 10–12 nucleosomes in average (supplemental Fig. S3). No differences in the biophysical properties between arrays reconstituted with unmodified H3 or H3K9me3 could be detected (supplemental Fig. S4 and data not shown) (27, 29, 41). To analyze the CDYL1 interaction biotinylated recombinant chromatin with or without the H3K9me3 modification was bound to streptavidin-coated beads. This matrix was used to pull out the binding proteins of HeLa S3 nuclear extracts. After extensive washing the bound fractions were analyzed by Western blotting. As Fig. 3D shows, HP1β and CDYL1 were clearly enriched on the H3K9me3 chromatin template compared with the unmodified control or the mock (beads only) reaction.
To further analyze chromatin binding of CDYL1, we in vitro transcribed and translated CDYL1 splicing variants a, b, and c and repeated the analysis using recombinant proteins. Although CDYL1b was able to bind to the H3K9me3 chromatin template, no interaction could be detected for CDYL1a or CDYL1c (Fig. 3E).
Heterochromatic Localization of CDYL1b Is Dependent on H3K9me3
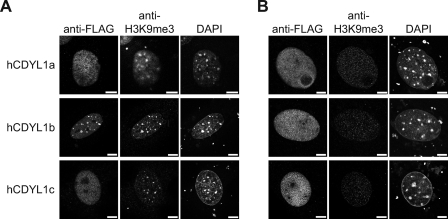
Next, we asked whether CDYL1 splicing variants localize to heterochromatic, H3K9me3-enriched loci in vivo. Because the commercially available antibodies as well as our specifically raised antisera recognizing CDYL1 failed to detect the protein in immunofluorescence experiments, we transiently expressed FLAG-tagged CDYL1 a, b, or c in MEF WT cells. Immunofluorescence analysis was carried out using antibodies against the FLAG tag and compared with the distribution of H3K9me3 and DNA (Fig. 4A). Whereas CDYL1a and CDYL1c were found diffusely distributed throughout the whole cell nucleus only CDYL1b showed a dotted distribution pattern. The regions enriched in CDYL1b-FLAG co-localized with the DNA-dense regions of pericentric heterochromatin as well as with the spots highly enriched in the H3K9me3 modification.
FIGURE 4.
CDYL1b subnuclear localization is dependent on H3K9me3. A, MEF WT cells were transfected with FLAG-tagged CDYL1a, -b, or -c and stained with the indicated antibodies. DNA was visualized using 4′,6-diamidino-2-phenylindole (DAPI). Bars represent 7.5 μm. B, MEF Suv3–9h1/h2 (−/−) double knock-out cells were transfected and analyzed as in A.
To find out if this localization to areas of heterochromatin is indeed dependent on H3K9me3 modification, we used MEF Suv3–9h1/h2 (−/−) double knock-out cells. Suv3–9h1 and Suv3–9h2 are known as main enzymes establishing the H3K9me3 modification (42). Therefore, MEF Suv3–9h1/h2 (−/−) double knock-out cells show a strongly reduced amount of H3K9me3. The cells nevertheless, sustain the dotted structure of DNA-dense regions corresponding to pericentric heterochromatin (43). We transiently expressed the different CDYL1 splicing variants in MEF Suv3–9h1/h2 (−/−) cells (Fig. 4B). Although the absence of Suv3–9h1/h2 neither impacted CDYL1a nor CDYL1c subnuclear localization, distribution of CDYL1b was significantly different compared with the wild type situation. CDYL1b did not localize to the DNA-dense heterochromatic areas, but showed a diffuse appearance in the nucleus. According to these observations, CDYL1b is a heterochromatin factor whose localization is dependent on the presence of H3K9me3.
CDYL1b Is Forming Multimers
An identical ECH-like domain is at the C terminus of all CDYL1 splicing variants. Enoyl-CoA hydratase enzymes are known to have besides hydratase activity also dehydrogenase and isomerase activity. During β-oxidation in mitochondria and peroxisomes enoyl-CoA hydratases add water to unsaturated acyl-CoA at the position between the second and third carbon atom within the fatty acid chain (25, 44). Sequence comparison of the CDYL1 ECH-like domain with the human peroxisomal (ECHP) and mitochondrial (ECHM) enoyl-CoA hydratase proteins revealed a good overall homology (Fig. 5A). BLAST alignment resulted in 39% identity, 58% homology to hECHM, and 24% identity, 47% homology to hECHP over a stretch of 167 aa. Three residues, one glycine and two glutamines are essential for ECH enzymatic activity (26, 45). In CDYL1 these three residues are exchanged against leucine, serine, and tyrosine, respectively, suggesting a different role for the CDYL1 ECH-like domain. In agreement, closer inspection of the structure of rat ECH with a bound acetoacetyl-CoA (Protein Data Bank code 1dub), which is a potent inhibitor of ECH enzymes (45), revealed that it is unlikely that the ECH-like domain of CDYL1 can catalyze a hydratase reaction (supplemental Fig. S5). Despite these amino acid exchanges, structural superposition of the CDYL1 ECH-like domain (PDB 1gtr) and the rat peroxisomal ECH (PDB 1dub) show that the overall trimeric-fold of these proteins remains unchanged (Fig. 5B).
We confirmed multimerization of CDYL1. Anti-FLAG immunoprecipitation of in vitro translated FLAG-tagged CDYL1b mixed with in vitro translated YFP-CDYL1b contained YFP-CDYL1b as analyzed by Western blotting using anti-green fluorescent protein antibodies (Fig. 5D). We then mapped the CDYL1 interaction regions using a series of CDYL1b-FLAG deletion constructs (see Fig. 5C). As Fig. 5D shows CDYL1 multimerization was depending on the C-terminal part of CDYL1b that contains the ECH-like domain. This region is identical in sequence to CDYL1c (Fig. 5D, CDYL1c). Multimerization of this 309-aa long region of CDYL1 (see also Fig. 2A) was abolished when the 73 aa C-terminal to the predicted ECH-like domain were missing (Fig. 5D, CDYL1cΔCterm). The isolated ECH-like region as determined by sequence alignment and domain prediction programs were not sufficient for CDYL1 multimerization (Fig. 5D, ECH). Additional self-binding of CDYL1b was mapped to the protein part covering a fragment of the hinge region and the chromodomain (Fig. 5D, chromohinge). We conclude that CDYL1 can interact with itself via a C-terminal region containing the ECH-like domain and an N-terminal part of the protein.
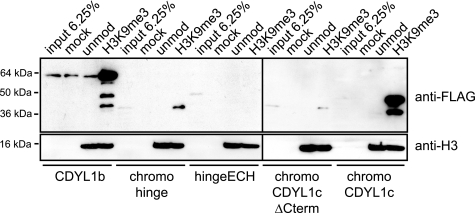
Multimerization Is Required for CDYL1b H3K9me3 Chromatin Binding
It has been suggested that simultaneous, multivalent histone modification binding is required to anchor chromatin proteins on their target sites (46, 47). Because CDYL1 shows a strong tendency to multimerize via a C-terminal region containing the ECH-like domain, we reasoned that multimeric presentation of the H3K9me3-binding N-terminal chromodomain might be a prerequisite for stable chromatin association. Reticulocyte extracts programmed to express CDYL1b and different deletion mutants (see Fig. 5C) were incubated with oligonucleosomal arrays reconstituted with unmodified H3 or H3 uniformly bearing the K9me3 modification. As Fig. 6 shows only mutant CDYL1 protein chromoCDYL1c, which contains a chromodomain directly linked to the C-terminal region containing the ECH-like domain, interacted robustly with H3K9me3 chromatin, like WT CDYL1b. The H3K9me3 interacting chromodomain is required for this binding as the multimerizing C-terminal region containing the ECH-like domain when linked to the hinge region showed no interaction (Fig. 6, hingeECH). Similarly, the chromodomain when linked to the C-terminal region containing the ECH-like domain that misses the very C-terminal part required for multimerization did not bind H3K9me3 chromatin in this assay (Fig. 6, chromoCDYL1cΔCterm). In agreement with the multimerization potential of the chromohinge domain mutant, this protein showed weak chromatin interaction (Fig. 6, chromohinge). From these experiments we conclude that multimerization is required for stable CDYL1 chromatin binding.
FIGURE 6.
CDYL1b multimerization is required for H3K9me3 chromatin interaction. The indicated FLAG-tagged XlCDYL1 proteins were produced by in vitro translation and incubated with recombinant 12-mer oligonucleosomal arrays reconstituted with the indicated H3 species. Recombinant chromatin was precipitated by addition of Mg2+ as described under “Experimental Procedures.” Western blot analysis of in vitro translated recombinant proteins (input) and material recovered after centrifugation using the indicated antibodies is shown. Mock, precipitation in the absence of added recombinant chromatin.
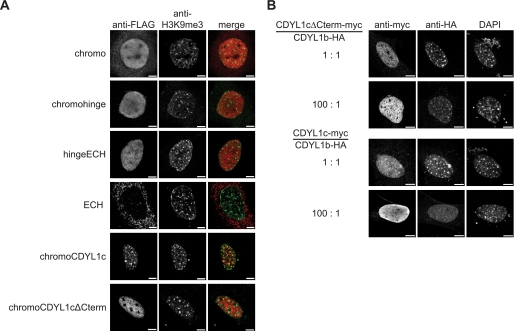
Multimerization Is Necessary for H3K9me3 Localization
To investigate if multimerization has any effect on localization of CDYL1, the subnuclear distribution of different FLAG-tagged truncated variants of CDYL1b were analyzed in MEF WT cells (Fig. 7A, for constructs, see Fig. 5C). With the exception of the ECH-like domain alone all mutant proteins showed accumulation in the cell nucleus. Although the chromodomain might accumulate in the nucleus indirectly, the results indicate a putative NLS at the end of the hinge region. Indeed, additional experiments verified a bona fide NLS at residues 235–298 of CDYL1b (supplemental Fig. S6).
FIGURE 7.
CDYL1b subnuclear localization is dependent on multimerization. A, the FLAG-tagged CDYL1 proteins indicated on the left were transiently expressed in MEF WT cells. Immunofluorescence analysis was carried out using the indicated antibodies. Images representative of a large number of analyzed cells are shown. B, MEF WT cells were transfected using the indicated ratios of CDYL1cΔNLS-myc and CDYL1b-HA or CDYL1c-myc and CDYL1b-HA plasmids. Immunofluorescence analysis was carried out using the indicated antibodies. DNA was visualized using 4′,6-diamidino-2-phenylindole (DAPI). Bars represent 7.5 μm.
Despite the interaction with H3K9me3, immunofluorescence analysis showed that the CDYL1b chromodomain alone is not able to localize to the regions containing pericentric heterochromatin. Also, the chromodomain connected to the following hinge region of the CDYL1 protein (chromohinge) showed no overlap with the DNA-dense regions enriched in H3K9me3. As predicted due to the absence of an H3K9me3 binding domain, the CDYL1b construct missing the chromodomain (hingeECH) showed no preferential localization to heterochromatin. In contrast, the chromodomain connected to the CDYL1c splicing variant (chromoCDYL1c) displayed a dotted localization within the nucleus overlapping with H3K9me3-dense areas. When this chromoCDYL1c construct lacks the very C-terminal part, the spotted distribution is abolished. This 73-aa region at the very C terminus of CDYL1c is necessary for multimerization (see Fig. 5D). From this analysis we conclude that the chromodomain of CDYL1b is necessary but not sufficient to localize the protein to H3K9me3 chromatin. Additionally, multimerization of the protein is required to anchor CDYL1b to heterochromatic regions.
Overexpression of CDYL1c Can Displace CDYL1b from H3K9me3 Heterochromatin
As shown in Figs. 5D and 7A, CDYL1c can interact with itself forming multimers but is not able to localize to H3K9me3 chromatin in vivo. We reasoned that CDYL1c could form complexes with CDYL1b thereby negatively regulating CDYL1b chromatin association. Therefore, we transfected myc-tagged CDYL1c and HA-tagged CDYL1b into MEF WT cells at different plasmid ratios. As Fig. 7B shows coexpression of CDYL1c at a 1:1 ratio did not impair CDYL1b localization to the DNA-dense foci of pericentric heterochromatin. In contrast, 80–90% of MEF cells transfected with CDYL1c:CDYL1b at a 100:1 plasmid ratio showed delocalization of CDYL1b from heterochromatin displaying a diffuse CDYL1b signal in the nucleoplasm. Importantly, coexpression of CDYL1cΔCterm, which is neither able to localize to H3K9me3 nor to multimerize with full-length CDYL1b (see Fig. 5D), did not have a similar negative effect on CDYL1b localization to heterochromatin. These experiments further sustain the interpretation that an intact chromodomain and multimerization is required for CDYL1b heterochromatin localization.
DISCUSSION
Our detailed analysis of CDYL1 function in the context of H3K9me binding has the following implications: (i) different splicing variants a, b, and c of CDYL1 bind differentially to methylated lysine residues in histones and other proteins. (ii) CDYL1b chromodomain/H3K9me3 interaction is not sufficient for association of the factor with heterochromatin. (iii) Multimerization of CDYL1b is necessary for stable H3K9me3 chromatin association in vitro and for H3K9 heterochromatin localization in vivo.
The originally described CDYL1 polypeptide corresponds to the less abundant CDYL1a splicing variant (15, 33). CDYL1 was identified and described in different experiments interacting with methylated lysine residues in histones and G9a (18, 19, 48). Although the CDYL1 splicing variant was not specified, interaction in the case of methylated G9a could not be verified using recombinant proteins (19). We think that it is likely that CDYL1a and not CDYL1b was used in these studies. Indeed, the chromodomains of both CDYL1b and CDYL1a after restoration of the aromatic cage by mutagenesis bind a methylated G9a peptide with affinity comparable with methylated histone peptides in vitro (this study and see Ref. 17). At present, it is unclear whether the N-terminal extension of CDYL1a compared with CDYL1b brings additional functionality to the protein, potentially establishing a different role for the methyl-lysine binding deficient chromodomain.
Biochemical evidence has been provided that places CDYL1 as a bridge between G9a and REST in a WIZ and histone modifying enzymes containing corepressor complex (21, 22). Although direct interaction of CDYL1 and G9a could be demonstrated using recombinant proteins, it is tempting to speculate that this interaction is in vivo further controlled by G9a automethylation and CDYL1b chromodomain binding. Methylation of CDYL1 might also play a role because in vitro G9a can target CDYL1 for methylation at a lysine residue C-terminal to the chromodomain. It has been suggested that this post-translational modification might negatively regulate the CDYL1/H3K9me3 interaction (18). However, as the observed effect was only 2-fold and as CDYL1a was used for these studies, the exact relation of G9a, CDYL1, and their methylation within the REST-CoREST complex needs further investigation. In any case, it has to be noted that a large fraction of CDYL1b exists outside of the REST-CoREST complex, likely targeting other methyl-lysine residues (see Ref. 22).5
Besides the chromodomain the ECH-like domain is likely central to CDYL1 and CDY protein family function. These regions show a very high degree of conservation, whereas the linking hinge region is more variable. It was originally suggested that the ECH-like domain of CDY and CDYL1 might have intrinsic histone acetyltransferase activity (24). However, structural analysis and modeling makes it unlikely that acetyl-CoA is indeed a substrate of the CDYL1 ECH-like domain (49). Indeed, we have failed to reproduce histone acetyltransferase activity of CDY family proteins.5 Nevertheless, it was found that the CDYL1 ECH-like domain interacts with CoA. Although CoA can indeed be modeled onto the structure of CDYL1 it remains to be determined whether CoA is a substrate, substrate carrier, or possibly an allosteric regulator.
Our studies indicate that a major function of the ECH-like domain resides in multimerization. Indeed, only CDYL1 that can interact with itself is directed to H3K9me3 heterochromatin, but a functional chromodomain on itself is not capable of this targeting. Although the CDYL1b chromodomain/H3K9me3 interaction at 2 μm is comparable with other chromodomain methylhistone peptide interactions (12, 34), it is nevertheless, weak. We hypothesize that CDYL1b protein multimerization generates a multivalent binding mode that significantly enhances interaction (46). Similar behavior has been suggested for HP1, which dimerizes via a C-terminal chromoshadow domain. Mutations that affect HP1 dimerization as well as overexpression of the chromoshadow domain alone cause dissociation of HP1 from heterochromatin (50, 51). Further studies are required to directly analyze and quantify the impact of ECH-like domain multimerization onto CDYL1b/H3K9me3 chromatin binding.
Our results also show that multimerization of CDYL1 might not be limited to the ECH-like domain, but could also be mediated by additional regions of the protein. Although it is currently unclear how many CDYL1 polypeptides do associate to form chromatin binding complexes, structural analysis of the homologous CDY has found evidence for hexamerization beyond trimerization (PDB 2fw2) (49). Obviously, stronger chromatin interaction will result from the presence of extra H3K9me3 binding sites if additional associations are sterically possible. Indeed, multiple histone modification binding domains in complexes or multimerization of individual chromatin readout factors could be key in reading and translating histone modifications by establishing robust interaction. Enhancement of binding via multimerization could possibly explain the obvious strong targeting of CDYL1b to sites of H3K9me3 in vivo compared with not much weaker binding to H3K27me3 in vitro.
Another aspect of CDYL1b multimerization besides generating binding strength could be affecting chromatin structure and behavior by binding multiple regions simultaneously. Although multivalent binding of chromatin factors to histone modifications has not yet been demonstrated and whereas effects of multiple binding events have not been analyzed, we wonder whether such interaction could be involved in establishing or maintaining higher order chromatin structures. In this sense CDYL1 might act as a chromatin architectural protein enhancing the transition of 10-nm fibers to 30-nm structures and higher order chromatin arrangements (52).
Acknowledgments
We thank Winfried Lendeckel for technical help and members of the Fischle laboratory for stimulating discussions. We also thank Monika Raabe and Uwe Plessmann for excellent mass spectrometric analysis. We are grateful to Dr. Daniela Rhodes (MRC, Cambridge, MA) for providing 12 × 200 × 601 DNA template for chromatin reconstitution, Dr. Thomas Jenuwein (IMP, Vienna) for MEF WT and Suv3–9h1/h2 (−/−) double knock-out cells, and Dr. Thomas Giger (Max Planck Institute for Evolutionary Anthropology, Leipzig) for human tissue mRNAs.
This work was supported in whole or in part by the Max Planck Society and the European Union FP6, Network of Excellence, “The Epigenome.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and “Materials and Methods.”
H. Franz and W. Fischle, unpublished observations.
- HP1
- heterochromatin protein 1
- CDYL
- chromodomain on the Y chromosome
- WT
- wild type
- MEF
- mouse embryonic fibroblast
- NLS
- nuclear localization signal
- aa
- amino acid
- ECH
- enoyl-CoA hydratase
- HA
- hemagglutinin
- MS
- mass spectrometry
- YFP
- yellow fluorescent protein
- PBS
- phosphate-buffered saline
- PDB
- Protein data bank
- HEK
- human embryonic kidney.
REFERENCES
- 1.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 2.Sims R. J., 3rd, Reinberg D. (2008) Nat. Rev. Mol. Cell Biol. 9, 815–820 [DOI] [PubMed] [Google Scholar]
- 3.Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 4.Lachner M., Sengupta R., Schotta G., Jenuwein T. (2004) Cold Spring Harbor Symp. Quant. Biol. 69, 209–218 [DOI] [PubMed] [Google Scholar]
- 5.Sims R. J., 3rd, Nishioka K., Reinberg D. (2003) Trends Genet. 19, 629–639 [DOI] [PubMed] [Google Scholar]
- 6.Elgin S. C., Grewal S. I. (2003) Curr. Biol. 13, R895–R898 [DOI] [PubMed] [Google Scholar]
- 7.Grewal S. I., Jia S. (2007) Nat. Rev. Genet. 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 8.Shogren-Knaak M., Peterson C. L. (2006) Cell Cycle 5, 1361–1365 [DOI] [PubMed] [Google Scholar]
- 9.Luger K., Hansen J. C. (2005) Curr. Opin. Struct. Biol. 15, 188–196 [DOI] [PubMed] [Google Scholar]
- 10.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel J. A., Pray-Grant M. G., Grant P. A. (2005) Cell Cycle 4, 919–926 [DOI] [PubMed] [Google Scholar]
- 12.Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., Khorasanizadeh S. (2003) Genes Dev. 17, 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahn B. T., Page D. C. (1997) Science 278, 675–680 [DOI] [PubMed] [Google Scholar]
- 14.Dorus S., Gilbert S. L., Forster M. L., Barndt R. J., Lahn B. T. (2003) Hum. Mol. Genet. 12, 1643–1650 [DOI] [PubMed] [Google Scholar]
- 15.Lahn B. T., Page D. C. (1999) Nat. Genet. 21, 429–433 [DOI] [PubMed] [Google Scholar]
- 16.Bhowmick B. K., Takahata N., Watanabe M., Satta Y. (2006) Genet. Mol. Res. 5, 696–712 [PubMed] [Google Scholar]
- 17.Fischle W., Franz H., Jacobs S. A., Allis C. D., Khorasanizadeh S. (2008) J. Biol. Chem. 283, 19626–19635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathert P., Dhayalan A., Murakami M., Zhang X., Tamas R., Jurkowska R., Komatsu Y., Shinkai Y., Cheng X., Jeltsch A. (2008) Nat. Chem. Biol. 4, 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampath S. C., Marazzi I., Yap K. L., Sampath S. C., Krutchinsky A. N., Mecklenbräuker I., Viale A., Rudensky E., Zhou M. M., Chait B. T., Tarakhovsky A. (2007) Mol. Cell 27, 596–608 [DOI] [PubMed] [Google Scholar]
- 20.Lakowski B., Roelens I., Jacob S. (2006) J. Mol. Neurosci. 29, 227–239 [DOI] [PubMed] [Google Scholar]
- 21.Kuppuswamy M., Vijayalingam S., Zhao L. J., Zhou Y., Subramanian T., Ryerse J., Chinnadurai G. (2008) Mol. Cell. Biol. 28, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulligan P., Westbrook T. F., Ottinger M., Pavlova N., Chang B., Macia E., Shi Y. J., Barretina J., Liu J., Howley P. M., Elledge S. J., Shi Y. (2008) Mol. Cell 32, 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caron C., Pivot-Pajot C., van Grunsven L. A., Col E., Lestrat C., Rousseaux S., Khochbin S. (2003) EMBO Rep. 4, 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahn B. T., Tang Z. L., Zhou J., Barndt R. J., Parvinen M., Allis C. D., Page D. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8707–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J. J., Battaile K. P. (2002) Curr. Opin. Struct. Biol. 12, 721–728 [DOI] [PubMed] [Google Scholar]
- 26.Hamed R. B., Batchelar E. T., Clifton I. J., Schofield C. J. (2008) Cell Mol. Life Sci. 65, 2507–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 28.Shogren-Knaak M. A., Peterson C. L. (2004) Methods Enzymol. 375, 62–76 [DOI] [PubMed] [Google Scholar]
- 29.Huynh V. A., Robinson P. J., Rhodes D. (2005) J. Mol. Biol. 345, 957–968 [DOI] [PubMed] [Google Scholar]
- 30.Leuba S. H., Bustamante C. (1999) Methods Mol. Biol. 119, 143–160 [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 32.Jacobs S. A., Fischle W., Khorasanizadeh S. (2004) Methods Enzymol. 376, 131–148 [DOI] [PubMed] [Google Scholar]
- 33.Li X., Liang J., Yu H., Su B., Xiao C., Shang Y., Wang W. (2007) Trends Genet. 23, 427–431 [DOI] [PubMed] [Google Scholar]
- 34.Jacobs S. A., Khorasanizadeh S. (2002) Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 35.Vagnarelli P., Earnshaw W. C. (2004) Chromosoma 113, 211–222 [DOI] [PubMed] [Google Scholar]
- 36.Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 37.Hirota T., Lipp J. J., Toh B. H., Peters J. M. (2005) Nature 438, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 38.Fischle W., Wang Y., Allis C. D. (2003) Nature 425, 475–479 [DOI] [PubMed] [Google Scholar]
- 39.Eskeland R., Eberharter A., Imhof A. (2007) Mol. Cell. Biol. 27, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muralidharan V., Muir T. W. (2006) Nat. Methods 3, 429–438 [DOI] [PubMed] [Google Scholar]
- 41.Lowary P. T., Widom J. (1998) J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 42.Völkel P., Angrand P. O. (2007) Biochimie 89, 1–20 [DOI] [PubMed] [Google Scholar]
- 43.Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 44.Palosaari P. M., Vihinen M., Mäntsälä P. I., Alexson S. E., Pihlajaniemi T., Hiltunen J. K. (1991) J. Biol. Chem. 266, 10750–10753 [PubMed] [Google Scholar]
- 45.Engel C. K., Mathieu M., Zeelen J. P., Hiltunen J. K., Wierenga R. K. (1996) EMBO J. 15, 5135–5145 [PMC free article] [PubMed] [Google Scholar]
- 46.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruthenburg A. J., Allis C. D., Wysocka J. (2007) Mol. Cell 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 48.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H., Min J., Antoshenko T., Plotnikov A. N. (2009) Proteins 76, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 50.Mateos-Langerak J., Brink M. C., Luijsterburg M. S., van der Kraan I., van Driel R., Verschure P. J. (2007) Mol. Biol. Cell 18, 1464–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiru A., Nietlispach D., Mott H. R., Okuwaki M., Lyon D., Nielsen P. R., Hirshberg M., Verreault A., Murzina N. V., Laue E. D. (2004) EMBO J. 23, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McBryant S. J., Adams V. H., Hansen J. C. (2006) Chromosome Res. 14, 39–51 [DOI] [PubMed] [Google Scholar]