Abstract
Bone marrow stromal antigen 2 (BST-2, also known as tetherin) is a recently identified interferon-inducible host restriction factor that can block the production of enveloped viruses by trapping virus particles at the cell surface. This antiviral effect is counteracted by the human immunodeficiency virus type 1 (HIV-1) accessory protein viral protein U (Vpu). Here we show that HIV-1 Vpu physically interacts with BST-2 through their mutual transmembrane domains and leads to the degradation of this host factor via a lysosomal, not proteasomal, pathway. The degradation is partially controlled by a cellular protein, β-transducin repeat-containing protein (βTrCP), which is known to be required for the Vpu-induced degradation of CD4. Importantly, targeting of BST-2 by Vpu occurs at the plasma membrane followed by the active internalization of this host protein by Vpu independently of constitutive endocytosis. Thus, the primary site of action of Vpu is the plasma membrane, where Vpu targets and internalizes cell-surface BST-2 through transmembrane interactions, leading to lysosomal degradation, partially in a βTrCP-dependent manner. Also, we propose the following configuration of BST-2 in tethering virions to the cell surface; each of the dimerized BST-2 molecules acts as a bridge between viral and cell membranes.
Introduction
Viral protein U (Vpu)2 is an 81-amino acid type I integral membrane phosphoprotein expressed by human immunodeficiency virus type 1 (HIV-1) (1, 2) and several simian immunodeficiency viruses (3–6). Vpu is not incorporated into virus particles (7), indicating that it acts exclusively in virus-producer cells. Indeed, Vpu is known to play two distinct roles during the later stages of infection. First, Vpu interacts with newly synthesized CD4 molecules complexed with the gp160 envelope glycoprotein precursor in the endoplasmic reticulum (8, 9) and recruits the β-transducin repeat-containing protein 1 (βTrCP-1) subunit of the Skp1-Cullin1-F-box ubiquitin ligase complex (10) as well as βTrCP-2 (11) through its phosphoserine residues at positions 52 and 56 in the cytoplasmic (CT) domain (12, 13). This event results in proteasome-mediated degradation of CD4 (10, 14, 15) allowing gp160 to resume transport toward the cell surface for virion incorporation. Second, Vpu mediates the enhancement of virion release (16–18) in a cell type-dependent manner (e.g. HeLa cells require Vpu, whereas COS7 cells do not (19, 20)), and its absence leads to the accumulation of viral particles at the cell surface (21).
In contrast to the effect of Vpu on CD4 degradation, little had been known about the mechanism by which Vpu enhances the release of virions. The finding that HeLa-COS7 heterokaryons exhibited HeLa-type properties suggested that Vpu-responsive HeLa cells might harbor endogenous a restriction factor(s) that could be counteracted by this viral protein (22), as seen in Vif-responsive cells harboring the antiretroviral factor APOBEC3G counteracted by Vif (23). Neil et al. (24) showed that Vpu-deficient viral particles accumulated at the cell surface could be released after subtilisin protease treatment, suggesting that the endogenous factor blocked by Vpu is a cell-surface-associated protein. They also showed that this endogenous factor was interferon-α-inducible and indeed overcome by Vpu (25) and, based on microarray analyses of messenger RNAs in interferon-α-treated and untreated cells, identified the host restriction factor and termed it tetherin (26), a transmembrane (TM) protein previously known as bone marrow stromal antigen 2 (BST-2), CD317, or HM1.24 (27–29). Subsequently, Van Damme et al. (30) demonstrated that the Vpu-induced inhibition of tetherin (referred to hereafter as BST-2) was due to down-regulation of BST-2 by Vpu.
BST-2 regulates the growth and development of B cells and is highly expressed in human myeloma cells (27–29). This protein has an unusual topology, harboring an N-terminal CT domain followed by a TM domain, an extracellular coiled-coil domain, and a glycosylphosphatidylinositol (GPI) anchor at the C terminus (31, 32). At the cell surface, BST-2 resides in cholesterol-rich lipid microdomains (also called lipid rafts) through the GPI anchor, whereas its TM domain apparently lies outside the lipid rafts (31, 32), indirectly interacting with the actin cytoskeleton (33). Based on this topology of BST-2, Neil et al. (26) speculated on several configurations, such as forms tethering virions to cell membranes, and to each other. However, the actual configuration remains unknown. As expected from the formation of tethers to capture enveloped viruses, BST-2 indeed shows broad-spectrum inhibition of the release of not only animal retroviruses, but also Ebola, Lassa, and Marburg viruses (34–36).
Herein, we show that the Vpu primary site of action is the plasma membrane, where this protein targets cell-surface BST-2 through their mutual TM-to-TM binding, leading to lysosomes, partially dependent on βTrCP. We also propose a possible configuration model of BST-2 to tether virions to the plasma membranes.
EXPERIMENTAL PROCEDURES
DNA Construction
The vesicular stomatitis virus glycoprotein expression vector pHIT/G (37), HIV-1 proviral construct pNL-E− (38), an HIV-1 proviral indicator construct pNL-Luc-E−, and a CD4 expression vector pNL-CD4 (39) have been described previously. The lentiviral plasmids psPAX2 and pLVTHM (40) were kindly provided by D. Trono. The HIV-1 Vpu expression plasmid pCA-Vpu-RRE and HIV-1 Rev expression plasmid pCA-Rev were created by inserting a PCR-amplified NL4-3-derived vpu gene (nucleotide 6061–6306) together with a Rev-responsive element (RRE; nucleotide 7759–7992) and a rev gene (nucleotide 5969–6044, 8369–8643), respectively, into a mammalian expression plasmid pCAGGS (41). A CT deletion mutant of Vpu pCA-VpuΔCT-RRE was created by replacing the full-length Vpu of pCA-Vpu-RRE with a PCR-amplified TM domain of Vpu (amino acid residues 1–27). A chimeric mutant comprising the human CD4 TM and Vpu CT domains, pCA-CD4tmVpu-RRE, was created by replacing the TM domain of Vpu with that of CD4 (amino acid residues 398–420), using overlapping PCR-based cloning. Hemagglutinin (HA)- or enhanced green fluorescent protein (EGFP)-tagged Vpu plasmids (pCA-Vpu-HA-RRE, pCA-VpuΔCT-HA-RRE, pCA-CD4tmVpu-HA-RRE, pCA-Vpu-EGFP-RRE, and pCA-CD4tmVpu-EGFP-RRE) were generated by inserting amplified Vpu fragments into a modified pCAGGS carrying a C-terminal HA tag with a RRE downstream of HA or into a modified pCAGGS carrying C-terminal EGFP tag with a RRE downstream of EGFP. A Vpu phosphorylation mutant pCA-Vpu2/6-RRE was created as previously described (12), by using pCA-Vpu-RRE as a template with QuikChange site-directed mutagenesis (Stratagene). A proviral Vpu phosphorylation mutant pNL-Vpu2/6-E- was similarly created except using pNL-E− as a template. Vpu- and envelope-deficient proviral clones, pNL-U−E− and pNL-Luc-U−E−, were generated by introducing a HpaI site at the vpu initiation codon of pNL-E− and pNL-Luc-E− using QuikChange site-directed mutagenesis. To create a BST-2 expression plasmid pCA-BST-2, total RNA was isolated from HeLa cells using a RNAqueous Kit (Ambion) and was subjected to reverse transcription followed by amplification with specific oligonucleotides. The amplified fragments were cloned into pCAGGS. Extracellular FLAG- and Myc-tagged BST-2 expression plasmids (pCA-BST-2-exFLAG and pCA-BST-2-exMyc) were generated by inserting the FLAG or Myc fragments into an AflII site (nucleotide 429 of BST-2 gene) in pCA-BST-2. An endocytosis mutant (pCA-BST-2-Y6A/Y8A) was generated by using pCA-BST-2 as a template with QuikChange site-directed mutagenesis. Chimeric versions between BST-2 and transferrin receptor (TfR) (42), pCA-TfRct/tm-BST-2, pCA-TfRct-BST-2, and pCA-TfRtm-BST-2 were created by replacing the N-terminal CT (amino acid residues 1–21) and TM domains (amino acid residues 22–43) of pCA-BST-2 with those of pTfR (kindly provided by Y. Takai; CT, amino acid residues 1–62; TM, 63–88), by replacing the CT domain of pCA-BST-2 with that of pTfR, and by replacing the TM domain of pCA-BST-2 with that of pTfR, respectively, using overlapping PCR-based cloning. A chimeric mutant of the human CD4 signal peptide and BST-2, pCA-BST-2-CD4sig, was generated by exchanging the CT with the TM domains of pCA-BST-2 for the CD4 signal peptide (amino acid residues 1–25) and an additional 10 residues of pNL-CD4 using the overlapping PCR-based cloning. A BST-2 mutant deleted of the GPI modification signal pCA-BST-2ΔGPI was created by inserting amplified BST-2 fragments deleted of the signal (amino acid residues 161–180) into pCAGGS. With regard to the mutant BST-2 plasmids described above, extracellular FLAG- and Myc-tagged versions were also constructed. To create βTrCP-1 and -2 expression plasmids (pCA-βTrCP-1-FLAG and pCA-βTrCP-2-FLAG), total RNA from MOLT-4 cells was subjected to reverse transcription followed by amplification with specific oligonucleotides. Amplified fragments were cloned into a modified pCAGGS carrying a C-terminal FLAG-tag. A CD4 expression plasmid pCA-CD4 was created by inserting a CD4 fragment amplified from pNL-CD4 into pCAGGS. To generate a dynamin-2 (Dyn2) expression plasmid (pCA-Dyn2), total RNA isolated from 293T cells was subjected to reverse transcription-PCR amplification of the Dyn2 gene using specific oligonucleotides. Amplified Dyn2 fragments were cloned into pCAGGS. A dominant-negative mutant of Dyn2 (pCA-Dyn2-K44A) was created by using pCA-Dyn2 as a template with QuikChange site-directed mutagenesis. EGFP-tagged Dyn2 and Dyn2-K44A expression plasmids (pCA-Dyn2-EGFP and pCA-Dyn2-K44A-EGFP) were generated by introducing wild-type (WT) and mutant-derived Dyn2 fragments into a modified pCAGGS carrying the C-terminal EGFP tag. An EGFP expression plasmid, pCA-EGFP, was created by inserting a PCR-amplified EGFP fragment into pCAGGS. Plasmids expressing short hairpin RNA (shRNA) against βTrCP-1 and βTrCP-2 were generated by inserting fragments containing sequences targeting βTrCP-1 (GCGTTGTATTCGATTTGATAA) and βTrCP-2 (GTGTCATTGTAACTGGCTCTT) into pLVTHM. All constructs were verified by DNA sequencing.
Cell Maintenance, Transfection, and Protein Analyses
HeLa, 293T, and COS7 cells were maintained under standard conditions. 293T cells were transfected with a BST-2 (WT or mutants) expression plasmid or HIV-1 proviral construct (Vpu-WT, -2/6, or -deficient) using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's instructions. Cell extracts from transfected cells were subjected to gel electrophoresis and then transferred to a nitrocellulose membrane. The membranes were probed with an anti-BST-2 mouse polyclonal antibody (Abnova) or an anti-Vpu serum (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health; HIV-1 NL4-3 Vpu antiserum was from F. Maldarelli and K. Strebel (43)). Reacted proteins were visualized by chemiluminescence using an ECL Western blotting detection system (GE Healthcare) and monitored using a LAS-3000 imaging system (FujiFilm).
shRNA Lentiviral Transduction
7 × 105 293T cells were cotransfected with 0.95 μg of βTrCP-1 or βTrCP-2 shRNA, 0.95 μg of psPAX2, and 0.1 μg of pHIT/G by using FuGENE6. After 48 h, the supernatants were harvested, and the amount of p24 antigen was measured by using an HIV-1 p24-antigen capture enzyme-linked immunosorbent assay (ELISA) (Advanced BioScience Laboratories). 1.25 × 105 HeLa cells were transduced with 1 μg of a lentivirus carrying βTrCP-1 and/or βTrCP-2 shRNA.
Virion Production Assay
1.75 × 105 293T cells were cotransfected with the proviral construct (0.5 μg) pNL-Luc-U−E− together with 25 ng of pCA-Vpu-RRE (WT or mutants), 25 ng of pCA-Rev, 2.5 ng of pCA-BST-2 (WT or mutants), and 947.5 ng of the empty vector by using FuGENE 6. After 48 h the supernatants were harvested and subjected to p24-antigen capture ELISA. To normalize transfection efficiency, cells were lysed in 75 μl of lysis buffer, and firefly luciferase activities were determined by using a firefly Luciferase Assay System (Promega) with a Centro LB960 luminometer (Berthold). Alternatively, 7 × 105 293T cells were cotransfected with the proviral construct (1 μg) pNL-E−, pNL-Vpu2/6-E−, or pNL-U−E− together with 0.1 μg of pHIT/G and 0.9 μg of the empty vector by using FuGENE 6. After 48 h the supernatants were harvested and subjected to p24-antigen capture ELISA. βTrCP-1 and/or βTrCP-2-knockdown HeLa cells seeded at 6.25 × 104 cells were infected with 12.5 ng each of vesicular stomatitis virus glycoprotein-pseudotyped HIV-1. Sixteen hours later cells were washed with phosphate-buffered saline, and 1 ml of fresh complete medium was added. After 24 h supernatants were harvested and subjected to HIV-1 p24-antigen capture ELISA.
Immunoprecipitations
7 × 105 293T cells were cotransfected with 0.4 μg of pCA-Rev, either 0.8 μg of pCA-Vpu-RRE or pCA-Vpu-HA-RRE (WT or mutants), and either 0.8 μg of pCA-BST-2 (WT, or mutants) or pCA-BST-2-exMyc using FuGENE 6. Alternatively, 0.5 μg of βTrCP-1 or-2 plasmid was added to 0.5 μg each of the plasmids as described above (in control experiments, 0.5 μg of pCA-CD4 was used instead of pCA-BST-2). At 48 h after transfection, the cells were suspended in 500 μl of lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% digitonin, and Complete protease inhibitor mixture (Roche Applied Science)). The resultant lysates were clarified by brief centrifugation, precleared with 30 μl of protein G-agarose beads (Sigma) for 1 h at 4 °C, incubated with the anti-BST-2 mouse polyclonal antibody the Vpu antiserum, an anti-Myc polyclonal antibody (Sigma) or an anti-CD4 monoclonal antibody (Santa Cruz Biotechnology) for 1 h at 4 °C, and then added to 30 μl of protein G-agarose beads. After 1 h at 4 °C, the immune complexes were extensively washed with lysis buffer, and equal aliquots of the total and bound fractions were subjected to gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were probed with the anti-BST-2 mouse polyclonal antibody, the Vpu antiserum, an anti-FLAG rabbit polyclonal antibody (Sigma), or an anti-HA mouse monoclonal antibody (Sigma).
Flow Cytometry
1.75 × 105 293T cells were cotransfected with 2.5 ng of either the control vector or extracellular FLAG-tagged series of BST-2 expression plasmids, 25 ng of pCA-Vpu-RRE, 25 ng of pCA-Rev, 50 ng of pCA-EGFP (in some experiments together with 25 ng of the dynamin-2 WT or mutant expression plasmid) and the empty vector up to 1 μg of total DNA. After 48 h, transfected cells were incubated with an anti-FLAG M2 mouse monoclonal antibody (Sigma) or an isotype control antibody (Immunotech) followed by staining with a goat anti-mouse IgG conjugated to R-phycoeythrin (Molecular Probes) for 30 min on ice. Cells were then washed extensively with phosphate-buffered saline plus 0.1% bovine serum albumin, fixed with 4% formaldehyde in phosphate-buffered saline, and analyzed by fluorescence-activated cell sorting on a CyFlow (Partec). The data were analyzed by using FlowJo software (Tree Star, Inc.).
Immunofluorescence
COS7 cells were plated on 13-mmΦ glass coverslips, cotransfected with the indicated plasmids, and cultured for 24 h before fixation. To evaluate the Vpu-induced down-regulation of BST-2, cells were cotransfected with 0.5 μg of pCA-Vpu-EGFP-RRE (WT or CD4tm chimera), 0.5 μg of pCA-Rev, and 50 ng of Myc-tagged BST-2 (WT or mutants) using FuGENE 6. Alternatively, cells were cotransfected with 0.3 μg of pCA-Vpu-HA-RRE (WT or CD4tm chimera), 0.3 μg of pCA-Rev, 50 ng of pCA-BST-2-exMyc together with 0.4 μg of either pCA-Dyn2-EGFP or pCA-Dyn2-K44A-EGFP. To detect Myc-tagged BST-2 accumulated in lysosomes, the transfected cells were cultured for 16 h in complete medium in the presence of lysosomal protease inhibitors (40 μm of leupeptin and pepstatin A; Peptide Institute Inc., 4 μg/ml E64d; Sigma). To evaluate the involvement of proteasomal degradation in the Vpu-induced down-regulation of BST-2, transfected cells were cultured for 12 h in complete medium in the presence of a proteasome inhibitor (0.8 μm of MG-132; Sigma). The transfected cells were fixed with 4% paraformaldehyde at room temperature for 30 min, permeabilized with 0.05% saponin for 10 min, and immunostained with an anti-cathepsin D polyclonal antibody (DAKO) and an anti-Myc monoclonal antibody (9E10; Sigma). To distinguish cell-surface and intracellular BST-2 proteins, cells were incubated in complete medium in the presence of the anti-Myc mouse monoclonal antibody at 4 °C for 5 min, washed in phosphate-buffered saline at 4 °C, and then fixed with 4% paraformaldehyde at room temperature for 30 min. The fixed cells were permeabilized with 0.05% saponin for 10 min and immunostained with the anti-Myc polyclonal antibody. Secondary goat anti-mouse and anti-rabbit antibodies that had been conjugated with Cy3 or Cy5 (Jackson ImmunoResearch Laboratories, Inc.) were used at 5 μg/ml. DNA staining with Hoechst (Molecular Probes) was performed at 0.5 μg/ml. All immunofluorescence images were observed on a Leica DMRB microscope (Wetzlar) equipped with a 100× 1.32 NA oil immersion lens (PL APO), acquired through a cooled CCD camera, MicroMAX (Princeton Instruments), and digitally processed using IPlab Software (Scanalytics).
RESULTS
Vpu Physically Interacts with BST-2
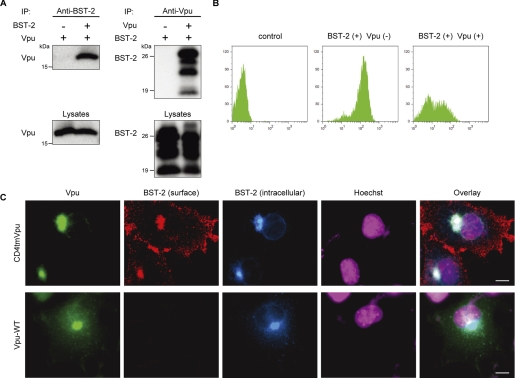
We first investigated the possible interaction between Vpu and BST-2. Precleared cell lysates from 293T cells cotransfected with Vpu and BST-2 expression plasmids were incubated with the anti-BST-2 antibody, and the resulting complexes were analyzed by Western blotting using an antibody to Vpu. An antibody against BST-2 was able to specifically coimmunoprecipitate Vpu protein from 293T cells (Fig. 1A, left panel). In the converse experiment cell lysates from 293T cells expressing Vpu and BST-2 proteins were incubated with the anti-Vpu antibody, and the precipitates were analyzed using the anti-BST-2 antibody. BST-2 protein was also able to be coimmunoprecipitated with Vpu (Fig. 1A, right panel). We, therefore, conclude that Vpu and BST-2 can physically interact.
FIGURE 1.
Vpu physically interacts with BST-2 and reduces its cell-surface expression. A, shown is Vpu-BST-2 interaction. Precleared cell extracts from 293T cells expressing Vpu with or without BST-2 were immunoprecipitated (IP) with an anti-BST-2 antibody, then immunoblotted with an antibody to Vpu (upper left), or the reciprocal experiment was done by using the antibodies to Vpu for immunoprecipitation and to BST-2 for immunoblotting, respectively (upper right). Aliquots of cell lysates were also analyzed by immunoblotting in parallel for Vpu (lower left) and BST-2 (lower right). B, shown is cell-surface expression of BST-2 in the presence and absence of Vpu. 293T cells transiently expressing extracellularly FLAG-tagged BST-2 and EGFP together with (right) or without Vpu (middle) and the control cells (left) were stained for cell-surface BST-2 by using an anti-FLAG monoclonal antibody and analyzed by two-color flow cytometry. The cells were gated for EGFP-positive cells. C, COS7 cells transiently expressing Myc-tagged BST-2 together with Vpu/EGFPs (WT; lower panels, CD4tm/Vpu chimera; upper panels) were processed for cell-surface and intracellular immunofluorescence staining for BST-2 as described under “Experimental Procedures.” Bars, 10 μm.
Cell-surface Expression of BST-2 Is Reduced by Vpu
To dissect the mechanisms by which Vpu counteracts BST-2 protein, we examined the cell-surface expression of BST-2 in the presence and absence of Vpu by flow cytometry using optimal doses of BST-2 and Vpu expression plasmids based on levels of endogenous expression in HeLa cells and of physiological expression from Vpu-positive NL4-3 proviral DNA, respectively (supplemental Fig. S1, A and B). Cell-surface expression of BST-2 in 293T cells transfected with the expression plasmid encoding BST-2 was readily detected by using the anti-BST-2 antibody. When Vpu was coexpressed with BST-2 in 293T cells, expression of BST-2 on the cell surface was remarkably diminished (Fig. 1B), as recently reported (30). BST-2-specific cell-surface and intracellular immunofluorescence staining confirmed that Vpu was able to block cell-surface expression of BST-2 (Fig. 1C, lower panels), although a control Vpu-CD4TM hybrid protein, which is unable to enhance virion release (44), did not affect BST-2 expression (Fig. 1C, upper panels). These results suggest that Vpu down-regulates cell-surface BST-2, thereby probably leading to the lysosomal degradation of this host protein. Alternatively, Vpu might prevent de novo BST-2 expression intracellularly by inducing proteasomal degradation, similar to the mechanism by which it acts on CD4 (10, 11) or by inducing lysosomal degradation before BST-2 reaches the plasma membrane.
BST-2 Is Degraded in the Lysosome by Vpu
We next attempted to determine the pathway of the Vpu-induced down-regulation of BST-2. To examine whether BST-2 undergoes lysosomal degradation, we transfected COS7 cells with Myc-tagged BST-2 expression plasmid and either the WT Vpu or CD4tmVpu expression plasmid in the presence or absence of a mixture of lysosomal protease inhibitors (leupeptin, pepstatin A, and E64d (45)) and then observed the subcellular localization of BST-2 protein. The coexpression of Vpu in the cells treated with lysosomal protease inhibitors led to its colocalization with BST-2 and a lysosome marker, cathepsin D (Fig. 2B, right panels), whereas the expression of Vpu-CD4TM protein (Fig. 2B, left panels) and the absence of lysosomal protease inhibitors (Fig. 2A) did not. Importantly, magnified images of selected areas (Fig. 2B lower panels) revealed clear outlines (possible lysosomal membranes) composed of the colors of Vpu (green) and BST-2 (blue) proteins, in which the lysosomal protease cathepsin D (red) was harbored within vesicle lumens (Fig. 2B, bottom right), whereas cathepsin D without any visible outlines was observed in the presence of non-functional Vpu (Fig. 2B, bottom left). These results suggest that lysosomes (represented by cathepsin D) can carry BST-2 in the presence of a functional Vpu. By treatment with the proteasome inhibitor, BST-2 expressions were rescued in both the presence and absence of Vpu protein (supplemental Fig. S2A), implying that BST-2 might physiologically undergo proteasomal degradation independently of the presence of Vpu. We, therefore, conclude that BST-2 is degraded by Vpu through a lysosomal degradation pathway.
FIGURE 2.
BST-2 is degraded by Vpu through the lysosomal degradation pathway. COS7 cells transiently expressing Myc-tagged BST-2 together with Vpu/EGFP (right panels in A and B) or with a control CD4 tmVpu/EGFP (left panels in A and B) were cultured in the absence (A) and presence (B) of a mixture of lysosome protease inhibitors (LPI; containing leupeptin, pepstatin A, and E64d). The cells were processed for immunofluorescence staining for total BST-2 and a lysosome marker cathepsin D, as described under “Experimental Procedures.” Squares indicate magnified regions. Bars, 10 μm.
Vpu-induced BST-2 Degradation Is Partially Dependent on βTrCP Proteins
In the case of proteasomal degradation of CD4, Vpu specifically recruits the βTrCP-1 and -2 subunits of the Skp1-Cullin1-F-box ubiquitin ligase complex. This is achieved by the interaction of phosphorylated serine residues at positions 52 and 56 in the CT domain of Vpu (12, 13) with seven C-terminal WD repeats in βTrCP (10). Because βTrCP is known to control not only proteasomal but also lysosomal degradation (46), we next examined whether the Vpu-induced lysosomal degradation of BST-2 would involve βTrCP proteins. Control lysates from 293T cells expressing CD4, either WT or βTrCP interaction-defective (52/56 mutant, 2/6) Vpu protein, and either FLAG-tagged βTrCP-1 or -2, were subjected to immunoprecipitation with an anti-CD4 antibody. As expected and previously reported (10, 11), βTrCP-1 and -2 proteins were specifically coimmunoprecipitated with CD4 in the presence of Vpu-WT but not Vpu2/6 (Fig. 3A, left). When BST-2 was expressed instead of CD4 and immunoprecipitated with the anti-BST-2 polyclonal antibody, both βTrCP-1 and -2 were also coimmunoprecipitated with similar efficiencies in the presence of WT Vpu but not Vpu2/6 (Fig. 3A, right). The results indicate that, as is the case of CD4, Vpu assembles a ternary complex with BST-2 and βTrCP between which no direct interaction occurs. Importantly, Vpu2/6 was still able to partially enhance virion production in HeLa cells (Fig. 3B). Supporting these results, knockdown experiments using shRNAs, which efficiently targeted βTrCP-1 and/or βTrCP-2 (supplemental Fig. S3), revealed a partial but additive reduction in the Vpu-mediated effect on virion release (Fig. 3C), which was comparable with that in Vpu2/6 treated with a control shRNA. Overall, these experiments indicate that the effect of Vpu on the lysosomal degradation of BST-2 is partially dependent on βTrCP.
FIGURE 3.
Vpu-induced BST-2 degradation is partially βTrCP-dependent. A, Vpu connects BST-2 with either βTrCP-1 or -2. Precleared cell extracts from 293T cells transiently expressing CD4, Vpu (WT or 2/6 mutant), and either FLAG-tagged βTrCP-1 or -2 were immunoprecipitated (IP) with an anti-CD4 antibody (left half). Similarly, precleared cell extracts from 293T cells transiently expressing BST-2, Vpu (WT or 2/6 mutant), and either FLAG-tagged βTrCP-1 or -2 were immunoprecipitated with the anti-BST-2 antibody (right half). The resulting complexes were analyzed by immunoblotting with antibodies to Vpu and FLAG (upper two panels). Aliquots of cell lysates were also analyzed by immunoblotting in parallel for Vpu and FLAG (lower two panels). B, βTrCP interaction-defective mutant of Vpu retains some activity to enhance virion release. HeLa cells were infected with vesicular stomatitis virus glycoprotein-pseudotyped WT, Vpu2/6, and ΔVpu viruses. After 40 h, viral supernatants were harvested and subjected to p24 ELISA. Data are presented as a percentage of the amount of WT release and shown as the mean ± S.D. Vpu expressions from the proviral DNAs were confirmed by immunoblotting. C, shown is partial inhibitory effect of βTrCP knockdown on Vpu activity. HeLa cells were transduced with a control lentiviral vector or with vectors producing βTrCP-1 and/or -2-specific shRNA. After 48 h, transduced cells were infected as described in B. Data are presented as a percentage of the amount of WT release in the presence of control shRNA and are representative of three independent experiments.
Vpu Targets and Internalizes Cell-surface BST-2
It was still unclear whether cell-surface BST-2 or intracellular de novo BST-2 would undergo Vpu-induced lysosomal degradation partially in a βTrCP-dependent manner. We, therefore, examined whether BST-2 expression reduced by Vpu was rescued by inhibiting endocytosis. To analyze this we performed flow cytometry by using 293T cells transiently expressing BST-2, Vpu, and either WT dynamin-2 (Dyn2-WT) or its dominant-negative form (Dyn2-K44A), the latter of which inhibits both clathrin-dependent and -independent, but not caveolae/lipid raft-dependent endocytosis (for review, see Ref. 47). Dyn2-K44A was able to drastically and specifically block Vpu-induced BST-2 down-regulation, whereas Dyn2-WT did not show any effect (Fig. 4A). This was also proved by the cell-surface/intracellular-staining immunofluorescence assay (Fig. 4B). These results suggest that cell-surface BST-2 undergoes lysosomal degradation by Vpu. Because a very recent report has suggested that Vpu would act after the physiological endocytosis of BST-2 via AP-2 (48), we examined whether or not a BST-2 mutant deficient in constitutive endocytosis could be insensitive to Vpu. To do this we introduced mutations into a non-canonical tyrosine-based motif containing two tyrosine residues at positions 6 and 8 in the CT domain of BST-2 (Y6A/Y8A), which has been known to be essential for clathrin-mediated internalization of this protein (32, 49). Protein expressions were confirmed by immunoblotting, showing more intensive bands in the endocytosis mutant Y6A/Y8A than in WT as expected (Fig. 4C). A virion production assay showed that the Y6A/Y8A mutant was still sensitive to Vpu (Fig. 4D). Consistent with this, flow cytometry revealed the efficient down-regulation of the mutant BST-2 Y6A/Y8A by Vpu (Fig. 4E). The fact that, without the ability of BST-2 to be endocytosed, Vpu is able to actively induce internalization of this host protein, strongly suggests that the actual site of action of Vpu is not post-endocytic but pre-endocytic. Taken together, we conclude that Vpu targets and internalizes BST-2 at the plasma membrane.
FIGURE 4.
Vpu actively internalizes cell-surface BST-2. A, inhibition of endocytosis by dominant-negative dynamin rescues Vpu-induced reduction of cell-surface expression of BST-2 is shown. 293T cells transiently expressing EGFP and FLAG-tagged BST-2 with (lower panels) or without Vpu (upper panels) together with a control vector (left), dynamin-2-WT (middle; Dyn2-WT), or dominant-negative dynamin-2 (right; Dyn2-K44A) were stained for cell-surface BST-2 using the anti-FLAG monoclonal antibody and analyzed by two-color flow cytometry. B, COS7 cells transiently expressing Myc-tagged BST-2 and either HA-tagged Vpu or a control HA-tagged CD4tmVpu together with Dyn2-WT-EGFP (left half) or with Dyn2-K44A-EGFP (right half) were processed for cell-surface and intracellular immunofluorescence staining by using anti-Myc monoclonal (cell-surface) and anti-HA polyclonal (intracellular) antibodies, respectively. Squares indicate magnified regions in which the implications for the section indicated by arrows are considered under “Discussion.” Bars, 10 μm. C–E, shown is the endocytosis mutant of BST-2 remains sensitive to Vpu. C, expressions of FLAG-tagged BST-2 WT and its endocytosis mutant (Y6A/Y8A) were confirmed by immunoblotting using the anti-FLAG monoclonal antibody. D, 293T cells were transfected with Vpu-deficient HIV-1 proviral DNA and either a control or Vpu expression plasmid together with BST-2 WT or Y6A/Y8A expression plasmid. After 48 h viral supernatants were harvested and subjected to p24 ELISA. Transfection efficiencies were normalized with the luciferase activity. Data shown are the mean ± S.D. E, experiments were the same as Fig. 1B, except that a FLAG-tagged BST-2 Y6A/Y8A mutant (right) was also analyzed.
TM Domain of BST-2 Is Specifically Recognized by Vpu
To determine the domains of BST-2 involved in the interaction with Vpu, we created FLAG-tagged chimeric constructs with BST-2 and TfR, which has a type II topology, by replacing the CT and/or TM domains of BST-2 with the corresponding domains of TfR (Fig. 5A). Expressions of all chimeric BST-2 proteins in the transfected cells were confirmed by immunoblotting using the anti-BST-2 antibody (Fig. 5B). Flow cytometry analysis (Fig. 5C, upper panels) verified that all of these constructs were expressed on the cell surface at equivalent levels. By performing both double-staining immunofluorescence (supplemental Fig. S4) and flow cytometry assays (Fig. 5C, lower panels), it was revealed that the cell-surface expressions of chimeric BST-2 constructs with the CT-TM and with the TM domains of TfR were retained even in the presence of Vpu, whereas those of WT and a chimeric BST-2 carrying TfR CT domain were efficiently reduced, suggesting that BST-2 is down-regulated from the cell surface through the specific interaction of Vpu with the TM domain of BST-2. Supporting the results, immunoprecipitation and virion production assays proved that the TM domain of BST-2 was indeed required for the interaction with Vpu and thereby determined the sensitivity to Vpu (Fig. 5, D and E).
FIGURE 5.
TM domain of BST-2 is specifically recognized by Vpu. A, shown is the predicted topology of BST-2 chimeric proteins. The CT, TM, and extracellular (EC) domains of BST-2 are represented in white, and the CT and TM domains of TfR are in black. Plasma membranes are depicted in gray. B, expressions of WT and chimeric BST-2 proteins were confirmed by immunoblotting using the anti-BST-2 polyclonal antibody, which recognizes the extracellular domain. C–E, the TM domain of BST-2 is required for interaction with Vpu and thereby determines the sensitivity to Vpu. C, cell-surface expressions of chimeric BST-2 are shown. 293T cells transiently expressing EGFP, with (lower panels) or without Vpu (upper panels) together with FLAG-tagged BST-2 WT, -TfRct/tm, -TfRct, or TfRtm were stained for cell-surface BST-2 using the anti-FLAG monoclonal antibody and analyzed by two-color flow cytometry. D, shown is the interaction of Vpu with the BST-2 TM domain. Immunoprecipitations (IP) were performed as described in the legend for Fig. 1A, right panels, except that chimeric BST-2 proteins were analyzed. Aliquots of the cell lysates were also analyzed by immunoblotting in parallel for BST-2 (middle panels) and Vpu (lower panels). Arrows indicate the specific bands corresponding to the chimeric BST-2 proteins. Asterisks indicate the positions of immunoglobulin G light chains. E, inhibition of virion release by BST-2 chimeric proteins and their sensitivity to Vpu is shown. The assay was performed as described in Fig. 4D, except that chimeric BST-2 proteins were analyzed. Data are presented as a percentage of the amount of Vpu (−) virion release in the absence of BST-2 and are shown as the mean ± S.D.
TM Domain of Vpu Specifically Interacts with That of BST-2
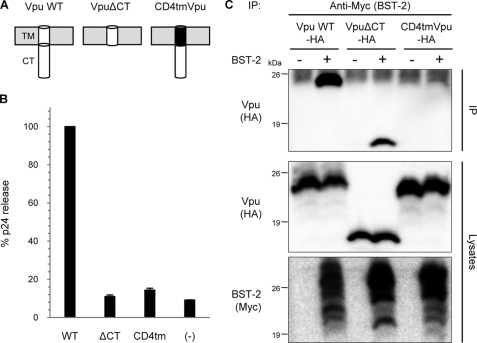
Used as a negative control of Vpu in our experiments (as described above), Vpu-CD4 TM hybrid protein is known not to enhance the release of virions (44), implying the TM domain of Vpu to be critical for the interaction with BST-2. In contrast, the CT domain has been thought to be dispensable for the virion production, as a Vpu mutant lacking this domain retained its biological activity for virus release (50). To assess the effects of those mutants on the production of virions, we performed the virion release assay using Vpu-WT, Vpu-CD4TM, and VpuΔCT (which has no single residue of CT domain; depicted in Fig. 6A) in the presence of BST-2. Interestingly, unlike the previously reported Vpu CT mutant comprising the TM domain and the N-terminal six residues of the CT domain (50), VpuΔCT failed to augment virion production as well as Vpu-CD4TM (Fig. 6B), implying that both the CT and TM domains might be involved in the interaction with BST-2. To test this, we performed immunoprecipitation assays using cells transiently expressing BST-2 and either Vpu-WT or mutants. Results obtained showed that VpuΔCT, but not Vpu-CD4TM, was specifically coimmunoprecipitated in the presence of BST-2 (Fig. 6C), indicating that only the TM domain of Vpu is required for the physical interaction with BST-2. Thus, we conclude that the TM domains of Vpu and BST-2 specifically interact with each other.
FIGURE 6.
TM domain of Vpu is specifically recognized by BST-2. A, shown is the predicted topology of Vpu mutant proteins. The TM and CT domains of Vpu are represented in white, and the TM domain of CD4 is in black. Plasma membranes are depicted in gray. B, CT domain of Vpu is also required to enhance virion release. The assay was performed as described in Fig. 4D, except that Vpu mutants were analyzed in the presence of BST-2 WT. Data are presented as a percentage of the amount of WT virion release and are shown as the mean ± S.D. C, the TM domain of Vpu interacts with BST-2. Precleared cell extracts from 293T cells expressing HA-tagged Vpu (WT or mutants) with or without Myc-tagged BST-2 were immunoprecipitated (IP) with an anti-Myc antibody followed by immunoblotting with an antibody to HA (upper). Aliquots of the cell lysates were also analyzed by immunoblotting for Vpu (middle) and BST-2 (lower).
The Inhibition of Virion Release by BST-2 Requires Its Anchoring to the Cell Membrane at Both Ends
To further investigate whether the antiviral activity of BST-2 would require its anchoring to the plasma membrane at one or both ends, we created GPI-anchor-deleted and CD4 signal peptide chimeric versions of BST-2, both of which anchor to the plasma membrane at one end (Fig. 7A). Neither the GPI-anchor deletion mutant nor the CD4 signal peptide chimera supported a reduction in virion production (Fig. 7B). We, therefore, conclude that BST-2 must be anchored to the cell membrane at both ends to have an inhibitory effect on virion production. It should be noted that the GPI-anchor-deleted mutant carrying the TM of BST-2 was able to interact with Vpu and thereby was Vpu-sensitive in its down-regulation from the cell surface by Vpu, whereas the CD4 signal peptide chimera lacking TM was unable to interact with Vpu and thereby was insensitive to Vpu (Fig. 7, C and D), consistent with Fig. 5D and supplemental Fig. S4.
FIGURE 7.
Anchoring of BST-2 to the cell membrane at both ends is required for the antiviral activity. A, predicted topology of BST-2 mutants is shown. Plasma membranes are depicted in gray. B, shown is the lack of an inhibitory effect of BST-2 mutants on virion release. The assay was performed as described in Fig. 4D, except that mutant BST-2 proteins were analyzed. Data are shown as described in Fig. 5E. C, immunoprecipitations (IP) were performed as described in the legend for Fig. 1A, right panels, and Fig. 5D, except that mutant BST-2 proteins were analyzed. Arrows indicate the specific bands corresponding to the BST-2 mutant. Asterisks indicate the positions of immunoglobulin G light chains. D, immunofluorescence was performed as described in Fig. 1C, except that mutant BST-2 proteins were also analyzed. Bars, 10 μm. E, models depicting potential configurations of BST-2 in tethering virions to cell membranes are shown.
DISCUSSION
In this study we present that Vpu physically interacts with BST-2 through TM-to-TM binding and targets and internalizes cell-surface BST-2 through an endocytic pathway, leading to lysosomal degradation partially dependent on βTrCP. Here, we found that Vpu degraded BST-2 through a lysosomal degradation pathway, based on experiments using lysosomal protease inhibitors. Goffinet et al. (51) recently showed that treatment with a proteasome inhibitor abrogated the Vpu-mediated enhancement of virion production, suggesting that Vpu suppresses BST-2 by accelerating its degradation via the proteasome. In contrast, we found that proteasomal inhibitors reduced virion production with or without BST-2 expression (supplemental Fig. S2B). This treatment was unable to rescue the cell-surface expression of BST-2 reduced by Vpu (supplemental Fig. S2C), suggesting that the reduced virion production in the presence of the protease inhibitor is not due to a recovery of the cell-surface expression of BST-2 but probably due to, as previously reported, the proteasome-inhibitor-induced reduction of virion release independently of Vpu, resulting from the rapid depletion of the free ubiquitin pool with prolonged drug treatment (52). Because of the unavailability of antibodies against cytoplasmic proteasome markers for immunofluorescence staining, we are unable to perform parallel experiments with treatment of lysosome inhibitors that showed colocalization of Vpu, BST-2, and the lysosome marker cathepsin D. We, therefore, cannot rule out that some fractions of BST-2 might be proteasomally degraded by Vpu. Despite this, our results are in agreement with two recent observations showing that Vpu antagonizes BST-2 via an endo-lysosomal degradation pathway (48, 53).
Vpu has been known to interact with the cellular F-box proteins βTrCP-1 (10) and βTrCP-2 (11) to induce CD4 degradation. This interaction is mediated by two phosphoserine residues in the CT domain of Vpu and C-terminal WD repeats in βTrCP. We herein attempted to determine whether the degradation of BST-2 by Vpu could be explained by the same mechanism. βTrCP-1 and -2 proteins were indeed coimmunoprecipitated with BST-2 in the presence of WT Vpu but not Vpu2/6. This indicates that Vpu acts as a linker molecule between BST-2 and either βTrCP-1 or -2, as is the case of the ternary complex consisting of CD4, Vpu, and βTrCP (10). Our data showed that Vpu2/6 retained partial antiviral activity, the level of which was nearly equivalent to that of WT Vpu in the cells in which both the βTrCP-1 and -2 genes were simultaneously silenced. These suggest that inhibition of BST-2 by Vpu is partially dependent on βTrCP proteins. This is partly consistent with two recent papers, one of which showed that the silencing of both βTrCP-1 and -2 inhibited the Vpu-mediated down-regulation of BST-2 whereas the knockdown of just one of these genes had little or no effect (48), and the other of which showed that the silencing of βTrCP-2, but not βTrCP-1, was effective (53). Although the differences might reflect the knockdown efficiency, these results need to be reexamined. One key question is why the βTrCP interaction-defective Vpu2/6 protein retained some activity to enhance the virion release at the level of WT Vpu in the βTrCP-1 and -2 knockdown cells. Two possible explanations for this are (i) interaction of Vpu with a unknown host factor(s), which is required for the degradation of BST-2, and (ii) the existence of another host restriction factor antagonized by Vpu. Regarding the latter possibility, it is tempting to speculate that calcium-modulating cyclophilin ligand recently, identified as a Vpu-sensitive restriction factor (54), regulates the cell-surface expression of BST-2. Because calcium-modulating cyclophilin ligand plays a role in the recycling of the epidermal growth factor receptor through recycling endosomes (55) to which BST-2 is localized (49), calcium-modulating cyclophilin ligand might be also required for the recycling of BST-2.
Our data also showed that inhibition of the broad endocytic pathway by the dominant-negative dynamin mutant led to a recovery from the Vpu-induced reduction of BST-2 expression. Additionally, when the dominant-negative dynamin was coexpressed in the cells, WT Vpu, normally distributed diffusely throughout the intracellular compartments, was also found to be expressed at the plasma membrane with a punctate pattern (Fig. 4B, row 2, right panel). This is probably because the endocytosis-free Vpu-BST-2 complex retained at the plasma membrane was able to avoid rapid degradation, resulting in visible signals. These results suggest that the BST-2 lysosomally degraded by Vpu is derived from that in the plasma membranes. In a very recent paper it was concluded that Vpu action against BST-2 is post-endocytic, based on the absence of an effect of Vpu on the rate of endocytosis and on the inhibition of down-regulation by bafilomycin A1, which inhibits pH gradient-dependent trafficking to late endosomes and lysosomes (48). The report also included data showing that the Vpu-reduced reduction of cell-surface BST-2 expression was recovered by silencing the μ2-subunit of the AP-2 complex, which regulates clathrin-mediated endocytosis (for review, see Ref. 56). Although this result itself could suggest that Vpu directly induces endocytosis of cell-surface BST-2, the authors concluded that Vpu targets BST-2, which has been endocytosed constitutively via AP-2 for the reasons described above. Importantly, another recent report suggests that BST-2 is physiologically endocytosed through interaction with α-adaptin, but not the μ2-subunit, of the AP-2 complex (49). This implies that what was blocked by silencing of the μ2-subunit described by Mitchell et al. (48) might be Vpu-induced internalization of BST-2, not the physiological endocytosis of BST-2. This hypothesis is indeed supported by our observation that an endocytosis-defective BST-2 mutant protein (α-adaptin interaction-defective mutant (49)) could still be internalized from the plasma membrane by Vpu. Thus, we conclude that the plasma membrane is the primary site of action of Vpu, where this small viral protein targets and internalizes cell-surface BST-2 presumably through a clathrin-mediated, but α-adaptin-independent endocytic pathway, partially in a βTrCP-dependent fashion.
Importantly, this study provides evidence of the physical interaction of Vpu and BST-2 through mutual TM-to-TM binding, obtained using chimeric constructs with BST-2 and the type II TM protein TfR. The requirement of the TM domain of BST-2 for the sensitivity to Vpu was also demonstrated recently (51, 57–59) using chimeric and point mutant BST-2 proteins based on the difference between human and other primate versions. Those observations, however, do not exclude the possibility that the CT domain of BST-2 might be additionally required for the interaction with Vpu. In contrast, our experiments revealed that BST-2 carrying the CT domain of the type II TM protein TfR retained the ability to bind Vpu and was Vpu-sensitive, whereas BST-2 carrying the TM and/or CT domains of TfR lost this ability and sensitivity to Vpu. These results suggest that the interaction of BST-2 with Vpu requires only the TM domain, not the CT domain, of BST-2. In terms of Vpu, Schubert et al. (50) reported that Vpu TM is sufficient to enhance HIV-1 virion production. However, we found that deletion of the entire CT (VpuΔCT), which could still bind BST-2, totally abolished the ability of Vpu to enhance virion release, suggesting that Vpu CT might recruit not only βTrCP but also an unknown cofactor(s). The discrepancy between their and our observations could be because their Vpu CT mutant retained six amino acid residues of the CT domain, which might be a site of interaction with unknown cellular cofactor(s). This needs to be elucidated with further experiments.
Based on the unusual topology of BST-2 (31, 32), Neil et al. (26) have proposed several configurations of this protein in tethering virions to cell membranes. These include (i) bridging between viral and cellular membranes with dimerization, (ii) simple dimerization of both viral and cellular sides of BST-2 proteins, and (iii) mediation of unknown virion and cell components, the latter two of which seemingly do not necessarily require BST-2 insertion into the cell membrane at both ends. In that paper a GPI-anchor mutant that anchors to the plasma membrane at one end was incapable of inhibiting virion production. In this case, however, deletion of the GPI modification signal abolishes the localization of BST-2 at lipid rafts (31, 32) at which HIV-1 and other enveloped viruses preferentially assemble and bud (60–62). We, therefore, created another chimeric version of BST-2 carrying the CD4 signal peptide in place of the CT and TM domains of BST-2. This chimeric protein, which has an intact GPI modification signal and thereby singly anchors at the lipid-raft end at the cell surface, could be exposed for interaction with the viral side of this protein or with, if any, unknown virion and cell components. Nevertheless, the one-end-anchored BST-2 completely lost the inhibitory activity toward virion production. We, therefore, propose that the antiviral activity of BST-2 requires the following configuration, i.e. both ends of the dimerized proteins unidirectionally bridge virions and cell membranes (Fig. 7E).
Another level of complexity about Vpu inhibitory effect has recently been added by the finding that, in CD4-positive transformed cell lines, very little effect of Vpu on the cell-surface expression of BST-2 was observed, whereas Vpu was able to enhance the release of HIV-1 virions (63). In this regard, our speculation is as follows; CD4-negative cells like the HeLa and 293T used in our and other studies allow Vpu to target only BST-2 (or another factor as well). On the other hand, CD4-postive cells present Vpu with an additional target that is CD4. Because of this, higher levels of CD4 in the transformed T-cell lines could overwork Vpu to capture CD4 in the endoplasmic reticulum, resulting in insufficient plasma membrane transport of Vpu, which is unable to efficiently down-regulate BST-2. Taken together with the findings that cell-surface CD4 itself interferes with not only viral infectivity but also the release of virions from Jurkat T cells (64), which do not endogenously express BST-2 (63), we speculate that the ultimate role of Vpu might simply be to enhance virion release by blocking the cell-surface expressions of both CD4 and BST-2 proteins. More studies will be needed to address this issue.
Acknowledgments
We thank D. Trono for the lentiviral vector plasmids pLVTHM and psPAX2 and K. Strebel for HIV-1 NL4-3 Vpu antiserum obtained through the National Institutes of Health AIDS Research and Reference Reagent Program.
This work was supported in part by a grant from the Ministry of Health, Labor, and Welfare of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- Vpu
- viral protein U
- HIV-1
- human immunodeficiency virus type 1
- βTrCP-1
- β-transducin repeat-containing protein 1
- CT
- cytoplasmic
- TM
- transmembrane
- BST-2
- bone marrow stromal antigen 2
- GPI anchor
- glycosylphosphatidylinositol anchor
- RRE
- Rev-responsive element
- HA
- hemagglutinin
- EGFP
- enhanced green fluorescent protein
- TfR
- transferrin receptor
- Dyn2
- dynamin-2
- WT
- wild-type
- shRNA
- short hairpin RNA
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Cohen E. A., Terwilliger E. F., Sodroski J. G., Haseltine W. A. (1988) Nature 334, 532–534 [DOI] [PubMed] [Google Scholar]
- 2.Strebel K., Klimkait T., Martin M. A. (1988) Science 241, 1221–1223 [DOI] [PubMed] [Google Scholar]
- 3.Huet T., Cheynier R., Meyerhans A., Roelants G., Wain-Hobson S. (1990) Nature 345, 356–359 [DOI] [PubMed] [Google Scholar]
- 4.Courgnaud V., Salemi M., Pourrut X., Mpoudi-Ngole E., Abela B., Auzel P., Bibollet-Ruche F., Hahn B., Vandamme A. M., Delaporte E., Peeters M. (2002) J. Virol. 76, 8298–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santiago M. L., Rodenburg C. M., Kamenya S., Bibollet-Ruche F., Gao F., Bailes E., Meleth S., Soong S. J., Kilby J. M., Moldoveanu Z., Fahey B., Muller M. N., Ayouba A., Nerrienet E., McClure H. M., Heeney J. L., Pusey A. E., Collins D. A., Boesch C., Wrangham R. W., Goodall J., Sharp P. M., Shaw G. M., Hahn B. H. (2002) Science 295, 465. [DOI] [PubMed] [Google Scholar]
- 6.Barlow K. L., Ajao A. O., Clewley J. P. (2003) J. Virol. 77, 6879–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strebel K., Klimkait T., Maldarelli F., Martin M. A. (1989) J. Virol. 63, 3784–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willey R. L., Maldarelli F., Martin M. A., Strebel K. (1992) J. Virol. 66, 7193–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willey R. L., Maldarelli F., Martin M. A., Strebel K. (1992) J. Virol. 66, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margottin F., Bour S. P., Durand H., Selig L., Benichou S., Richard V., Thomas D., Strebel K., Benarous R. (1998) Mol. Cell 1, 565–574 [DOI] [PubMed] [Google Scholar]
- 11.Butticaz C., Michielin O., Wyniger J., Telenti A., Rothenberger S. (2007) J. Virol. 81, 1502–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert U., Henklein P., Boldyreff B., Wingender E., Strebel K., Porstmann T. (1994) J. Mol. Biol. 236, 16–25 [DOI] [PubMed] [Google Scholar]
- 13.Friborg J., Ladha A., Göttlinger H., Haseltine W. A., Cohen E. A. (1995) J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8, 10–22 [PubMed] [Google Scholar]
- 14.Fujita K., Omura S., Silver J. (1997) J. Gen. Virol. 78, 619–625 [DOI] [PubMed] [Google Scholar]
- 15.Schubert U., Antón L. C., Bacík I., Cox J. H., Bour S., Bennink J. R., Orlowski M., Strebel K., Yewdell J. W. (1998) J. Virol. 72, 2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klimkait T., Strebel K., Hoggan M. D., Martin M. A., Orenstein J. M. (1990) J. Virol. 64, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Göttlinger H. G., Dorfman T., Cohen E. A., Haseltine W. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7381–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao X. J., Göttlinger H., Haseltine W. A., Cohen E. A. (1992) J. Virol. 66, 5119–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geraghty R. J., Talbot K. J., Callahan M., Harper W., Panganiban A. T. (1994) J. Med. Primatol. 23, 146–150 [DOI] [PubMed] [Google Scholar]
- 20.Sakai H., Tokunaga K., Kawamura M., Adachi A. (1995) J. Gen. Virol. 76, 2717–2722 [DOI] [PubMed] [Google Scholar]
- 21.Yao X. J., Garzon S., Boisvert F., Haseltine W. A., Cohen E. A. (1993) J. Acquir. Immune Defic. Syndr. 6, 135–141 [PubMed] [Google Scholar]
- 22.Varthakavi V., Smith R. M., Bour S. P., Strebel K., Spearman P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15154–15159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 24.Neil S. J., Eastman S. W., Jouvenet N., Bieniasz P. D. (2006) PLoS Pathog. 2, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neil S. J., Sandrin V., Sundquist W. I., Bieniasz P. D. (2007) Cell Host Microbe 2, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neil S. J., Zang T., Bieniasz P. D. (2008) Nature 451, 425–430 [DOI] [PubMed] [Google Scholar]
- 27.Goto T., Kennel S. J., Abe M., Takishita M., Kosaka M., Solomon A., Saito S. (1994) Blood 84, 1922–1930 [PubMed] [Google Scholar]
- 28.Ishikawa J., Kaisho T., Tomizawa H., Lee B. O., Kobune Y., Inazawa J., Oritani K., Itoh M., Ochi T., Ishihara K., et al. (1995) Genomics 26, 527–534 [DOI] [PubMed] [Google Scholar]
- 29.Ohtomo T., Sugamata Y., Ozaki Y., Ono K., Yoshimura Y., Kawai S., Koishihara Y., Ozaki S., Kosaka M., Hirano T., Tsuchiya M. (1999) Biochem. Biophys. Res. Commun. 258, 583–591 [DOI] [PubMed] [Google Scholar]
- 30.Van Damme N., Goff D., Katsura C., Jorgenson R. L., Mitchell R., Johnson M. C., Stephens E. B., Guatelli J. (2008) Cell Host Microbe 3, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupzig S., Korolchuk V., Rollason R., Sugden A., Wilde A., Banting G. (2003) Traffic 4, 694–709 [DOI] [PubMed] [Google Scholar]
- 32.Rollason R., Korolchuk V., Hamilton C., Schu P., Banting G. (2007) J. Cell Sci. 120, 3850–3858 [DOI] [PubMed] [Google Scholar]
- 33.Rollason R., Korolchuk V., Hamilton C., Jepson M., Banting G. (2009) J. Cell Biol. 184, 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jouvenet N., Neil S. J., Zhadina M., Zang T., Kratovac Z., Lee Y., McNatt M., Hatziioannou T., Bieniasz P. D. (2009) J. Virol. 83, 1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaletsky R. L., Francica J. R., Agrawal-Gamse C., Bates P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuma T., Noda T., Urata S., Kawaoka Y., Yasuda J. (2009) J. Virol. 83, 2382–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouchier R. A., Meyer B. E., Simon J. H., Fischer U., Malim M. H. (1997) EMBO J. 16, 4531–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinomoto M., Yokoyama M., Sato H., Kojima A., Kurata T., Ikuta K., Sata T., Tokunaga K. (2005) J. Virol. 79, 5996–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tokunaga K., Greenberg M. L., Morse M. A., Cumming R. I., Lyerly H. K., Cullen B. R. (2001) J. Virol. 75, 6776–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 42.Schneider C., Owen M. J., Banville D., Williams J. G. (1984) Nature 311, 675–678 [DOI] [PubMed] [Google Scholar]
- 43.Maldarelli F., Chen M. Y., Willey R. L., Strebel K. (1993) J. Virol. 67, 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul M., Mazumder S., Raja N., Jabbar M. A. (1998) J. Virol. 72, 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volanti C., Gloire G., Vanderplasschen A., Jacobs N., Habraken Y., Piette J. (2004) Oncogene 23, 8649–8658 [DOI] [PubMed] [Google Scholar]
- 46.Kumar K. G., Tang W., Ravindranath A. K., Clark W. A., Croze E., Fuchs S. Y. (2003) EMBO J. 22, 5480–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols B. (2003) J. Cell Sci. 116, 4707–4714 [DOI] [PubMed] [Google Scholar]
- 48.Mitchell R. S., Katsura C., Skasko M. A., Fitzpatrick K., Lau D., Ruiz A., Stephens E. B., Margottin-Goguet F., Benarous R., Guatelli J. C. (2009) PLoS Pathog. 5, e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masuyama N., Kuronita T., Tanaka R., Muto T., Hirota Y., Takigawa A., Fujita H., Aso Y., Amano J., Tanaka Y. (2009) J. Biol. Chem. 284, 15927–15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schubert U., Bour S., Ferrer-Montiel A. V., Montal M., Maldarell F., Strebel K. (1996) J. Virol. 70, 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goffinet C., Allespach I., Homann S., Tervo H. M., Habermann A., Rupp D., Oberbremer L., Kern C., Tibroni N., Welsch S., Krijnse-Locker J., Banting G., Kräusslich H. G., Fackler O. T., Keppler O. T. (2009) Cell Host Microbe 5, 285–297 [DOI] [PubMed] [Google Scholar]
- 52.Schubert U., Ott D. E., Chertova E. N., Welker R., Tessmer U., Princiotta M. F., Bennink J. R., Krausslich H. G., Yewdell J. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13057–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douglas J. L., Viswanathan K., McCarroll M. N., Gustin J. K., Früh K., Moses A. V. (2009) J. Virol. 83, 7931–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varthakavi V., Heimann-Nichols E., Smith R. M., Sun Y., Bram R. J., Ali S., Rose J., Ding L., Spearman P. (2008) Nat. Med. 14, 641–647 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Tran D. D., Russell H. R., Sutor S. L., van Deursen J., Bram R. J. (2003) Dev. Cell 5, 245–256 [DOI] [PubMed] [Google Scholar]
- 56.Benmerah A., Lamaze C. (2007) Traffic 8, 970–982 [DOI] [PubMed] [Google Scholar]
- 57.Gupta R. K., Hué S., Schaller T., Verschoor E., Pillay D., Towers G. J. (2009) PLoS Pathog. 5, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McNatt M. W., Zang T., Hatziioannou T., Bartlett M., Fofana I. B., Johnson W. E., Neil S. J., Bieniasz P. D. (2009) PLoS Pathog. 5, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rong L., Zhang J., Lu J., Pan Q., Lorgeoux R. P., Aloysius C., Guo F., Liu S. L., Wainberg M. A., Liang C. (2009) J. Virol. 83, 7536–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheiffele P., Rietveld A., Wilk T., Simons K. (1999) J. Biol. Chem. 274, 2038–2044 [DOI] [PubMed] [Google Scholar]
- 61.Nguyen D. H., Hildreth J. E. (2000) J. Virol. 74, 3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ono A., Freed E. O. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13925–13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyagi E., Andrew A. J., Kao S., Strebel K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cortés M. J., Wong-Staal F., Lama J. (2002) J. Biol. Chem. 277, 1770–1779 [DOI] [PubMed] [Google Scholar]