Abstract
Many RNA nucleases and helicases participate in ribosome biogenesis, but how they cooperate with each other is largely unknown. Here we report that in vivo cleavage of the yeast pre-rRNA at site D, the 3′-end of the 18 S rRNA, requires functional interactions between PIN (PilT N terminus) domain protein Nob1 and the DEAH box RNA helicase Prp43. Nob1 showed specific cleavage on a D-site substrate analogue in vitro, which was abolished by mutations in the Nob1 PIN domain or the RNA substrate. Genetic analyses linked Nob1 to the late pre-40 S-associated factor Ltv1, the RNA helicase Prp43, and its cofactor Pfa1. In strains lacking Ltv1, mutation of Prp43 or Pfa1 led to a striking accumulation of 20 S pre-rRNA in the cytoplasm due to inhibition of site D cleavage. This phenotype was suppressed by increased dosage of wild-type Nob1 but not by Nob1 variants mutated in the catalytic site. In ltv1/pfa1 mutants the 20 S pre-rRNA was susceptible to 3′ to 5′ degradation by the cytoplasmic exosome. This degraded into the 3′ region of the 18 S rRNA, strongly indicating that the preribosomes are structurally defective.
Introduction
Eukaryotic ribosome formation is a complex process that requires coordination of rRNA synthesis and processing with the subsequent incorporation of the ribosomal (Rpl and Rps) proteins (reviewed in Refs. 1–5). In the yeast Saccharomyces cerevisiae, ribosome synthesis starts with the transcription of a 35 S pre-rRNA, which is the common precursor to the mature 18, 5.8, and 25 S rRNAs. This pre-rRNA is co-transcriptionally bound by ribosomal proteins as well as many non-ribosomal trans-acting factors to form a 90 S preribosomal particle. Next, this 90 S intermediate undergoes a series of early pre-rRNA processing and modification steps in the nucleolus before an endonucleolytic cleavage at site A2 in the pre-rRNA separates the pathways for 60 and 40 S subunit assembly.
Pre-60 S particles undergo further maturation steps in the nucleus, whereas pre-40 S particles contain few non-ribosomal proteins and are rapidly exported to the cytoplasm. The factors involved in 40 S export are largely unknown, but the non-essential pre-40 S factor Ltv1 was suggested to play a role in this process (6). Following nuclear export, the pre-40 S subunit undergoes cytoplasmic maturation, involving two major events. (i) 20 S pre-rRNA cleavage at site D generates the 3′-end of the mature 18 S rRNA, and (ii) structural reorganization forms the characteristic beak structure of the mature 40 S subunit, resulting in exposure of RNA helix 33 within the 18 S rRNA (7).
The sites and the order of endonuclease and exonuclease processing events that convert the 35 S pre-rRNA into the mature rRNA species are well characterized in yeast (Fig. 3A) (reviewed in Refs. 4 and 8). However, although most or all of the exonucleases participating in rRNA processing are known, several predicted endonucleases remain to be identified. Two proteins, Fap7 and Nob1, were proposed to cleave site D (9–11), but for neither was endonuclease activity shown. Fap7 is a putative NTPase that transiently associates with preribosomes, whereas Nob1 is a PilT N terminus (PIN)4 domain protein that stably associates with late pre-40 S particles (10–12). Several proteins were recently shown to have endonuclease activity that was dependent on an intact PIN (13–18) domain.
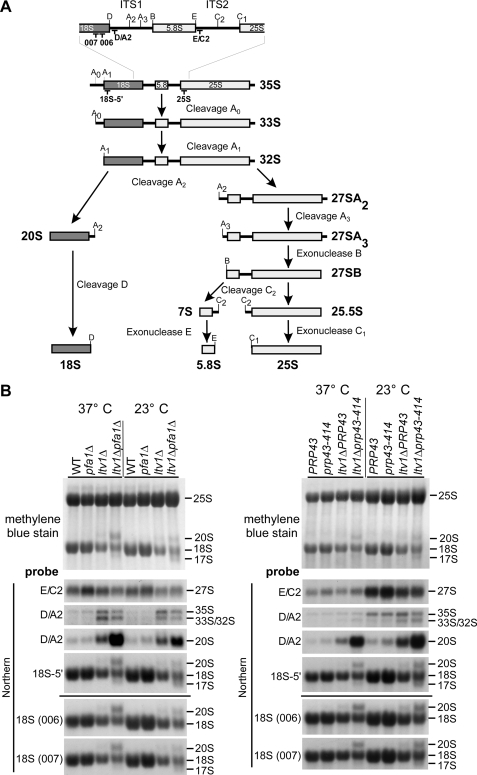
FIGURE 3.
Ltv1, Pfa1, and Prp43 are required for 20 to 18 S rRNA processing. A, simplified rRNA processing pathway in yeast. Only the major pathway for generation of the 5′-end of the 5.8 S rRNA is shown. The rRNA cleavage sites and the binding sites of the probes used for Northern blotting are indicated. ITS1 and ITS2, internal transcribed spacers 1 and 2. In the course of pre-rRNA processing, the 35 S pre-rRNA undergoes a series of endonucleolytic processing events at sites A0, A1, and A2 that lead to the separation into the 20 S and 27 S A2 pre-rRNAs. Endo- and exonucleolytic processing steps of the 27 S A2 pre-rRNA finally yield the mature 25 and 5.8 S rRNAs contained in 60 S subunits. In the cytoplasm, the final processing step in 40 S maturation takes place when the 20 S pre-rRNA is converted into the 18 S rRNA by endonucleolytic cleavage at processing site D. B, rRNA steady state levels in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants. Cells were either grown at 37 °C to an A600 of 0.8 (37 °C) or grown at 37 °C to an A600 of 0.1, transferred to 23 °C, and further grown to an A600 of 0.8 (three cell divisions). RNA was isolated, separated by agarose gel electrophoresis, and transferred to a nylon membrane that was stained with methylene blue. rRNA processing intermediates were detected by Northern blotting using the indicated probes. WT, wild type.
Assembly of preribosomes also involves 19 putative ATP-dependent RNA helicases. When considering the highly structured nature of the rRNA, the requirement of RNA helicases in ribosome biogenesis could involve the conformational rearrangement of RNA secondary structures, recruitment of additional factors, release of small nucleolar RNAs and proteins from the preribosomal RNAs, or stimulation of endo- and exonuclease activities (19). The direct substrates of the RNA helicases in ribosome biogenesis are largely unknown, and for only few of them (Dbp3, Dbp4, Mtr4, and Rok1) possible substrates were suggested (20–23).
Here, we report that the late pre-40 S factor Ltv1 is genetically linked to the RNA helicase Prp43, its co-factor Pfa1/Sqs1 (24), and to Nob1. 20 to 18 S rRNA processing was strongly impaired in double mutants between ltv1 and prp43 or ltv1 and pfa1. However, this defect was rescued by overexpression of Nob1. To test if Nob1 is the D-site-specific RNA processing enzyme, we developed an in vitro assay using a model RNA substrate. Nob1 purified from yeast exhibited highly specific endonuclease activity, cleaving precisely at the location corresponding to cleavage site D. Our data indicate that Prp43 and Pfa1 act together with Ltv1 to promote D-site cleavage by the endonuclease Nob1.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains used in this study are listed in supplemental Table S1. Deletion disruption and C-terminal tagging at the genomic locus were performed as described previously (25–27). The synthetic lethal screen with the ltv1Δ strain based on red/white colony sectoring was performed at 30 °C as described previously (28). Site-directed mutagenesis of Nob1 was performed by fusion PCR. All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in supplemental Table S2.
Fluorescence in Situ Hybridization
Fluorescence in situ hybridization was carried out as described previously (29), using a Cy3-labeled ITS1 (internal transcribed spacer)-specific probe (5′-Cy3-ATG CTC TTG CCA AAA CAA AAA AAT CCA TTT TCA AAA TTA TTA AAT TTC TT-3′) for detection of ITS1-containing pre-rRNAs. Cells were examined by fluorescence microscopy using an Imager Z1 microscope (Carl Zeiss) with a ×100, numerical aperture 1.4 Plan-Apo-Chromat oil immersion lens and a DICIII, 4′,6-diamidino-2-phenylindole, or HECy3 filter, respectively.
Sucrose Gradient Analysis and Fractionation
Cell extracts for polysome profile analyses were prepared as described previously (30). Sucrose gradients were analyzed and fractionated using an UA-6 system (Teledyne ISCO) with continuous monitoring at A254 nm. For Northern analysis of sucrose gradient fractions, six A260 nm units of cell extract were layered onto 7–50% sucrose gradients, and the gradients were centrifuged at 38,000 rpm in a SW40 rotor (Beckman Coulter) for 4 h and 30 min. Fractions were collected and adjusted to 10 mm Tris-HCl, pH 7.5, 10 mm EDTA, 0.5% SDS, and RNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1; two times) and chloroform/isoamyl alcohol (24:1), followed by ethanol precipitation.
RNA Isolation and Northern Blotting
Total RNA preparations were performed from 40 A600 units using the mechanical disruption protocol of the RNeasy minikit (Qiagen). 3 μg of RNA per sample were separated on 1.5% MOPS-agarose gels as described in the manual for the RNeasy minikit. The RNA was transferred overnight onto a Hybond N nylon membrane (Amersham Biosciences) and afterward cross-linked to the membrane by UV. RNA on the membrane was stained with 0.02% methylene blue (in 0.3 m sodium acetate, pH 5.5). Hybridization was done overnight at 42 °C in 500 mm NaPO4 buffer, pH 7.2, 7% SDS, 1 mm EDTA using 5′-32P-labeled oligonucleotides with the following sequences: D/A2, 5′-GAC TCT CCA TCT CTT GTC TTC TTG-3′; E/C2, 5′-GGC CAG CAA TTT CAA GTT A-3′; 25 S, 5′-CTC CGC TTA TTG ATA TGC-3′; 18 S-5′, 5′-CAT GGC TTA ATC TTT GAG AC-3′; 18 S (006), 5′-GTA CTA GCG ACG GGC GGT GTG-3′; 18 S (007), 5′-GCG GTG TGT ACA AAG GGC AG-3′. The membranes were washed three times for 20 min at 42 °C in 40 mm NaPO4 buffer, pH 7.2, 1% SDS, and radioactivity was detected by exposing x-ray films. Membranes were regenerated by washing in 1% SDS.
Tandem Affinity Purification (TAP) and Mass Spectrometry
TAP purifications of TAP-tagged bait proteins were, unless otherwise indicated, performed in a buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2, and 0.075% Nonidet P-40 essentially as described previously (27). Tobacco etch virus (TEV) protease was preincubated with RiboLock RNase inhibitor (Fermentas), and for cleavage, dithiothreitol was added to a final concentration of 1 mm to the buffer. For analysis of the protein composition of the purified material, the eluates were trichloroacetic acid-precipitated and dissolved in SDS sample buffer. The samples were then separated on NuPAGE SDS 4–12% gradient polyacrylamide gels (Invitrogen) and stained with colloidal Coomassie (Sigma). Mass spectrometric identification of the proteins contained in Coomassie-stained bands was performed as described previously (31). RNA extractions from the eluates were performed as described above for sucrose gradient analysis.
Preparation of Total Yeast Protein Extracts and Western Analysis
Total yeast protein extracts were prepared as described previously (32). Western blot analysis was performed using the following antibodies: anti-Arc1 (1:20,000) (33) and secondary goat anti-rabbit horseradish peroxidase-conjugated antibody (1:7,500) (Bio-Rad); anti-FLAG monoclonal, peroxidase-conjugated (1:1,000; Sigma). For detection of Protein A-tagged proteins, the peroxidase anti-peroxidase complex antibody was used at a dilution of 1:10,000 (Dako). Proteins were visualized using enhanced chemiluminescence detection kits (Immobilon Western (Millipore) and Amersham Biosciences ECL (GE Healthcare)).
Nob1 Purification for Nuclease Assays
For the nuclease assays, ProtA-TEV-FLAG-Nob1 wild type and D15N were expressed in yeast from plasmids pADH111-ProtA-TEV-FLAG-Nob1 or pADH111-ProtA-TEV-FLAG-Nob1-D15N (ADH1 promoter). Because a nob1 D15N mutant is not viable (Fig. 9A), we overexpressed the protein in a strain that still contained the wild-type copy of the protein. Because we noticed that Nob1 forms multimers and wild-type Nob1 co-purified with the D15N mutant (data not shown), we used a His6-tagged wild-type copy that was under the control of a GAL promoter (pRS313 pGAL-His6-Nob1) when purifying the D15N mutant protein. Cells were grown to an A600 of 0.2 in synthetic medium containing 2% raffinose and 0.2% galactose to avoid overexpression of wild-type Nob1 from the GAL promoter. Subsequently, expression of the wild-type Nob1 protein was repressed by adding glucose to a final concentration of 2%. Cells were depleted of wild-type Nob1 for ∼20 h and harvested at an A600 of 2. Cells expressing the wild-type Nob1 protein were grown in glucose-containing synthetic medium and also harvested at an A600 of 2. Proteins were affinity-purified using a buffer containing 750 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1.5 mm MgCl2, and 0.075% Nonidet P-40. Briefly, cells were lysed by bead beating (Pulverissete 6, Fritsch) and Nob1 was bound to IgG-Sepharose (Amersham Biosciences) via the ProtA tag. IgG slurry was washed, and proteins were eluted by cleavage with the TEV-protease and incubated with anti-FLAG M2-agarose (Sigma). Anti-FLAG slurry was washed, and the bound material was eluted with FLAG peptide according to the manufacturer's instructions (Sigma). To remove residual His6-Nob1, the eluates were incubated with Ni2+-nitrilotriacetic acid-agarose (Qiagen) in the presence of 10 mm imidazole. The unbound material was concentrated, and the buffer was exchanged into (150 mm NaCl, 50 mm Tris-HCl, 0.5 mm dithiothreitol, 10% glycerol) on Ultrafree-0.5 centrifugal filter devices (10K NMWL, Millipore).
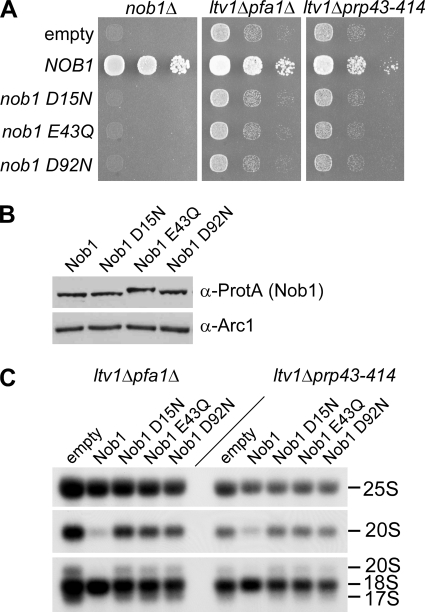
FIGURE 9.
Suppression of the rRNA processing defects in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants requires the endonuclease activity of Nob1. A, to test whether the nob1 mutant alleles are functional in vivo, plasmids expressing Nob1 mutants that were N-terminally tagged with ProtA-FLAG were transformed into a NOB1 shuffle strain, and growth was tested by spotting cells in 10-fold dilution steps onto plates containing 5-fluoroorotic acid (5-FOA) and incubated at 30 °C for 2 days. Growth indicates complementation of the nob1 deletion. To test for suppression, plasmids were transformed into the double mutants ltv1Δ pfa1Δ and ltv1Δ prp43–414, and transformants were spotted onto SDC−leu plates. Plates were incubated at 30 °C for 4 days. B, expression of the indicated Nob1 mutant proteins was tested by analyzing whole cell lysates from a NOB1 shuffle strain transformed with the respective plasmids and Western blotting against the ProtA tag. The Western blot using an antibody against Arc1 serves as a loading control. Note that the electrophoretic mobility of the mutant variants is slightly changed, which could be due to the removed negative charge. C, Northern blotting of ltv1Δ pfa1Δ and ltv1Δ prp43–414 strains transformed with plasmids carrying the indicated ProtA-FLAG-tagged Nob1 variants. Cells were grown at 30 °C to an A600 of 0.8. RNA was isolated and analyzed by Northern blotting using the 25 S probe to detect 25 S rRNA, the D/A2 probe to detect 20 S pre-rRNA, and the 18 S 5′ probe to detect 20, 18, and 17 S rRNAs.
In Vitro Nuclease Assays
Nuclease activity tests of wild-type or mutant (D15N) Nob1 proteins purified from yeast (see above) on 5′-radiolabeled RNAs were performed at 30 °C in 25 mm Tris/HCl (pH 7.6), 75 mm NaCl, 2 mm dithiothreitol, 100 μg/ml bovine serum albumin, 0.8 unit/μl RNasin, 4.5% glycerol, 0.05% Nonidet P-40, 0.3 μm Escherichia coli tRNA. Unless otherwise indicated, 5 mm MnCl2 was used. Prior to the addition of the labeled RNA substrate, the 10-μl reactions containing the protein were preincubated for 5 min at 30 °C and then subsequently (unless otherwise indicated) incubated for 60 min at 30 °C in the presence of the RNA. After a proteinase K digest for 30 min at 37 °C, the RNA was then extracted, precipitated, and resuspended in formamide RNA loading buffer. Reaction products were resolved on a denaturing 12% polyacrylamide, 8 m urea gel and visualized by autoradiography.
5′-End-labeled substrates were stem D-site WT or stem D-site “U17G” RNAs (see Fig. 7B; Dharmacon). 5′-End labeling and gel purification of the labeled RNAs was performed as described (34).
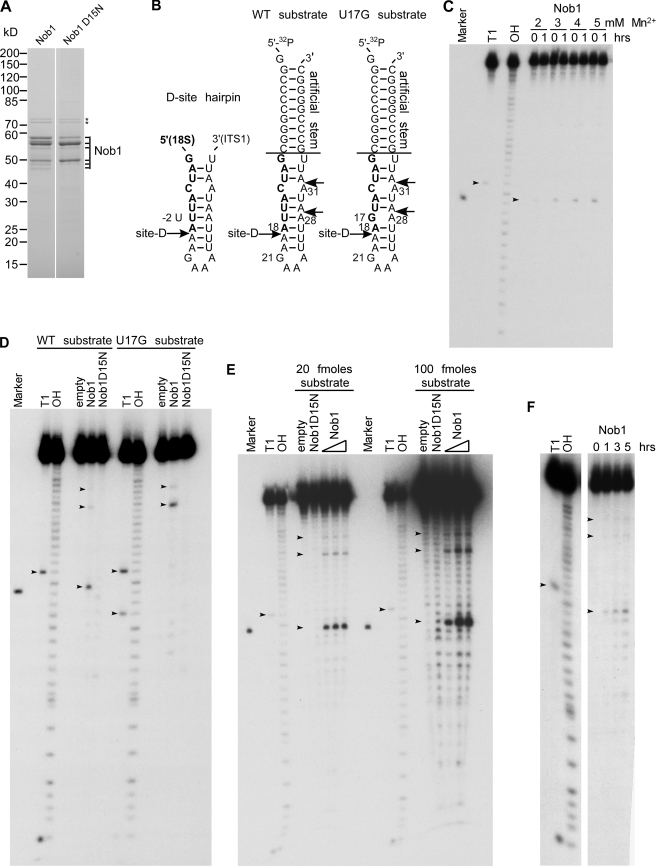
FIGURE 7.
Nob1 has D-site-specific endonuclease activity in vitro. A, purified wild-type Nob1 and mutant Nob1-D15N used in the in vitro nuclease assay. The proteins were affinity-purified from yeast and analyzed by SDS-PAGE and Coomassie staining. All bands labeled with Nob1 were identified by mass spectrometry. Hsp70 contaminants are indicated by stars. B, model of the cleavage site D within the 20 S pre-rRNA. Left sequence, authentic D-site hairpin ranging from 5′ (part of mature 18 S rRNA; shown in boldface type) to 3′ (part of ITS1, shown in normal type) (53); middle and right sequences, RNA model substrates with engineered wild-type (middle) and mutated cleavage site D (U17G mutation) (right). Both RNA substrates carry an additional stem sequence attached to the D-site hairpin. Alternative cleavage sites at positions A28 and A31 are indicated. C–F, Nob1 in vitro cleavage assays. Marker, 18-nucleotide RNA resembling the expected D-site cleavage product, ranging from nucleotide 1 to 18 of the used WT substrate. T1, substrate digested with T1 nuclease, which cleaves after single-stranded G residues (G1 and G21, as well as G17 in the U17G substrate). OH, partial alkaline hydrolysis of the RNA substrate. Due to the generation of 3′-phosphates upon hydrolysis by T1 nuclease or OH−, the products run slightly faster (∼1.5 nucleotides) than cleavage products with 3′-OH ends. 3′-OH ends are found in the marker and in cleavage products predicted to be generated by PIN endonucleases, as shown for the exosome subunit Rrp44 (14). Note that the cleavage product of Nob1 runs at a similar height as the A18 oligonucleotide, indicating that cleavage occurs after A18, corresponding to the D-site. Small variations in cleavage efficiencies in the assays shown are probably due to the use of different protein and/or RNA substrate preparations. C, manganese titration. 5′-[32P]-end-labeled RNA substrate was incubated with purified wild-type Nob1 at 30 °C for 0 or 1 h in the presence of increasing concentrations of manganese as indicated. ∼5 pmol of Nob1 and 10 fmol of substrate were used (500-fold molar excess). The arrows indicate the G21 cleavage product generated by T1 nuclease and the A18 cleavage product generated by Nob1. D, cleavage does not occur when either the catalytic site of Nob1 (Nob1-D15N) or the sequence of the substrate (U17G) is mutated. 10 fmol of 5′-32P-end-labeled RNA substrates (shown in B) were incubated without protein (empty) or with purified wild-type Nob1 or mutant Nob1-D15N (5 pmol each) at 30 °C for 1 h. The arrows indicate the G17 and G21 cleavage products generated by T1 nuclease and the A18, A28, and A31 cleavage products generated by Nob1. E, dependence of the cleavage reaction on enzyme and substrate concentration. 20 (left) and 100 (right) fmol of RNA substrate were incubated with 1.25, 2.5, or 5 pmol of Nob1 protein for 1 h at 30 °C. In the case of the Nob1-D15N protein, 5 pmol were used. The arrows indicate the G21 cleavage product generated by T1 nuclease and the A18, A28, and A31 cleavage products generated by Nob1. F, time course of Nob1 cleavage. 20 fmol of substrate were incubated with 10 pmol of Nob1 at 30 °C for the indicated time periods. The arrows indicate the G21 cleavage product generated by T1 nuclease and the A18, A28, and A31 cleavage products generated by Nob1. WT, wild type.
RESULTS
A Genetic Network between the Pre-40 S Factor Ltv1, the RNA Helicase Prp43, the G-patch Protein Pfa1, and the Nuclease Nob1
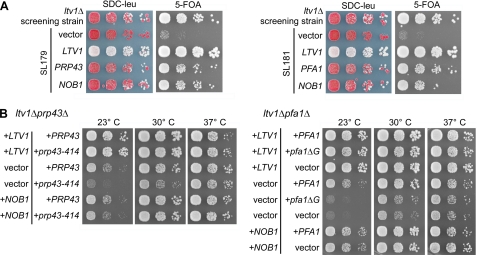
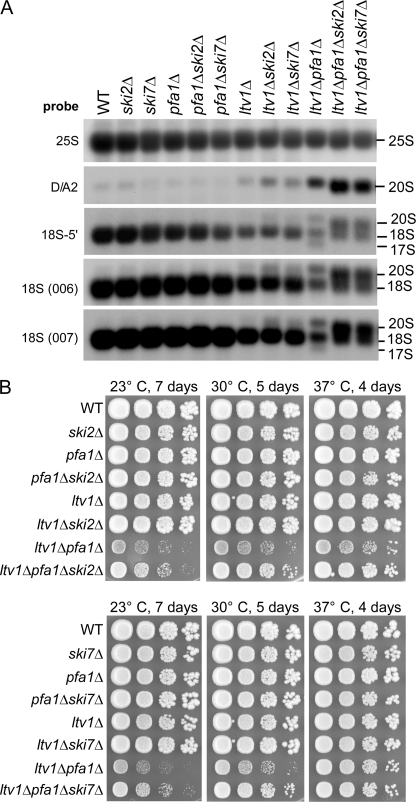
Ltv1 is a non-essential component of late pre-40 S particles that also contain Enp1, Hrr25, Nob1, Rio2, Tsr1, Dim1, and Dim2 (7). Within this particle, Ltv1 forms a stable complex with Enp1 and the ribosomal protein Rps3, which was previously proposed to be involved in 40 S beak formation (7). To identify pre-40 S factors that functionally interact with Ltv1, we performed a synthetic lethal screen with a strain harboring the ltv1 null allele. Haploid ltv1Δ cells are viable and exhibit only a slight growth impairment at 23 or 30 °C (Fig. 1). Our screen was performed at 30 °C, and eight mutants were identified that exhibited a synthetic enhanced phenotype in combination with deletion of LTV1.
FIGURE 1.
Genetic network between LTV1, PRP43, PFA1, and NOB1. A, identification of PRP43, PFA1, and NOB1 in a synthetic lethal screen with an ltv1 deletion strain. Mutants SL179 and SL181 isolated in the synthetic lethal (sl) screen were transformed with plasmids carrying the indicated genes and spotted in serial 10-fold dilution steps onto SDC−leu and SDC + 5-fluoroorotic acid (5-FOA) plates. As control, the non-mutagenized screening strain (ltv1Δ, pHT4467-CENΔ-LTV1) was spotted. Plates were incubated at 30 °C for 4 days. Red colony color on SDC−leu and slow growth on 5-fluoroorotic acid plates indicate a synthetic growth defect. Red/white sectoring on SDC−leu and growth on 5-fluoroorotic acid indicate complementation of the synthetic enhancement phenotype. B, temperature dependence of genetic interactions between LTV1 and PRP43 and between LTV1 and PFA1. The LTV1/PFA1 and LTV1/PRP43 shuffle strains were transformed with plasmids that carry the indicated wild-type and mutant alleles. pfa1ΔG is a PFA1 allele lacking the G-patch. After 5-fluoroorotic acid shuffling, cells were spotted in 10-fold serial dilution steps onto SDC−leu−trp plates to select for the transformed plasmids and incubated at 23, 30, and 37 °C for 4 days.
After transformation with a genomic library, we found that one of these mutants (SL179) was complemented by either NOB1 or PRP43, whereas the other seven mutants (including SL181) were complemented by either NOB1 or PFA1/SQS1 (Fig. 1A). DNA sequencing revealed that the NOB1 gene locus was not mutated in any of these synthetic enhanced mutants. SL179 carried a point mutation in PRP43 corresponding to a S414F amino acid exchange in the sequence between helicase motifs V and VI of the protein (hereafter referred to as prp43–414). The remaining seven synthetic enhanced strains carried mutations in the non-essential gene PFA1 (data not shown). In addition, synthetic growth inhibition was also observed between ltv1Δ and other prp43 mutant alleles mapping in several different motifs of the helicase fold (data not shown). Subsequently, the synthetic enhanced genetic interactions were verified by generating the corresponding double mutants, ltv1Δ prp43–414 and ltv1Δ pfa1Δ (Fig. 1B). Notably, the synthetic growth defects, which were very pronounced at 23 °C, were gradually reduced at higher temperatures (Fig. 1B, compare 23, 30, and 37 °C). As predicted from the genetic screen, the presence of the NOB1 gene on a centromeric plasmid was sufficient to suppress the synthetic enhanced growth phenotypes of both double mutant strains (Fig. 1B). Altogether, these data suggest that ltv1Δ exhibits a synthetic enhanced genetic interaction with PRP43 and PFA1 that is suppressed by increased dosage of Nob1.
The DEAH-box RNA helicase Prp43 is a characterized splicing factor (35, 36) and is also required for early steps in ribosome biogenesis (24, 37, 38) and for the release of small nucleolar RNAs from the pre-60 S ribosomes (39). Prp43 is associated with several pre-rRNAs, including 20 S (24, 37, 38), and binds the pre-rRNA at multiple sites, including 18 S helix 44, close to cleavage site D (39). In the splicing pathway, Prp43 forms a complex with the G-patch protein Ntr1/Spp382 that stimulates its helicase activity (40–42). The G-patch domain is an ∼50-amino acid-long motif that is found in several proteins linked to RNA processing (43). Two other G-patch proteins, Pxr1/Gno1 and Pfa1/Sqs1, were reported to interact with Prp43 (24). The interaction between Prp43 and Pfa1 is most likely direct, because both proteins can be co-purified from yeast in a salt-resistant complex that is stable even after treatment with RNase A (Fig. 2A) (data not shown). Moreover, the G-patch domain of Pfa1 is of functional importance, because expression of Pfa1 lacking this motif (Pfa1ΔG) did not complement the growth defect of the ltv1Δ pfa1Δ mutant (Fig. 1B). This truncated version of Pfa1 was expressed similarly to the full-length protein and was still able to interact with Prp43, suggesting that removal of the G-patch disturbs a function of Pfa1 other than interaction with Prp43 (Fig. 2B) (data not shown). By analogy to the helicase-activating function of Ntr1, our data suggest that the G-patch of Pfa1 might stimulate Prp43 helicase activity during a late step in 40 S subunit biogenesis.
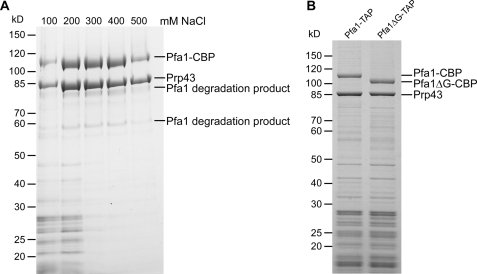
FIGURE 2.
The interaction between Prp43 and Pfa1 is independent of the G-patch and resistant to increased salt concentrations. A, Pfa1-TAP was purified from yeast cells using increasing salt concentrations, ranging from 100 to 500 mm NaCl. Samples were analyzed by SDS-PAGE and Coomassie staining. The indicated proteins were identified by mass spectrometry. Note that at higher salt concentrations, only Pfa1 (full-length and degradation products) and Prp43 were recovered, indicating that Pfa1 and Prp43 directly interact in vivo. Pfa1-CBP, Pfa1 with the C-terminal calmodulin-binding protein tag. B, Pfa1- and Pfa1ΔG-TAP purifications. Note that the C-terminally truncated version of Pfa1 co-purified amounts of Prp43 similar to those provided by the full-length protein. All bands appearing in only one of the two purifications could be identified by mass spectrometry as N-terminal degradation products of Pfa1 that differed in size due to the different C termini of the bait proteins.
Ltv1, Prp43, and Pfa1 Are Required for 20 to 18 S rRNA Processing in the Cytoplasm
To investigate whether Pfa1 and Prp43, in conjunction with Ltv1, control the late 20 to 18 S rRNA processing step in the cytoplasm, we performed Northern analyses using the probes depicted in Fig. 3A. Robust accumulation of 20 S pre-rRNA and concomitant reduction in mature 18 S rRNA was observed in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants, whereas the single mutants were unaffected (pfa1Δ and prp43–414) or only slightly impaired (ltv1Δ) in this processing step (Fig. 3B). Unexpectedly, although the synergistic growth defects of the double mutants were less obvious at 37 °C (see Fig. 1B), 20 S pre-rRNA accumulation was even stronger at 37 °C than at 23 °C (Fig. 3B; see also “Discussion”).
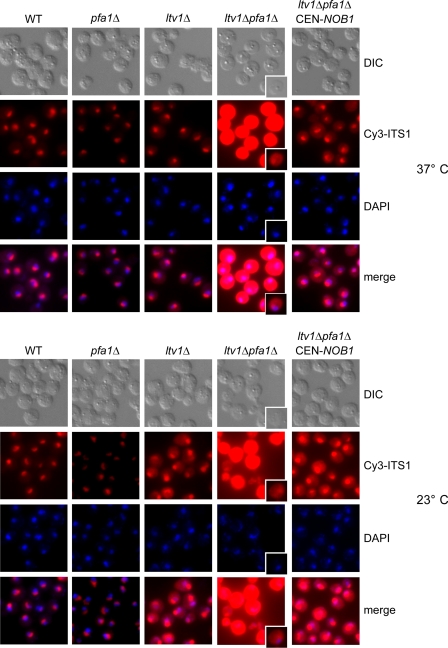
To determine where 20 S pre-rRNA accumulated in the double mutants, we performed in situ hybridization using a fluorescently labeled oligonucleotide complementary to a sequence between cleavage sites D and A2 in ITS1. This probe detects the 20 S pre-rRNA and larger precursors but not mature 18 S rRNA (Fig. 3A). Consistent with the Northern data, the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants, but not the single mutants, massively accumulated pre-rRNAs harboring ITS1 in the cytoplasm at both 37 and 23 °C (Fig. 4 and supplemental Fig. S1). Shorter exposure also revealed some ITS1 accumulation in the nucleolus (Fig. 4). This could, however, be due to increased levels of 35 S and 32/33 S pre-rRNAs in these mutants, which were clearly visible in the Northern analysis (Fig. 3B). Notably, the strong cytoplasmic ITS1 signal and the accumulation of 20 S pre-rRNA and an abnormal 17 S RNA species (see below) could be reverted by increasing the dosage of Nob1 (Figs. 4 and 9C). Together, these studies demonstrate that combined mutations in LTV1 and PFA1 or LTV1 and PRP43 cause a robust inhibition of cytoplasmic 20 to 18 S rRNA processing, which can be suppressed by overexpression of Nob1.
FIGURE 4.
20 S pre-rRNA accumulates in the cytoplasm in ltv1Δ pfa1Δ mutants. ltv1Δ pfa1Δ cells transformed with empty plasmids or plasmids carrying LTV1, PFA1, or NOB1 wild-type alleles were either grown at 37 °C to an A600 of 0.7–1 (37 °C) or grown to an A600 of 0.3, and shifted to 23 °C for 3 h (23 °C). Cells were fixed with formaldehyde, spheroplasted, and subjected to fluorescence in situ hybridization using a Cy3-labeled probe complementary to a sequence in the D/A2 segment of ITS1. To visualize the nucleoplasm, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI). Due to the strong signal in the ltv1Δ pfa1Δ cells, also single cells with a shorter exposure of the Cy3 signal are shown. WT, wild type.
An Abnormal 17 S RNA Product Generated by the Exosome Is Formed in ltv1Δ pfa1Δ and ltv1Δ prp43–414 Double Mutants
In addition to the 20 S pre-rRNA, we observed the formation of a 17 S RNA species in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants (Figs. 3B and 6 and supplemental Fig. S2). As shown by using specific Northern probes (see Figs. 3A and 10A), this abnormal 17 S RNA species is a 3′-truncated form of 18 S rRNA that was already described in rps14 and fap7 mutants (9, 44). Notably, the 17 S RNA accumulated more strongly at 23 °C than at 37 °C in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants (Fig. 3B).
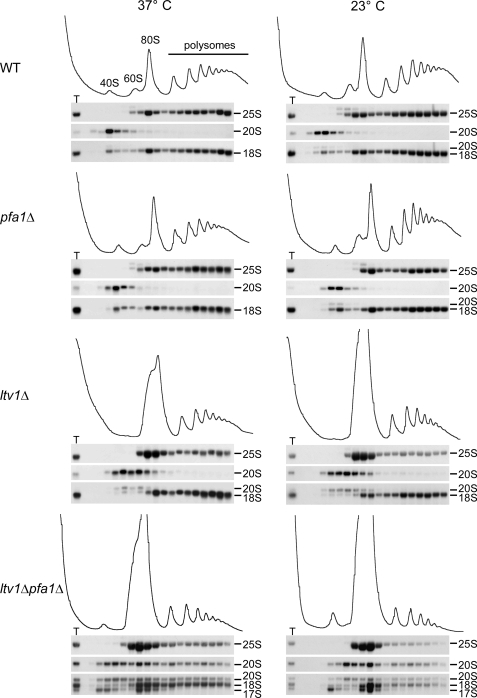
FIGURE 6.
17 S RNA co-sediments only with 40 and 80 S, whereas 20 S pre-rRNA is also found in polysomes. Cells were either grown at 37 °C to an A600 of 0.7–1 (37 °C) or grown to an A600 of 0.1, shifted to 23 °C, and incubated to an A600 of 0.7–1 (23 °C). Cell extracts were loaded onto 7–50% sucrose gradients and fractionated. RNA was extracted from each fraction and analyzed by Northern blotting using the probes 25 S, D/A2 (to detect 20 S pre-rRNA), and 5′-18 S (to detect 20, 18, and 17 S rRNAs). T, RNA preparation of the total extract. WT, wild type.
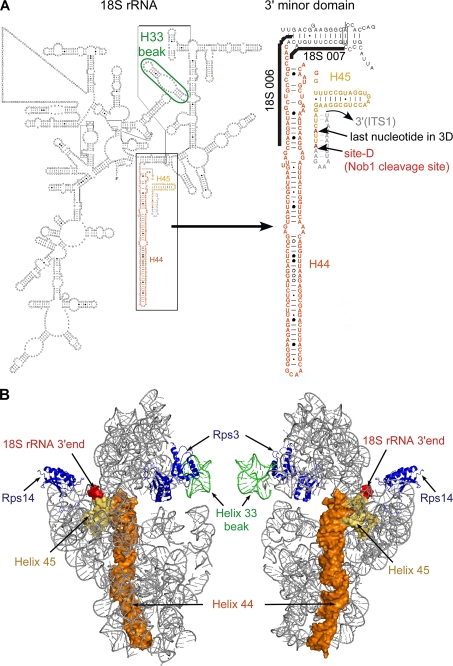
FIGURE 10.
Structure of the 3′-end of the 18 S rRNA. A, left, modeled secondary structure of the 18 S rRNA according to Refs. 54 and 55). Helix 33 is encircled in green. The box indicates the 3′ minor domain, consisting of helices 44 (orange) and 45 (yellow). Right, enlarged view of the 3′ minor domain. Binding sites of oligonucleotides 18 S 006 and 007 are indicated. The last nucleotide in the three-dimensional model shown in B and the actual 3′-end of the 18 S rRNA are displayed in red. The sequence of ITS1 within the predicted D-site hairpin (53) was added manually (gray). B, modeled structure of the yeast 40 S subunit, shown from the solvent (left) and intersubunit (right) sides. Helices 44 (orange) and 45 (yellow) corresponding to the last ∼160 nucleotides at the 3′-end of the 18 S rRNA are shown as a surface representation. The 3′-end of the 18 S rRNA is displayed in red, and helix 33 is shown in green. Of the ribosomal proteins, only Rps3 and Rps14 are shown (blue). The displayed images were generated in PyMOL (DeLano Scientific).
To determine whether the 17 S RNA was generated by endonuclease or exonuclease activity in the double mutants, we first searched for a hypothetical rRNA endonuclease cleavage product extending from the 3′-end of the 17 S RNA to cleavage site A2 by Northern analysis. However, we could not detect such a fragment even in a strain lacking the 5′–3′ exonuclease XrnI, which is responsible for degradation of the D/A2 spacer fragment generated by 20 S cleavage (data not shown) (45). We next tested whether the 3′ to 5′ exonuclease activity of the cytoplasmic exosome is involved in generating the 17 S RNA. Loss of cofactors for the cytoplasmic exosome Ski2 or Ski7 (46, 47) from the ltv1Δ pfa1Δ strain abolished formation of 17 S RNA, whereas levels of 20 S pre-rRNA were significantly increased (Fig. 5A). This suggests that the 17 S RNA was predominantly derived from 20 S pre-rRNA, although it remains possible that some 17 S RNA can be generated from mature 18 S rRNA. Ski2, Ski7, and the cytoplasmic exosome participate in several mRNA degradation pathways but were not previously reported to process or degrade rRNAs (reviewed in Ref. 48).
FIGURE 5.
17 S RNA is generated via an 3′–5′ exonucleolytic attack by the exosome. A, the abnormal 17 S RNA species is not formed when SKI2 or SKI7 is deleted. Cells were grown at 30 °C to an A600 of 0.8. RNA was isolated and analyzed by Northern blotting using the indicated probes. B, ltv1Δ pfa1Δ mutants grow faster when also SKI2 or SKI7 is deleted. Growth of ski2Δ (top) and ski7Δ (bottom) mutants is shown. Cells were spotted in 10-fold serial dilution steps onto YPD plates and incubated at the indicated temperatures for the indicated time periods. WT, wild type.
Analysis of the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants by sucrose density gradient centrifugation showed reduced levels of polysomes and 40 S subunits, whereas 60 and 80 S peaks were increased (Fig. 6 and supplemental Fig. S2). Subsequent Northern analysis of these fractions showed that 20, 18, and 17 S rRNAs were prominently enriched in the area corresponding to the 80 S peak. Notably, 20 S pre-rRNA was not only found in the 40 and 80 S peak but also in the polysomal fractions, implying that it may be present in functional polysomes. In contrast, 17 S RNA was not detected in the polysomal fractions, suggesting that 40 S subunits containing 17 S RNA are non-functional. Consistent with this hypothesis, the abundance of free 60 S subunits was reduced in an ltv1Δ pfa1Δ ski2Δ triple mutant as compared with the ltv1Δ pfa1Δ strain, suggesting that more 40 S subunits are capable of subunit joining when 17 S RNA formation is prevented (supplemental Fig. S3). Furthermore, ltv1Δ pfa1Δ mutants grew faster when SKI2 or SKI7 were also deleted, showing that the 17 S RNA accumulation and therefore the presence of aberrant 40 S subunits accounts for part of the growth defect observed in ltv1Δ pfa1Δ double mutants (Fig. 5B; also see “Discussion”).
Nob1 Has D-site-specific Endonuclease Activity in Vitro
Increased dosage of Nob1 suppressed both the slow growth phenotype (Fig. 1) and the 20 S pre-rRNA accumulation (Figs. 4 and 9C and supplemental Fig. S1) of the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants. This suggested that Nob1 participated directly in pre-rRNA cleavage at site D. Hence, we tested whether Nob1 exhibits in vitro site D cleavage activity.
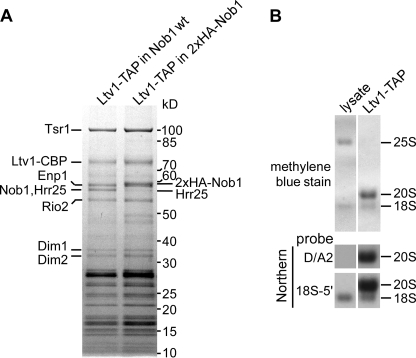
To obtain purified Nob1, we first tested recombinant expression in E. coli but failed to obtain pure and soluble protein. We then overexpressed ProtA-FLAG-tagged Nob1 in yeast and affinity-purified it under stringent, high salt conditions (Fig. 7A). As a control, we also affinity-purified a mutant Nob1-D15N protein, in which the catalytic nuclease activity of the PIN domain was predicted to be destroyed (10, 11). Since the Nob1-D15N mutant on its own does not support growth (Fig. 9A), it was expressed in a strain in which the endogenous NOB1 gene was under GAL regulation (for details regarding the purification procedure, see “Experimental Procedures”). When analyzed by SDS-PAGE and Coomassie staining, purified wild-type and mutant Nob1 proteins exhibited several bands. The major bands between 50 and 60 kDa were identified by mass spectrometry to be Nob1. Although we cannot fully exclude the possibility that the larger versions are modified forms of Nob1, we consider it unlikely because similar gel migration was observed with protein purified from E. coli (data not shown). In addition, mass spectrometric analysis suggested that the band sedimenting at 50 kDa was most likely a C-terminally truncated version of Nob1 (data not shown). For these reasons, we assume that the uppermost band (close to the 60 kDa band of the protein marker) corresponds to full-length Nob1 (with a predicted molecular mass of ∼52 kDa) that exhibits a slightly abnormal running behavior, whereas the faster migrating bands are most likely C-terminally truncated versions. The two bands above 70 kDa were identified as Hsp70 contaminants (Fig. 7A).
As an in vitro substrate for Nob1, we synthesized an RNA oligonucleotide comprising the RNA sequence surrounding cleavage site D (expected cleavage between A18 and A19). This was embedded in an artificial stem (Fig. 7B) to prevent 3′–5′ and 5′–3′ exonuclease degradation of the substrate and facilitate measurement of endonuclease activity. In addition, we synthesized the same RNA with a U17G point mutation at position −2 with respect to the predicted cleavage site (Fig. 7B). This mutation was previously shown to inhibit 20 S pre-rRNA processing in vivo (49).
Incubation of the 5′-radiolabeled RNA substrate with purified wild-type Nob1 in the presence of manganese resulted in one main product generated by cleavage between A18 and A19, the precise location of site D in vivo (Fig. 7, B and C). Longer exposure revealed two additional cleavage products, corresponding to cleavage at A28 and A31 (Fig. 7, B, D, E, and F). Cleavage efficiency improved with increasing concentrations of manganese, with an optimal efficiency at 5 mm (Fig. 7C), whereas no cleavage was observed in the presence of magnesium (data not shown). The requirement for manganese is consistent with observations from other PIN domain proteins, which also need manganese for cleavage activity and show inhibition by (13–18) magnesium.
Importantly, no cleavage occurred when the same amount of purified Nob1-D15N mutant protein was used in the in vitro assay (Fig. 7D). Moreover, cleavage at the authentic site D was abolished when wild-type Nob1 was tested with the RNA substrate containing the U17G mutation. Instead, Nob1 cleaved at the alternative sites A28 and A31. Interestingly, the sequence around A28 (UUAA) resembles that of the bona fide site D (Fig. 7, B and D). Cleavage at A28 was much stronger in the U17G substrate than in the wild-type substrate, suggesting that the altered structure at A28 (e.g. loss of the U17-A28 base pairing) allows more efficient cleavage at this site. Weak cleavage at A31 was also visible with the U17G substrate. An explanation for the lower cleavage efficiency could be the different sequence before the cleavage site (AUAA, compared with UUAA at site D).
The yield of site D cleavage product was improved by increasing the amount of substrate or protein used (Fig. 7E), or extending the incubation time (Fig. 7F). However, saturation of the reaction was observed at elevated protein concentrations, suggesting that either the amount of correctly folded RNA substrate is limiting or that the RNA substrate is fully bound by Nob1, but only a small fraction of the protein is cleavage-competent (Fig. 7E).
Together, these data show that Nob1 purified from yeast exhibits endonuclease activity in vitro and cleaves a model substrate at the cleavage site D, demonstrating that Nob1 is indeed the D-site endonuclease.
To test if in vivo Nob1 has exerted its cleavage activity on 20 S pre-rRNA in late pre-40 S particles, with which it is associated, we analyzed the RNA content of the late pre-40 S particle affinity-purified via Ltv1-TAP (7). This nascent 40 S subunit contained Nob1 and 20 S pre-rRNA with little mature 18 S rRNA (Fig. 8, A and B). These data imply that, although Nob1 was present in the purified pre-40 S particle, it had not significantly cleaved the 20 S pre-rRNA either in vivo or during purification. This observation suggests that Nob1 endonuclease activity is under regulation in the pre-40 S particle (see “Discussion”).
FIGURE 8.
pre-40 S particles purified via Ltv1-TAP contain Nob1 and unprocessed 20 S pre-rRNA. A, Nob1 is stably associated with the pre-40 S particle affinity-purified via Ltv1-TAP. The Ltv1-TAP preparation was analyzed by SDS-PAGE and Coomassie staining. Bands identified by mass spectrometry are indicated. Nob1 and Hrr25 co-migrate as one band. To estimate the proportion of Nob1 in this band, we performed another purification where the size of Nob1 was increased by fusion to an N-terminal hemagglutinin tag. B, the main RNA species in the Ltv1-TAP purification is 20 S pre-rRNA. RNA was extracted from the cell lysate and the pre-40 S subunit purified via Ltv1-TAP, separated on an agarose gel and transferred to a nylon membrane that was stained with methylene blue. The 20 and 18 S rRNAs were detected by Northern blotting with specific probes (see Fig. 3A).
Suppression of the 20 S Pre-rRNA Processing Defect in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 Mutants Requires the Endonuclease Activity of Nob1
To test whether the endonuclease activity of Nob1 is required for suppression of the phenotypes of ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants, we tested catalytically inactive Nob1-D15N. As anticipated, Nob1-D15N failed to suppress growth inhibition in the double mutants, although expression levels of mutant and wild-type Nob1 were similar (Fig. 9, A and B). Even when Nob1-D15N was overexpressed (either from the ADH1 or NOP1 promoter), no suppression was observed (data not shown). Moreover, expression of Nob1-E43Q or Nob1-D92N, which have mutations in other critical PIN domain residues (supplemental Fig. S4) (11) could neither complement the nob1Δ mutant nor suppress the growth defect of ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants (Fig. 9, A and B). Furthermore, an increased dosage of wild-type Nob1 but not of D15N, E43Q, or D92N mutant protein prevented accumulation of 20 S pre-rRNA and 17 S RNA in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants (Fig. 9C). These in vivo data demonstrate that the catalytic activity of Nob1 is essential for suppressing the 20 S pre-rRNA processing defect in ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants.
DISCUSSION
In this study, we identified a functional network linking the pre-40 S component Ltv1, the RNA helicase Prp43, the G-patch protein Pfa1, and the PIN domain protein Nob1. These function together in the processing of 20 S pre-rRNA to 18 S rRNA, which takes place in late, cytoplasmic pre-40 S subunits.
Up to now, the identity of the endonuclease catalyzing cytoplasmic D-site cleavage has been subject to speculation. Here, we developed an in vitro assay that faithfully reconstitutes Nob1-dependent endonuclease cleavage of a model substrate that mimics both the RNA sequence and the predicted secondary structure around cleavage site D of the 20 S pre-rRNA. Cleavage by Nob1 was specific because it mainly occurred at the predicted nucleotide, and cleavage at this model D-site was abolished by a single point mutation in the RNA substrate. The efficiency of cleavage was relatively low, perhaps reflecting the absence of auxiliary factors and/or the context of the preribosome in the in vitro assay system. Consistent with both possibilities, isolated nascent pre-40 S subunits purified via Ltv1-TAP contain high amounts of unprocessed 20 S pre-rRNA, despite the presence of Nob1 (see Fig. 8, A and B). Thus, D-site cleavage by Nob1 may only be allowed to occur at a defined step late during the 40 S maturation pathway. Regulation of cleavage could be achieved either by directly controlling the activity of Nob1, by restricting the access of Nob1 to the substrate, or by controlling the conformation of the substrate. In all cases, additional factors would be required to accomplish such a regulation.
Our genetic screen isolated factors that are required for 20 S processing in addition to the Nob1 endonuclease. Mutations in PRP43 and deletions of PFA1 resulted in a dramatic accumulation of 20 S pre-rRNA, when combined with the LTV1 deletion, indicating that Prp43, Pfa1, and Ltv1 are crucial for 20 to 18 S rRNA processing.
Despite the strong 20 S accumulation, the mutants were only slightly delayed in growth at higher temperatures, showing that high concentrations of 20 S pre-rRNA are not necessarily inhibitory to cell growth. This was supported by the co-sedimentation of 20 S pre-rRNA with polysomes in ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants, suggesting that 20 S pre-rRNA containing 40 S subunits can be tolerated in translating polysomes. This is in contrast to the wild-type strain and the prp43–414, pfa1Δ, and ltv1Δ strains, indicating that accumulated pre-40 S particles in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants are structurally different from the wild type or single mutants. Notably, co-sedimentation of 20 S pre-rRNA with polysomes was also described recently for an rps31 mutant (50). Based on these observations, it is conceivable that 20 to 18 S rRNA processing might occur after 40 S-60 S subunit joining or even after the first round of translation. In Dictyostelium, only 40 S subunits that are capable of translation undergo the final rRNA processing step, providing a precedent (51).
Apart from the 20 S pre-rRNA, ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants also accumulated 17 S RNA, a truncated version of the 18 S rRNA lacking ∼160 nucleotides of the 3′-end (corresponding to rRNA helices 44 and 45 of the 18 S rRNA; see also Fig. 10, A and B), that is generated by the cytoplasmic exosome. This suggests that in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants, the 3′ region of the 20 S pre-rRNA, including helices 44 and 45 of the 18 S rRNA, is not protected from exonuclease attack. This further supports the idea that a structural difference may exist between the incompletely matured 40 S particles in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants and wild-type pre-40 S and mature 40 S subunits.
Notably, helix 44 contributes to the decoding site of 40 S subunits, which is critical for translational fidelity (52). An aberrant structure in this region could be deleterious to accurate translation, especially at low temperatures, when the rRNA is probably less flexible. Indeed, the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants contained fewer polysomes and more 80 S monosomes at 23 than at 37 °C, suggesting that translation in the mutants is particularly impaired at lower temperatures (see also the distribution of 20 and 18 S rRNAs in the gradients; Fig. 6 and supplemental Fig. S2). Consistent with impaired translation, the ltv1Δ and ltv1Δ pfa1Δ strains showed increased sensitivity toward neomycin and paromomycin, both aminoglycosides known to bind to the upper part of helix 44.5 The combination of inefficient translation with the increased accumulation of non-functional 17 S RNA provides an explanation for the stronger growth defects of the double mutants at low temperatures.
Our results show that Ltv1, Pfa1, and Prp43 are involved in 20 S processing, but in what way do they promote the endonucleolytic cleavage by Nob1? Since the rRNA processing defects in ltv1Δ pfa1Δ and ltv1Δ prp43–414 mutants could be rescued by overexpression of the Nob1 endonuclease, the conditions for Nob1 RNA-substrate interaction or endonuclease activity are probably suboptimal in the ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants. These can, however, be compensated for by higher amounts of Nob1. This finding suggests that the correct functioning of Ltv1, Pfa1, and Prp43 is a prerequisite for efficient D-site cleavage by Nob1.
There is no evidence for a direct interaction between Nob1 and either Pfa1, Prp43, or Ltv1, making it unlikely that these proteins function as recruiting factors or allosteric activators of Nob1. We consider it more likely that Ltv1, Pfa1, and Prp43 regulate access of Nob1 to its substrate or prepare the substrate for efficient cleavage. In principal, this reorganization could take place in the nucleus. However, the finding that Ltv1, Pfa1, and Prp43 co-purify pre-40 S particles containing both 20 S pre-rRNA and 18 S rRNA makes it probable that they act in the cytoplasm, shortly before and during D-site cleavage (Fig. 8) (24).
A physical barrier preventing access of Nob1 to the cleavage site could be composed of either RNA or protein. A well characterized structural maturation step involving rRNA is exposure of helix 33 to generate the characteristic beak structure of the mature 40 S subunit. In the late pre-40 S particle, Ltv1 forms a complex with Enp1 and the ribosomal protein Rps3, which is implicated in this beak formation (7). In the modeled structure of the mature 40 S subunit, Rps3 is located close to the beak, suggesting that this is also likely to be the area where Ltv1 is bound (Fig. 10B). The position of helix 33 in the pre-40 S particle is not known; it is, however, possible that before being exposed to form the beak structure, this RNA is folded back on the particle and thereby hinders access of Nob1 to the cleavage site. The beak is formed concomitantly to or as a consequence of dissociation of Enp1 and Ltv1 from the particle (7). We speculate that this structural reorganization, which might be delayed in ltv1 mutants, could be a prerequisite for 20 S cleavage by Nob1. Considering the major conformational rearrangement of rRNA that has to occur during beak formation, it is possible that also an RNA helicase is required for this process. In this case, Pfa1 and Prp43 might contribute together with Ltv1 to formation of the beak structure and thereby influence 20 S processing.
Our observation that in ltv1Δ pfa1Δ mutants, the 3′-end of the 18 S rRNA (corresponding to helices 44 and 45) is accessible to the exosome makes it, however, more likely that this part of the rRNA is the direct target of Pfa1-Prp43. The action of these two proteins might be required to generate a stably structured, nuclease-resistant 3′-end of the 18 S rRNA. This idea is strongly supported by cross-linking experiments, which show that Prp43 binds to the 18 S rRNA at the base of helix 44, very close to cleavage site D (39). We speculate that the helicase activity of the Pfa1-Prp43 heterodimer acts to remodel the 18 S rRNA around helices 44 and/or 45 in order to prepare the RNA for cleavage by Nob1. Such a remodeling could be achieved by directly restructuring the rRNA but could also include the release of proteins bound in this region that might otherwise block access of Nob1 to the cleavage site.
We propose a model in which cytoplasmic 20 to 18 S rRNA processing is promoted by a number of interdependent steps: (i) beak formation and the respective structural rearrangement of the pre-40 S subunit, (ii) local restructuring of RNA helix 44 and/or 45 by Prp43-Pfa1 to allow access to cleavage site D, and (iii) cleavage at site D by the endonuclease Nob1. The combination of in vitro endonuclease assays with genetic analyses should allow the dissection of late steps in rRNA processing and their coordination with structural rearrangements.
Acknowledgments
We thank Dieter Kressler and Beate Schwer for providing plasmids. We also acknowledge the technical assistance of P. Ihrig and J. Reichert under the supervision of J. Lechner (Mass Spectrometry Unit, BZH Heidelberg). We thank D. Roser, D. Kressler, S. Ferreira-Cerca, A. Köhler, M. Skruzny, and H. Bergler for fruitful discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
B. Pertschy, unpublished results.
- PIN
- PilT N terminus
- MOPS
- 4-morpholinepropanesulfonic acid
- ProtA
- protein A
- SDC
- synthetic dextrose complete
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus.
REFERENCES
- 1.Fatica A., Tollervey D. (2002) Curr. Opin. Cell Biol. 14, 313–318 [DOI] [PubMed] [Google Scholar]
- 2.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 3.Granneman S., Baserga S. J. (2004) Exp. Cell Res. 296, 43–50 [DOI] [PubMed] [Google Scholar]
- 4.Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) Cell Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tschochner H., Hurt E. (2003) Trends Cell Biol. 13, 255–263 [DOI] [PubMed] [Google Scholar]
- 6.Seiser R. M., Sundberg A. E., Wollam B. J., Zobel-Thropp P., Baldwin K., Spector M. D., Lycan D. E. (2006) Genetics 174, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. (2006) Nature 441, 651–655 [DOI] [PubMed] [Google Scholar]
- 8.Venema J., Tollervey D. (1999) Annu. Rev. Genet. 33, 261–311 [DOI] [PubMed] [Google Scholar]
- 9.Granneman S., Nandineni M. R., Baserga S. J. (2005) Mol. Cell. Biol. 25, 10352–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica A., Oeffinger M., Dlakiæ M., Tollervey D. (2003) Mol. Cell. Biol. 23, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A., Tollervey D., Dlakiæ M. (2004) RNA 10, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003) EMBO J. 22, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider C., Leung E., Brown J., Tollervey D. (2009) Nucleic Acids Res. 37, 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., Sanchez-Rotunno M., Arraiano C. M., van Hoof A. (2009) Nat. Struct. Mol. Biol. 16, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skruzný M., Schneider C., Rácz A., Weng J., Tollervey D., Hurt E. (2009) PLoS Biol. 7, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebreton A., Tomecki R., Dziembowski A., Séraphin B. (2008) Nature 456, 993–996 [DOI] [PubMed] [Google Scholar]
- 17.Eberle A. B., Lykke-Andersen S., Mühlemann O., Jensen T. H. (2009) Nat. Struct Mol. Biol. 16, 49–55 [DOI] [PubMed] [Google Scholar]
- 18.Huntzinger E., Kashima I., Fauser M., Saulière J., Izaurralde E. (2008) RNA 14, 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleichert F., Baserga S. J. (2007) Mol. Cell 27, 339–352 [DOI] [PubMed] [Google Scholar]
- 20.Bohnsack M. T., Kos M., Tollervey D. (2008) EMBO Rep. 9, 1230–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Cruz J., Kressler D., Tollervey D., Linder P. (1998) EMBO J. 17, 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kos M., Tollervey D. (2005) Mol. Cell 20, 53–64 [DOI] [PubMed] [Google Scholar]
- 23.Weaver P. L., Sun C., Chang T. H. (1997) Mol. Cell. Biol. 17, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebaron S., Froment C., Fromont-Racine M., Rain J. C., Monsarrat B., Caizergues-Ferrer M., Henry Y. (2005) Mol. Cell. Biol. 25, 9269–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 26.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 27.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 28.Kressler D., Doère M., Rojo M., Linder P. (1999) Mol. Cell. Biol. 19, 8633–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosshans H., Hurt E., Simos G. (2000) Genes Dev. 14, 830–840 [PMC free article] [PubMed] [Google Scholar]
- 30.Kressler D., de la Cruz J., Rojo M., Linder P. (1997) Mol. Cell. Biol. 17, 7283–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. (2001) Mol. Cell 8, 517–529 [DOI] [PubMed] [Google Scholar]
- 32.Yaffe M. P., Schatz G. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simos G., Segref A., Fasiolo F., Hellmuth K., Shevchenko A., Mann M., Hurt E. C. (1996) EMBO J. 15, 5437–5448 [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell P., Petfalski E., Tollervey D. (1996) Genes Dev. 10, 502–513 [DOI] [PubMed] [Google Scholar]
- 35.Arenas J. E., Abelson J. N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin A., Schneider S., Schwer B. (2002) J. Biol. Chem. 277, 17743–17750 [DOI] [PubMed] [Google Scholar]
- 37.Combs D. J., Nagel R. J., Ares M., Jr., Stevens S. W. (2006) Mol. Cell. Biol. 26, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leeds N. B., Small E. C., Hiley S. L., Hughes T. R., Staley J. P. (2006) Mol. Cell. Biol. 26, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohnsack M. T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D. (2009) Mol. Cell., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon K. L., Auchynnikava T., Edwalds-Gilbert G., Barrass J. D., Droop A. P., Dez C., Beggs J. D. (2006) Mol. Cell. Biol. 26, 6016–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka N., Aronova A., Schwer B. (2007) Genes Dev. 21, 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai R. T., Tseng C. K., Lee P. J., Chen H. C., Fu R. H., Chang K. J., Yeh F. L., Cheng S. C. (2007) Mol. Cell. Biol. 27, 8027–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aravind L., Koonin E. V. (1999) Trends Biochem. Sci. 24, 342–344 [DOI] [PubMed] [Google Scholar]
- 44.Jakovljevic J., de Mayolo P. A., Miles T. D., Nguyen T. M., Léger-Silvestre I., Gas N., Woolford J. L., Jr. (2004) Mol. Cell. 14, 331–342 [DOI] [PubMed] [Google Scholar]
- 45.Stevens A., Hsu C. L., Isham K. R., Larimer F. W. (1991) J. Bacteriol. 173, 7024–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson J. S., Parker R. P. (1998) EMBO J. 17, 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T. (2001) EMBO J. 20, 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houseley J., Tollervey D. (2009) Cell 136, 763–776 [DOI] [PubMed] [Google Scholar]
- 49.van Beekvelt C. A., Jeeninga R. E., van't Riet J., Venema J., Raué H. A. (2001) RNA 7, 896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lacombe T., García-Gómez J. J., de la Cruz J., Roser D., Hurt E., Linder P., Kressler D. (2009) Mol. Microbiol. 72, 69–84 [DOI] [PubMed] [Google Scholar]
- 51.Mangiarotti G., Chiaberge S., Bulfone S. (1997) J. Biol. Chem. 272, 27818–27822 [DOI] [PubMed] [Google Scholar]
- 52.Ogle J. M., Brodersen D. E., Clemons W. M., Jr., Tarry M. J., Carter A. P., Ramakrishnan V. (2001) Science 292, 897–902 [DOI] [PubMed] [Google Scholar]
- 53.Yeh L. C., Thweatt R., Lee J. C. (1990) Biochemistry 29, 5911–5918 [DOI] [PubMed] [Google Scholar]
- 54.Spahn C. M., Beckmann R., Eswar N., Penczek P. A., Sali A., Blobel G., Frank J. (2001) Cell 107, 373–386 [DOI] [PubMed] [Google Scholar]
- 55.Cannone J. J., Subramanian S., Schnare M. N., Collett J. R., D'Souza L. M., Du Y., Feng B., Lin N., Madabusi L. V., Müller K. M., Pande N., Shang Z., Yu N., Gutell R. R. (2002) BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]