Abstract
The actin-binding protein filamin links membrane receptors to the underlying cytoskeleton. The cytoplasmic domains of these membrane receptors have been shown to bind to various filamin immunoglobulin repeats. Notably, among 24 human filamin repeats, repeat 17 was reported to specifically bind to platelet receptor glycoprotein Ibα and repeat 21 to integrins. However, a complete sequence alignment of all 24 human filamin repeats reveals that repeats 17 and 21 actually belong to a distinct filamin repeat subgroup (containing repeats 4, 9, 12, 17, 19, 21, and 23) that shares a conserved ligand-binding site. Using isothermal calorimetry and NMR analyses, we show that all repeats in this subgroup can actually bind glycoprotein Ibα, integrins, and a cytoskeleton regulator migfilin in similar manners. These data provide a new view on the ligand specificity of the filamin repeats. They also suggest a multiple ligand binding mechanism where similar repeats within a filamin monomer may promote receptor clustering or receptor cross-talking for regulation of the cytoskeleton organization and diverse filamin-mediated cellular activities.
Introduction
Human filamin (FLN)2 was discovered decades ago and is now known to consist of three isoforms, FLNa, FLNb, and FLNc (1–3). FLNa and FLNb are ubiquitously expressed, whereas FLNc is present predominantly in cardiac and muscle tissue. All three filamins are composed of two actin-binding calponin homology domains at their N termini followed by 24 Ig-like repeats of ∼95 amino acids. The last repeat 24 dimerizes so the full-length filamin is thought to exist as an extended dimer held together at its C terminus. The 24 repeats are interrupted by two hinge regions: one between repeats 15 and 16 and another between repeats 23 and 24. The regions 1–15 and 16–24 are referred to often as rod 1 and rod 2, respectively. A vast array of proteins including multiple membrane receptors has been found to bind the rod 2 segment, but few have been found to bind to the rod 1 segment (1, 2, 4). Interestingly, rod 1, which follows the N-terminal actin-binding calponin homology domains, was recently found to have weak actin binding activity (5).
The prominent role of the C terminus (rod 2) of filamin in protein-protein interactions has led to the extensive structural characterizations of repeat-target complexes including repeats 17 (6), 21 (7–10), 23 (11), and 24 (12). These studies suggested that while maintaining very similar immunoglobulin fold, the filamin repeats preferentially bind to different targets, notably, repeat 17 binds to glycoprotein Ibα (GPIbα) (6), repeat 21 binds to both integrin β cytoplasmic tails and migfilin (7–10), repeat 23 binds to FilGAP (11), and repeat 24 forms the filamin homodimer. In all of the ligand-bound structures of filamin repeats, the ligand is sandwiched between β strands C and D of the repeat, and the peptide ligand itself is a β strand and extends an existing β-sheet. A structure of multiple repeats, repeats 19–21, has been recently reported (13). Interestingly, the first strand of repeat 20 was found to act as a ligand by interacting with the groove between the β strands C and D of the adjoining repeat 21. This binding mode mimics that for integrin, and thus repeat 20 is presumed to block integrin binding to the repeat 21, suggesting an autoinhibition mechanism for regulating the FLNa/integrin interaction.
The highly conserved ligand binding mode observed in various reported structures of the filamin repeat-target complexes raises a fundamental question as to the basis of ligand specificity. For example, whereas repeat 17 was shown to recognize GPIbα and repeat 21 bind to integrin or migfilin (6, 7), no apparent sequence preferences on the ligand-binding sites of these two repeats can be derived from their structures. Furthermore, data on the previously reported ligand specificity of the filamin repeats is conflicting. For example, FLNc repeat 21 was shown to bind to migfilin (10, 14), but another study on FLNb showed that repeats 10–13 specifically recognize migfilin (15). The sequences of repeat 21 from FLNa, b, and c are very similar, and the ligand-contacting strands C and D have miniscule differences making it unlikely that FLNb repeat 21 does not bind migfilin.
In this study, we have undertaken a detailed bioinformatics analysis on all 24 filamin repeats and discovered that they can be divided into four subgroups. To our surprise, the distinct ligand-binding repeats 17 and 21 belong to the same subgroup A, which contains a total of seven repeats: 4, 9, 12, 17, 21, and 23. The ligand-binding sites within these repeats have a far greater degree of similarity to each other than the rest of the filamin repeats. Detailed binding analyses demonstrate that these repeats do bind similar ligands including GPIbα, integrins, and migfilin, with repeat 21 having the strongest affinity to the ligands. These new results significantly change the filamin literature landscape. They also provide mechanistic insight into how similar filamin repeats may utilize a multiple ligand binding mechanism to promote the clustering of the same receptor or cross-talk among different receptors during cellular signaling processes.
MATERIALS AND METHODS
Sequences and Constructs
Human FLNa full-length cDNA (NM_001456) clone was kindly provided by Dr. Fumihiko Nakamura (Harvard Medical School). The accession numbers for the chicken and Drosophila proteins used in our analysis are AAA58939 and AAG43431. Primers were designed to subclone repeats 4 (residues 574–668), 9 (residues1065–1157), 10 (residues1158–1252), 12 (residues1353–1445), 17 (residues1863–1956), 19 (residues2045–2140), 21 (residues2236–2329), and 23 (residues2427–2522) from human FLNa. All of the repeats were cloned into GST parallel-1 vector between BamHI and XbaI restriction sites for expression in Escherichia coli BL21(DE3). DNA sequencing confirmed the authenticity of the repeats cloned. Protein expression of unlabeled and uniformly 15N-labeled samples was induced at A600 of 0.7 with 0.3 mm isopropyl β-d-thiogalactopyranoside and allowed accumulate protein for a further 16 h at room temperature. E. coli was lysed in 50 mm Tris, pH 8.0, 1 mm DTT, 1 mm EDTA, and 10% glycerol with protease inhibitors using the French press/sonicator. The GST-fused repeat proteins were purified by standard methods and assessed for homogeneity by SDS-PAGE. Uniformly 15N-labeled and unlabeled GST filamin repeats were cleaved with tobacco etch virus protease to release the repeats from GST. The cleaved GST and repeat mixture was passed through a glutathione resin to remove GST, and a final size exclusion chromatography, using a S75 column (GE Healthcare) was incorporated to obtain homogeneous filamin repeat proteins. Filamin repeats were extensively buffer-exchanged into 25 mm sodium phosphate, pH 6.4, 5 mm NaCl, and 1 mm DTT and used for isothermal calorimetry (ITC) experiments. All of the repeats have an N-terminal GAMDP sequence derived from the vector. Of the eight repeats (including repeat 10, the negative control) used in 1H-15N heteronuclear single quantum spectra (HSQC) experiments, only repeat 4 gave extra number of peaks and was unstable during expression and purification. The conformational instability/heterogeneity problem was also detected during purification on the S75 size exclusion column where a fraction of the repeat 4 seems to be unfolded. This peculiarity of repeat 4 prevented good isothermal calorimetry data from being obtained.
Peptides
The following peptides were synthesized in the biotechnology core facility of the Lerner Research Institute at the Cleveland Clinic: peptide 1, MASKPEKRVASSVFITLAPPRRDV (Migfilin, residues 1–24); peptide 2, LRGSLPTFRSSLFLWVRPNGRV (GPIbα, residues 556–577); peptide 3, WKQDSNPLYKSAITTTINPRFQEADSPTL (β7, residues 769-798); peptide 4, IKRLLSEKKTCQCPHRFQKTCSPI (CD4, residues 435–458); and peptide 5, WNNDNPLFKSATTTVMNPKFAES (β2, residues 747–769). The lyophilized peptides were carefully weighed and dissolved in the water or the above mentioned phosphate buffer, assuming 75% purity. Peptides with a tryptophan residue were estimated by UV absorption, and the two methods gave very similar values.
NMR Spectroscopy
All of the HSQC spectra were recorded in the 600-MHz cryo-cooled Bruker spectrometer at the Cleveland Center for Structural Biology. After recording the free form spectra, each of the peptides were added to the repeat proteins, and the pH was readjusted to with 0.02 units. The proteins were in a final buffer containing 25 mm sodium phosphate, pH 6.4, 5 mm NaCl, and 1 mm DTT. All of the spectra recorded were at 30 °C. The ligand-bound spectra were recorded in an identical manner. All of the spectra were processed using NMRPipe and visualized using NMRdraw (16).
Isothermal Calorimetry
Purified filamin repeats were extensively buffer-exchanged into degassed 25 mm sodium phosphate, pH 6.4, 5 mm NaCl, and 1 mm DTT to mimic the buffer conditions used for magnetic resonance spectroscopy. GST-fused filamin repeats were estimated using UV absorbance. The potential ligand peptides (GPIbα, β2, β7, Mig-N, and CD4) were also made in an identical batch of buffer to minimize experimental variability. Protein concentration of 35–50 μm was used to titrate 1 mm ligand in the injection syringe. 2-μl injections of the ligand at 3-min intervals were done to estimate the heat changes on binding. The use of purified repeats eliminated the small precipitation observed during ITC titrations of GPIbα with GST-fused repeats. A single-site binding model with free stoichiometry was used to analyze the data.
RESULTS
Filamin Repeats Can Be Subdivided into Four Distinct Subgroups
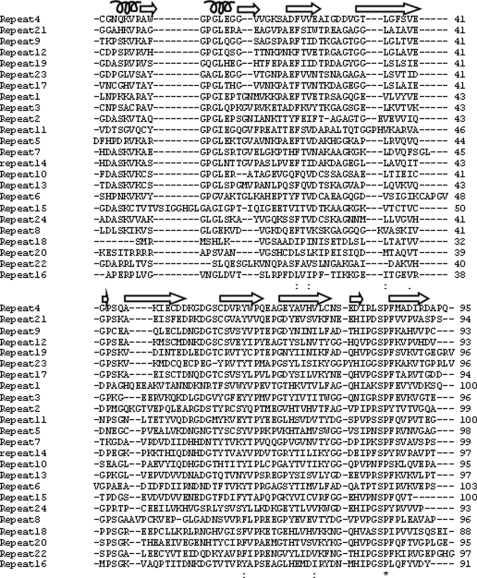
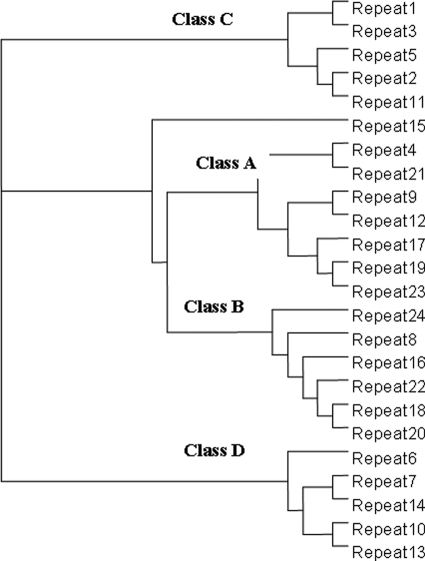
Human filamin has long been known to contain 24 Ig repeats, each of 90–100 amino acids. However, sequence alignment of these repeats in FLNa using the sequence alignment program ClustalW2 (17) shows that very few amino acids are identical throughout all of the repeats. A single proline is all that is absolutely conserved when all of the repeats are considered (Fig. 1). On the other hand, a phylogenetic tree of these repeats based on amino acid similarities revealed that the repeats can be divided into four subgroups (subgroups A, B, C, and D) (Fig. 2). Group A contains seven repeats: 4, 9, 12, 17, 19, 21, and 23, some of which were surprisingly extensively characterized as specific ligand-binding repeats including repeats 17, 21, and 23 (6–11). Compared with other subgroups, group A appears to have a well conserved ligand-binding site based on available structures (Fig. 3, red bar), suggesting that they can all bind the same ligand or simultaneously bind to different ligands. This prediction, if proven, would significantly alter the view of the specificity for the filamin repeats that were thought to be highly specific, e.g. repeat 17 specifically binds GPIbα (6), whereas repeat 21 recognizes integrins and migfilin (7–10). To experimentally verify this prediction, we decided to characterize the binding of the group A repeats to a set of known physiologically relevant ligands including the GPIbα cytoplasmic tail (residues 556–577, GPIbα), integrin β2 cytoplasmic tail (residues 747–769), integrin β7 cytoplasmic tail (residues 769–798), and the N terminus of migfilin 1–24 (Mig-N).
FIGURE 1.
Alignment of 24 immunoglobulin repeats of human filamin A using ClustalW2. Notice that there is only a single conserved proline residue (*) among the repeats, despite the fact that these repeats may adapt similar structures. Secondary structural motifs as in the repeat 21-integrinβ7 structure are shown above the sequences.
FIGURE 2.
Sequence-based relationships between filamin repeats in human filamin A. The classification is based on the sequence similarities in Fig. 1. Repeat 15 does not fall into any distinct class.
FIGURE 3.
FLNa repeat/ligand interface. A, alignment of the seven Class A repeats. The conserved residues are marked with asterisks. Similar amino acids in the alignment are marked with colons or periods. The ligand-binding site is highlighted in red, and this forms the β strands C and D that lock down the peptide ligand (integrin, migfilin, or GP1bα). The residues in blue are peculiar to repeat 23, which may explain its ligand affinity, which is lower than that of other repeats (see text). B, comparison of the five filamin repeat target sequences. Integrin, GPIbα, and migfilin have similarities but are completely different from CD4, which does not bind group A repeats (it binds to repeat 10, not a member of group A, see text).
Group A Repeats Bind GPIbα with the Repeat 21 Having the Highest Affinity
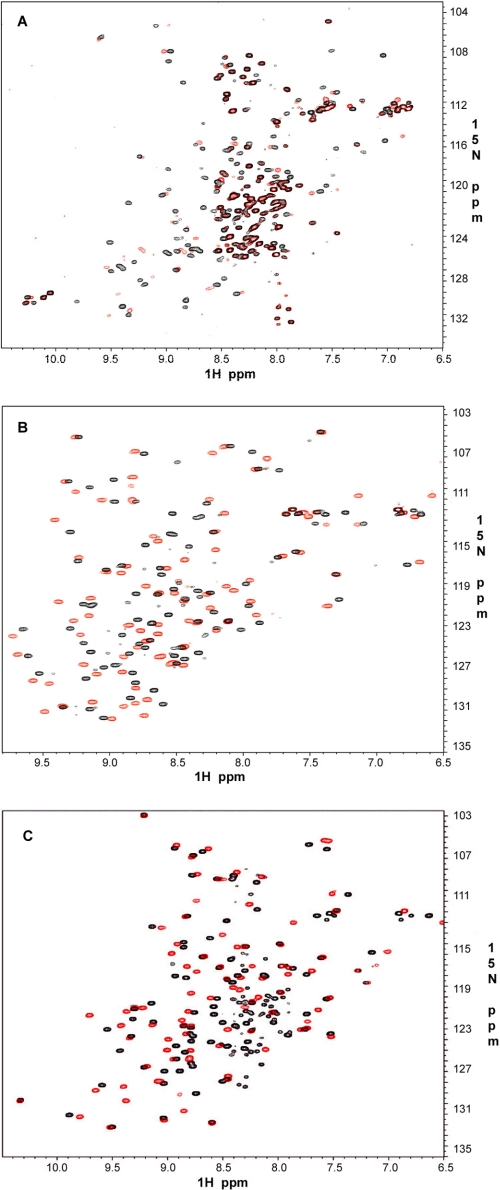
We started by assessing the binding of GPIbα to the group A repeats because this peptide was the first well characterized target bound to FLNa repeat 17 (6). Fig. 4 shows the ITC profiles of repeats 9, 12, 17, 19, 21, and 23 upon the addition of the GPIbα peptide. In addition to repeat 17, all other group A repeats bind GPIbα potently with affinities in the submicromolar to micromolar range. Surprisingly, the highest affinity is not repeat 17 but repeat 21 (KD = ∼0.13 μm), which is significantly higher than repeat 17 (KD = ∼0.71 μm).
FIGURE 4.
Isothermal calorimetric estimation of dissociation constants (Kd) for GPIbα binding to repeat 9 (A), repeat 12 (B), repeat 17 (C), repeat 19 (D), repeat 21 (E), and repeat 23 (F). All of the experiments were in degassed 25 mm sodium phosphate, pH 6.4, 5 mm NaCl, and 1 mm DTT at 30 °C. Note that repeat 4 was unstable, preventing the precise measurement of the affinity, but the quick HSQC experiment clearly shows that it binds to GPIbα (see Fig. 5A).
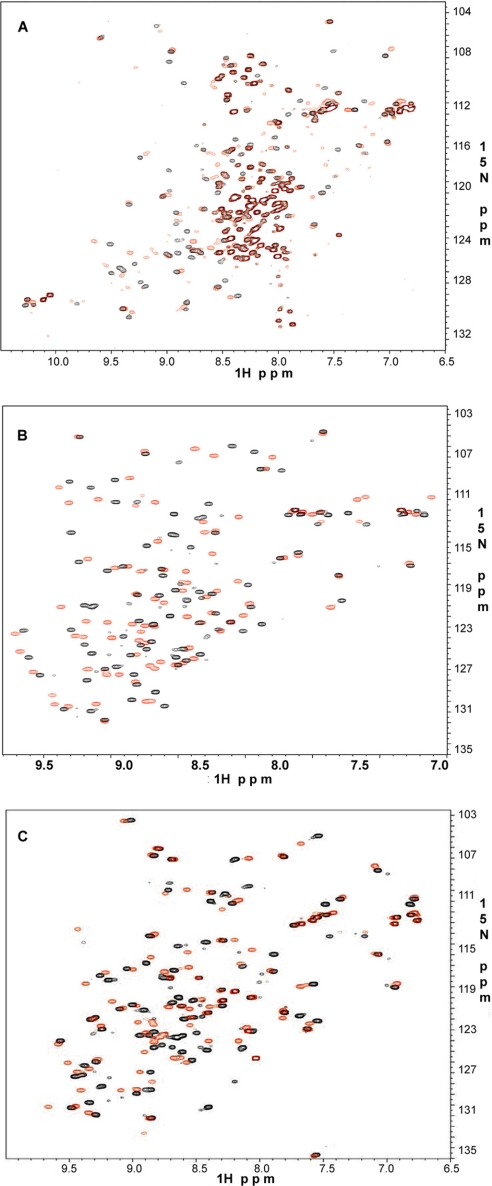
As an independent approach, we performed NMR-based binding experiments using quick two-dimensional 1H-15N HSQC. Fig. 5 shows selective cases involving repeat 4 (Fig. 5A), repeat 9 (Fig. 5B), and repeat 21 (Fig. 5C). It is clear that these repeats bind potently to GPIbα as indicated by the substantial chemical shift changes. The binding data of other group A repeats are provided in supplemental Figs. S1 and S2, respectively. The extent of the chemical shift change upon binding to GPIbα is similar among these repeats, indicating that the affinities should be similar, consistent with the ITC data in Fig. 4. Note that the repeat 4 binding to GPIbα seems to be of intermediate exchange with significant line broadening (Fig. 5A). However, the extent of the chemical shift change (based on the visible signals) is similar to other repeats. Repeat 4 clearly exhibits conformational heterogeneity as evidenced by the presence of five tryptophan side chain resonances present, when the protein itself has only two. Further, the presence of many strong peaks in the 7.8–8.4-ppm range suggests a fraction of unfolded protein being present. However, GP1bα binding does occur to at least one or possibly more species present in solution.
FIGURE 5.
GPIbα peptide binding to FLNa repeats. Shown are 600-MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 4 (A), repeat 9 (B), and repeat 21 (C) in the absence (black) and presence (red) of 0.3 mm unlabeled GPIbα peptide at pH 6.4 and 30 °C.
Group A Repeats Also Bind Integrins
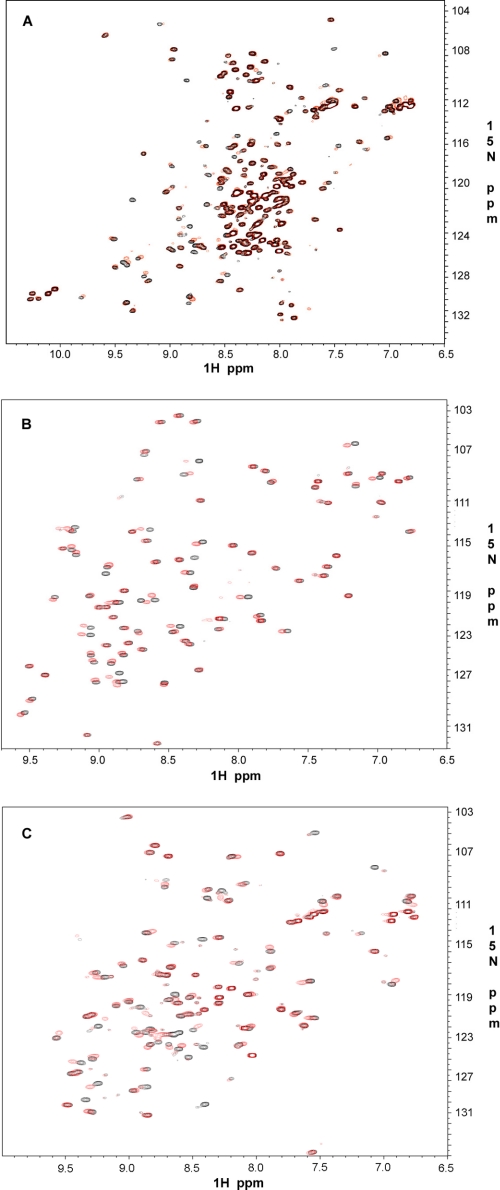
Next, we asked whether group A repeats bind integrin cytoplasmic tails. The binding of integrins to full-length filamin has been extensively characterized in vivo and in vitro (18, 19), and β7 cytoplasmic tail was shown to bind filamin repeat 21 with the highest affinity (7, 20, 21) at a KD level of 16–32 μm (Table 1). This affinity is much lower than that for repeat 21/GPIbα interaction. Other repeats bind even more weakly to integrin tails, which precluded precise ITC measurements. However, alternative NMR-based HSQC experiments for all seven repeats revealed appreciable chemical shift changes upon binding to β7 cytoplasmic tail, indicating that these repeats do bind integrins. Fig. 6 shows representative HSQC plots for repeats 4, 12, and 17. The extent of the chemical shift changes is either similar or less than previously reported repeat 21/β7 interaction (7, 10). Interestingly, repeat 17, which has been characterized to specifically bind GPIbα, also binds potently to the integrin β7 tail (Fig. 6C). Repeat 4, located in rod 1 segment and closest to the actin-binding domain, also binds integrin (Fig. 6A). Repeats 9 (not shown) and 12 (Fig. 6B) in the rod 1 segment of filamin bind integrins as well. These results significantly extend the number of integrin-binding sites in filamin. A similar phenomenon was also observed for integrin β2 cytoplasmic tail with the chemical shift changes being smaller than β7 (not shown). A spectral comparison of the strongest (GPIbα) versus weakest binding (β2) to repeat 21 is shown in supplemental Fig. S3. Thus, all seven group A repeats can bind various integrin cytoplasmic tails. Although single repeat/integrin tail may interact modestly, the full-length filamin may allow more extensive binding where multiple similar repeats can simultaneously bind to multiple integrins on the membrane surface in a high affinity clustered fashion.
TABLE 1.
Comparison of the affinities of different ligands for human FLNa repeat 21
| Ligand | Affinity (KD) |
|---|---|
| μm | |
| GPIbα | 0.15 |
| Migfilin | 2–4 |
| Integrin β7 | 16–32 |
| Integrin β2 | NDa |
a ND, not detectable; reported value is 525 μm (8).
FIGURE 6.
Integrin β7 binding to FLNa repeats. Shown are 600-MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 4 (A), repeat 12 (B), and repeat 17 (C) in the absence (black) and presence (red) of 0.3 mm unlabeled β7 at pH 6.4 and 30 °C.
Group A Repeats Also Bind Migfilin
Previous structural studies have shown that the cytoskeleton regulator migfilin and integrin β tail essentially occupy the same site on filamin repeat 21 (9, 10), and therefore we decided to determine whether other group A repeats also bind to Mig-N. As predicted from the above studies, we found that all filamin repeats in group A bind migfilin. Representative HSQC perturbations for repeats 4, 9, and 17 with Mig-N are shown in Fig. 7, respectively. A negative control experiment was performed using a repeat from another subgroup (see below).
FIGURE 7.
Migfilin-N binding to FLNa repeats. Shown are 600-MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 4 (A), repeat 9 (B), and repeat 17 (C) in the absence (black) and presence (red) of 0.3 mm unlabeled migfilin-N at pH 6.4 and 30 °C.
Promiscuity versus Specificity of the Group A Repeats
The above results demonstrate that the seven group A repeats all recognize the same set of target peptides including β7, β2, Mig-N, and GPIbα (Fig. 3B). Because the structures of some of the group A filamin repeats with these peptides are known, one can readily deduce that these peptides likely occupy a similar site on all seven repeats, i.e. straddled between strands C and D (Fig. 3A). As indicated above, the integrin cytoplasmic tails and migfilin appear to be weaker filamin ligands than GPIbα as judged by the extent of the chemical shift changes. This is also quantitatively shown for the representative repeat 21, which binds to different ligands with GPIbα having the highest affinity followed by migfilin and integrin respectively (Table 1). An extreme case was found for repeat 23, which binds to GPIbα very potently but to migfilin and integrins almost negligibly (supplemental Fig. S4). Repeat 23 does deviate the most around the ligand-binding site as compared with other group A repeats, which may contribute to this phenomenon (Fig. 3A). The precise basis as to why GPIbα generally binds stronger to group A repeats than other target peptides remains to be determined. Nevertheless, the affinities to the same ligand(s) within the seven repeats appear to be rather similar as indicated by the ITC data on GPIbα (KD range between submicromolar and micromolar) (Fig. 4).
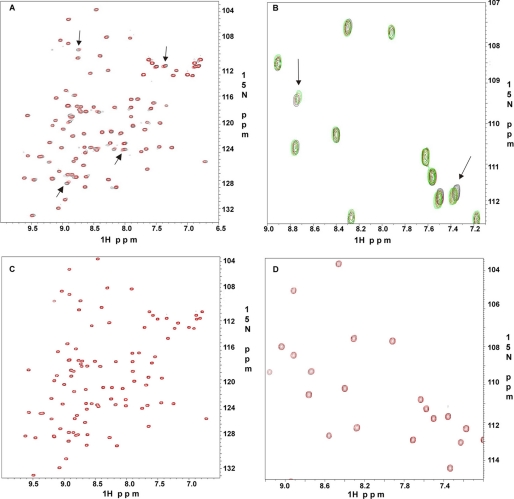
The multiple ligand binding (GPIbα, Mig-N, integrin β7, and integrin β2) to the group A repeats indicates that group A is quite promiscuous for selecting ligands. On the other hand, the repeats in this group, as noted above, do contain a specific ligand-binding site (Fig. 3). To further examine whether this group is specific to the above ligands, a control experiment is necessary. We selected repeat 10, which is outside the group A, for the test because this repeat has been recently shown to bind human immunodeficiency virus CD4 receptor at its cytoplasmic domain and play a role in human immunodeficiency virus entry into immune cells (22). As shown in Fig. 8A, the addition of CD4 caused small but specific chemical shift changes of 15N-labeled repeat 10 (see arrows), confirming the weak binding. We titrated repeat 10 with 3, 6, and 10 molar excess of CD4 to confirm the specificity of this interaction. A representative expanded region in shown in Fig. 8B substantiating the specificity. In contrast, migfilin, which caused dramatic chemical shift changes to group A repeats (Fig. 7), did not bind repeat 10 at all at large excess (1:5 protein:peptide ratios) (Fig. 8, C and D), providing evidence that other group repeats are indeed different from the group A repeats. Comparison of the amino acid sequences of integrin, GPIbα, and migfilin show critical conserved hydrophobic residues, but these ligands are completely different from CD4 that recognizes repeat 10 that is outside the group A, providing a basis for understanding the specificity for group A-target complexes. Note that integrin β2 has the lowest homology to GPIbα in terms of hydrophobicity (Fig. 3B) as compared with integrin β7 and Mig-N, which may explain why β2 binds to the group A repeats with the lowest affinity.
FIGURE 8.
Specificity of ligand binding to repeat 10 at pH 6.4 and 30 °C. A, 600-MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 10 with 1 mm CD4. B, expanded 600 MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 10 with 0.6 mm CD4 (red) and 1 mm CD4 (green). The arrows indicate specific peak perturbations, which are absent in C. C, 0.1 mm 15N-labeled repeat 10 with 0.5 mm migfilin-N. D, expanded 600-MHz two-dimensional 1H-15N HSQC of 0.1 mm 15N-labeled repeat 10 with 0.5 mm migfilin-N. There are no recognizable chemical shift changes in C and D.
Evolutionary Conservation of the Group A Repeats
Because filamin is a well conserved molecule from Drosophila to mammals, we examined the distribution of these group A repeats in Drosophila (AAG43431) and chicken (AAA58939) filamins. We noticed that the C terminus of both Drosophila and chicken filamin have the same alternating group A and group B repeats as in humans (see supplemental Figs. S5 and S6 for a full comparison). Furthermore, the rod domain 1 of both Drosophila and chicken also has the group A repeats in the same pattern as human filamin. The only difference is that the Drosophila filamin has only six of these repeats in total and not seven as in higher animals. Nevertheless, our data indicate that the group A repeats are evolutionarily conserved, which may be functionally important in terms of the ligand binding for regulating cytoskeleton and a variety of cellular activities.
DISCUSSION
In this study, we have performed the first systematic structure and sequence-based classification of filamin repeats in human FLNa. The major finding of the study was the identification and characterization of the distinct group A repeats that appear to interact with a set of biologically important ligands (Figs. 2 and 3). A comparison of three human filamin isoforms (1) reveals high similarity all through their sequences, and therefore the group A repeats are expected to have similar properties in all isoforms. Our finding significantly extends the view of the specificity of the filamin repeats. Contrasting the previous notion that one repeat binds to one individual ligand, e.g. repeat 17 to GPIbα (6) and repeat 21 to integrin (7), the whole group A can bind to a set of ligands including GPIbα and integrins. Our study also resolves the ambiguity originated from the previous cell-based studies. Specifically, migfilin was shown to bind repeat 21 of FLNa and FLNc (14), whereas Takafuta et al. (15) only detected the binding of FLNb repeat 11–13 to migfilin but not FLNb repeat 21. Our finding that both repeat 12 (contained in the repeat 11–13 construct of Takafuta et al. (15)) and repeat 21 belong to group A and both bind to migfilin now resolves this confusing issue. The results of the two studies (14, 15) were partially correct, but both were incomplete. The binding of repeat 12 to migfilin found in our study is also consistent with a recent study where rod 1 segment of filamin containing repeat 12 binds to migfilin (9).
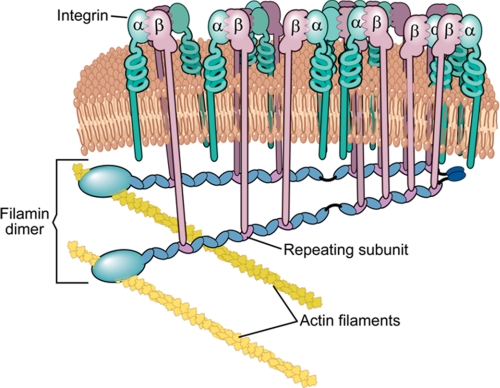
Because all of the group A repeats can bind the cytoplasmic tails of large membrane receptors such as integrins or GPIbα, it seems plausible that they can bind them simultaneously, and such interactions may promote clustering of these cell surface receptors. Based on our experimental evidence we propose a model (Fig. 9) where receptors like integrins are spatially clustered by filamin. Such clustering may be further modulated by the typical multimeric ligands that bind to the extracellular domain and/or by different conformational transitions that are induced during different stages of integrin signaling, and filamin likely facilitates this process. The clustering can of course be extended to other types of filamin group A repeat-binding receptors such as GPIbα. Our use of the term clustering does not imply integrin activation, a phenomenon where multivalent ligand binding to the extracellular domain of integrins bridges integrins together. Here we suggest that multiple integrins binding to a single filamin molecule (monomer/dimer) should promote the local enrichment of the integrins and prevent the free diffusion of integrins-GP1bα on the fluid plasma membrane. If this hypothesis turns out positive, filamin by engaging many membrane receptors may act as a raft shipping defined signaling complexes to various cellular docking sites to initiate signal transduction events. Although proving this hypothesis requires vigorous in vivo characterization, filamin-mediated ion channel receptor clustering has been reported as a result of filamin-HCN1 (ion channel receptor) interaction (23).
FIGURE 9.
A model for integrin receptor clustering mediated by seven filamin repeats.
Another extrapolation of our data is that filamin group A repeats may simultaneously bind to different receptors/proteins such as integrin αIIbβ3 and GPIbα, both of which are membrane receptors present at high density in platelets. This may allow these receptors to communicate with each other. Although filamin may be localized to specific cellular compartments for different interactions (24, 25), a single filamin molecule may also switch binding partners in the same cellular compartment to trigger signaling events. For example, as reported recently, migfilin competes with integrin (9, 10) in displacing filamin-integrin connections. By doing so, we showed that migfilin can promote integrin activation (10).
In conclusion our results provide new and surprising information on the ligand specificity of the filamin repeats. Our findings also suggest a multiple ligand binding mechanism for filamin, which may facilitate receptor clustering or receptor cross-talking, regulating the cytoskeleton and diverse filamin-mediated cellular activities.
Acknowledgments
We thank Dr. Saurav Misra, Dr. Witold Surewicz, Dr. Prasanta Hota, and Dr. Vytautas Smirnovas for help with ITC measurements and many stimulating discussions. We also acknowledge the assistance of Dr. Jun Yang and Jianmin Liu in NMR studies of the filamin repeats.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM62823 and P01 HL073311 (to J. Q. and E. F. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- FLN
- filamin
- GP
- glycoprotein
- ITC
- isothermal calorimetry
- GST
- glutathione S-transferase
- DTT
- dithiothreitol
- HSQC
- heteronuclear single quantum coherence
- Mig-N
- migfilin-N.
REFERENCES
- 1.van der Flier A., Sonnenberg A. (2001) Biochim. Biophys. Acta. 1538, 99–117 [DOI] [PubMed] [Google Scholar]
- 2.Robertson S. P. (2005) Curr. Opin. Genet. Dev. 15, 301–307 [DOI] [PubMed] [Google Scholar]
- 3.Feng Y., Walsh C. A. (2004) Nat. Cell Biol. 6, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 4.Popowicz G. M., Schleicher M., Noegel A. A., Holak T. A. (2006) Trends Biochem. Sci. 31, 411–419 [DOI] [PubMed] [Google Scholar]
- 5.Nakamura F., Osborn T. M., Hartemink C. A., Hartwig J. H., Stossel T. P. (2007) J. Cell Biol. 179, 1011–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura F., Pudas R., Heikkinen O., Permi P., Kilpeläinen I., Munday A. D., Hartwig J. H., Stossel T. P., Ylänne J. (2006) Blood 107, 1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., Campbell I. D., Ylänne J., Calderwood D. A. (2006) Mol. Cell 21, 337–347 [DOI] [PubMed] [Google Scholar]
- 8.Takala H., Nurminen E., Nurmi S. M., Aatonen M., Strandin T., Takatalo M., Kiema T., Gahmberg C. G., Ylänne J., Fagerholm S. C. (2008) Blood 112, 1853–1862 [DOI] [PubMed] [Google Scholar]
- 9.Lad Y., Jiang P., Ruskamo S., Harburger D. S., Ylänne J., Campbell I. D., Calderwood D. A. (2008) J. Biol. Chem. 283, 35154–35163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ithychanda S. S., Das M., Ma Y. Q., Ding K., Wang X., Gupta S., Wu C., Plow E. F., Qin J. (2009) J. Biol. Chem. 284, 4713–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura F., Heikkinen O., Pentikäinen O. T., Osborn T. M., Kasza K. E., Weitz D. A., Kupiainen O., Permi P., Kilpeläinen I., Ylänne J., Hartwig J. H., Stossel T. P. (2009) PLoS One. 4, e4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pudas R., Kiema T. R., Butler P. J., Stewart M., Ylänne J. (2005) Structure. 13, 111–119 [DOI] [PubMed] [Google Scholar]
- 13.Lad Y., Kiema T., Jiang P., Pentikäinen O. T., Coles C. H., Campbell I. D., Calderwood D. A., Ylänne J. (2007) EMBO J. 26, 3993–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Y., Wu S., Shi X., Chen K., Wu C. (2003) Cell 113, 37–47 [DOI] [PubMed] [Google Scholar]
- 15.Takafuta T., Saeki M., Fujimoto T. T., Fujimura K., Shapiro S. S. (2003) J. Biol. Chem. 278, 12175–12181 [DOI] [PubMed] [Google Scholar]
- 16.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 17.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.Bioinformatics 23, 947–2948 [DOI] [PubMed] [Google Scholar]
- 18.Sharma C. P., Ezzell R. M., Arnaout M. A. (1995) J. Immunol. 154, 3461–3470 [PubMed] [Google Scholar]
- 19.Goldmann W. H. (2000) Biochem. Biophys. Res. Commun. 271, 553–557 [DOI] [PubMed] [Google Scholar]
- 20.Travis M. A., van der Flier A., Kammerer R. A., Mould A. P., Sonnenberg A., Humphries M. J. (2004) FEBS Lett. 569, 185–190 [DOI] [PubMed] [Google Scholar]
- 21.Calderwood D. A., Huttenlocher A., Kiosses W. B., Rose D. M., Woodside D. G., Schwartz M. A., Ginsberg M. H. (2001) Nat. Cell Biol. 3, 1060–1068 [DOI] [PubMed] [Google Scholar]
- 22.Jiménez-Baranda S., Gómez-Moutón C., Rojas A., Martínez-Prats L., Mira E., Ana Lacalle R., Valencia A., Dimitrov D. S., Viola A., Delgado R., Martínez-A C., Mañes S. (2007) Nat. Cell Biol. 9, 838–846 [DOI] [PubMed] [Google Scholar]
- 23.Gravante B., Barbuti A., Milanesi R., Zappi I., Viscomi C., DiFrancesco D. (2004) J. Biol. Chem. 279, 43847–43853 [DOI] [PubMed] [Google Scholar]
- 24.Feng S., Lu X., Kroll M. H. (2005) J. Biol. Chem. 280, 6709–6715 [DOI] [PubMed] [Google Scholar]
- 25.van der Flier A., Kuikman I., Kramer D., Geerts D., Kreft M., Takafuta T., Shapiro S. S., Sonnenberg A. (2002) J. Cell Biol. 156, 361–376 [DOI] [PMC free article] [PubMed] [Google Scholar]