Abstract
Long QT syndrome (LQTS) is a hereditary arrhythmia caused by mutations in genes for cardiac ion channels, including a potassium channel, KvLQT1. Inheritance of LQTS is usually autosomal-dominant, but autosomal-recessive inheritance can be observed in patients with LQTS accompanied by hearing loss. In this study, we investigated the functional alterations caused by KCNQ1 mutations, a deletion (delV595) and a frameshift (P631fs/19), which were identified in compound heterozygous state in two patients with autosomal-recessive LQTS not accompanied by hearing loss. Functional analyses showed that both mutations impaired cell surface expression due to trafficking defects. The mutations severely affected outward potassium currents without apparent dominant negative effects. It was found that delV595 impaired subunit binding, whereas P631fs/19 was retained in endoplasmic reticulum due to the newly added 19-amino acid sequence containing two retention motifs (R633GR and R646LR). This is the first report of novel mechanisms for trafficking abnormality of cardiac ion channels, providing us new insights into the molecular mechanisms of LQTS.
Introduction
Long QT syndrome (LQTS)2 is characterized by prolongation of QT interval in electrocardiogram (ECG), syncope, and sudden death due to polymorphic ventricular tachyarrhythmia (1). From the genetic viewpoints, LQTS is classified into two subtypes, Jervell and Lange-Nielsen syndrome (Online Mendelian Inheritance in Man (OMIM) 220400) and Romano Ward syndrome (OMIM 192500). The Jervell and Lange-Nielsen syndrome is defined as LQTS of the autosomal-recessive genetic trait, which is often accompanied by sensorineural deafness, whereas Romano Ward syndrome defines LQTS without deafness, which is inherited as the autosomal-dominant trait. Recent genetic analyses have revealed that most of the Jervell and Lange-Nielsen syndrome patients carried mutations in KCNQ1 that encodes the α-subunit of a voltage-gated cardiac potassium channel, KvLQT1 (2). In contrast, Romano Ward syndrome can be caused by mutations in genes for potassium channels, including KvLQT1 and HERG, sodium channels, a calcium channel, and other molecules associated with cardiac ion channels (1, 3–6).
The α-subunit of KvLQT1 has the domain structure composed of six membrane-spanning segments (S1–S6) containing a pore region localized at cell membrane and the N-terminal and C-terminal regions that are localized in the cytoplasm. The KvLQT1 channel consists of four α-subunits (7) and two β-subunits (MinK) encoded by KCNE1 (8), which generates slow component of delayed rectifier potassium current (IKs). Many KCNQ1 mutations associated with Jervell and Lange-Nielsen syndrome and Romano Ward syndrome have been reported, most of which were found in the membrane-spanning segments and impaired the channel function (1). There are several other mutations not affecting the pore structure but causing functional impairment, most of which were identified in the C-terminal cytoplasmic region. Although the cytoplasmic region contained several functional domains (9–12), molecular mechanisms of KCNQ1 mutations in the cytoplasmic domain have not been fully elucidated.
On the other hand, more information is available about the functional alterations caused by mutations in KCNH2 that encodes pore-forming α-subunits of another voltage-gated cardiac potassium channel, HERG. The α-subunits form the tetrameric channel complex in the endoplasmic reticulum (ER) (13–15), and the tetramer is glycosylated in ER and Golgi body (16) and then transported to and expressed at the cell membrane. The HERG channel generates rapid component of the delayed rectifier potassium current (IKr) (5). With regard to the functional alterations caused by KCNH2 mutations associated with Romano Ward syndrome, most mutations in the cytoplasmic regions caused trafficking defects due to misfolding in ER (17) or the failure of transport from ER to Golgi body (18). These functional abnormalities decreased the IKr current in the cardiomyocytes and led to the prolongation of repolarizing phase of action potential, manifested with prolonged QT interval in the ECG (19).
In this study, we investigated functional alterations caused by two mutations, delV595 and P631fs/19, in the C-terminal region of the KvLQT1 channel, which were found in an LQTS family of autosomal-recessive inheritance. It was demonstrated that both mutations lead to trafficking defects, resulting in the loss of channel function. This is the first report on two novel mechanisms of channel-trafficking abnormalities: an impaired subunit binding and a newly generated ER retention signal.
EXPERIMENTAL PROCEDURES
Case Presentation
The proband of the LQTS family was a 10-year-old girl with a history of cardiopulmonary arrest during exercise. Ventricular fibrillation was confirmed when she was revived by cardiopulmonary resuscitation. Her ECG showed a wide T-wave and prolonged corrected QT (QTc) interval (0.55 s). Her eldest brother also suffered from LQTS. He experienced syncope during exercise at the age of 13. His ECG after the event showed T-wave alternans, and prolongation of QTc interval (0.48 s) was found even at rest. Neither patient had any history of hearing loss. Prolongation of QTc interval was not found in the ECGs of either the parents or the second eldest brother. The other relatives of this family had no history of syncope or sudden death.
Genetic Analysis
The research protocol was approved by the Ethics Reviewing Committee of the Medical Research Institute, Tokyo Medical and Dental University. Genetic screening was performed for the proband and her family members. The control subjects were 180 genetically unrelated Japanese individuals who were selected at random. Blood samples were obtained from each subject, who provided informed consent for the clinical and genetic studies. Genomic DNA was extracted from peripheral blood samples by conventional methods. Each exon of KCNQ1, KCNH2, SCN5A, KCNE1, KCNE2, and KCNJ2 genes was amplified by PCR. The PCR products were analyzed by direct sequencing using Big Dye Terminator chemistry (Applied Biosystems) according to the manufacturer's instructions. Sequences of primers used in the genetic analyses are available on request.
Alignment of Amino Acid Sequences and Prediction of Tertiary Structure
Amino acid sequences of human KvLQT1 protein predicted from nucleotide sequences (GenBankTM NM_000218) were aligned with those of bovine (XM_001252337), dog (XM_540790), rat (NM_032073), mouse (NM_008434), and chicken (XM_421022), using GENETIX version 8.1.2 software (Genetics, Tokyo, Japan). The tertiary structure of coiled-coil was predicted by using COILS (available on the World Wide Web) (20).
Electrophysiological Studies
Wild-type (WT) full-length KCNQ1 (NM_000218) and KCNE1 (NM_000219) cDNA fragments corresponding to aa 1–676 and aa 1–129, respectively, were amplified by reverse transcription-PCR from human heart cDNA. Mutant KCNQ1 cDNA fragments carrying deletion of GTA (delV595 mutation) or insertion of C (P631fs/19 mutation) were generated by the primer-directed mutagenesis method. The KCNQ1 cDNA fragments were cloned in the bicistronic plasmid pIRES2-EGFP (Clontech, Mountain View, CA) to make pIRES2-EGFP-KvLQT1 constructs, whereas KCNE1 cDNA was inserted into the bicistronic plasmid pCD8-IRES to make the pCD8-IRES-MinK construct. All of the constructs used in this study were sequenced to ensure that no error was introduced. Sequences of the primers used in the plasmid construction are listed in Table 1.
TABLE 1.
Primers used for cloning
| Production | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| KvLQT1-WT | GCTCGGATCCGTTATGGCCGC | GGATATCTGCAGAATTCGGC |
| L1-Myc-KvLQT1 | GCTGTCCACCATCGAGGAGCAGAAACTCATCTCTGA AGAGGATCTGCAGTATGCCGCCCTGGC | GCCAGGGCGGCATACTGCAGATCCTCTTCAGAGATGAG TTTCTGCTCCTCGATGGTGGACAGC |
| C terminus KvLQT1 | GGGGATCCATCGGGCCACCATTAAGGTCATT | CTTGCGGCCGCCAGCCCCATCCCCTCCTCA |
| KvLQT1-delV595 | CGCCTGAACCGAGAAGAC | CGTCACCTTGTCTTCTCGG |
| KvLQT1-P631fs/19 | GCGGCCCCCCCCAGAGAG | CCCTCTCTGGGGGGGGCC |
| KvLQT1-AAA/RLR | CCCCAGGCAGCTGCGGGCCCACATCAC | GTGGGCCCGCAGCTGCCTGGGGGGGGCC |
| KvLQT1-RGR/AAA | CCTGCGGCAGTGGGCTGCAGCTCGACC | GCTCAGGGTCGAGCTGCAGCCCACTGC |
| KvLQT1-P631stop | GCTCGGATCCGTTATGGCCGC | CCGAATTCTCAGGGGGGGCC |
| KvLQT1-P631fs2/34 | GCGGCCCCCCCCCAGAGAG | CCCTCTCTGGGGGGGGGCC |
| KvLQT1-R594Q | CGCCCGCCTGAACCAAGTAGAAG | ACCTTGTCTTCTACTTGGTTCAG |
CHO-K1 cells were transiently transfected using Lipofectamine (Invitrogen) with a pIRES2-EGFP-KvLQT1 plasmid (0.75 μg) of either WT or mutant in combination with the pCD8-IRES-MinK plasmid (0.75 μg) to visually identify the cells expressing heterologous KvLQT1 with EGFP under a fluorescent microscope and MinK with Dynabeads M-450 CD8 (Invitrogen), respectively. Potassium currents were recorded from cells that were positive for both EGFP and CD8 using the whole-cell patch clamp techniques as described previously (21). Currents were recorded using an Axopatch 200A amplifier (Axon Instruments, Foster City, CA), and series resistance errors were reduced by 60–70% using electronic compensation. Holding potential was −80 mV. All of the signals were acquired at 5–500 kHz (Digidata 1322, Axon Instruments) with a personal computer running Clampex 9 software (Axon Instruments) and filtered at 5 kHz with a 4-pole Bessel low pass filter. Membrane currents were analyzed with Clampfit 9 software (Axon Instruments).
Co-immunoprecipitation (co-IP) Assay
The cDNA fragments corresponding to the C terminus of KvLQT1 (aa 353–676) were generated for WT, delV595, and P631fs/19 by PCR from the KCNQ1 plasmids described earlier. Another mutant cDNA containing a substitution of G with A (R594Q mutation) was obtained from the KvLQT1-WT construct by the primer-directed mutagenesis method. The cDNA fragments corresponding to the C terminus of KvLQT1 were cloned into pCMV-Tag3C (Stratagene, Cedar Creek, TX) and pBIND (Promega, Madison, WI).
The co-IP assays were performed as previously described (22, 23). Briefly, COS-7 cells or HEK293 cells were co-transfected with a combination of pCMV-Tag3C-KvLQT1 (Myc-tagged C terminus KvLQT1) and pBIND-KvLQT1 (GAL4-tagged C terminus KvLQT1) constructs to analyze the binding of KvLQT1 subunits. Aliquots of the cellular extracts were collected for assessing the expression levels, and the remaining supernatants containing equal amount of proteins were used for the co-IP assay using the Catch and Release version 2.0 reversible immunoprecipitation system, according to the manufacturer's instructions (Millipore, Billerica, MA), with mouse anti-GAL4 monoclonal antibody (Ab) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Eluted samples were separated by SDS-PAGE, transferred to a nitrocellulose membrane, preincubated with 5% skimmed milk in phosphate-buffered saline (PBS), and incubated with primary rabbit anti-c-Myc polyclonal Ab (1:100; Santa Cruz Biotechnology, Inc.) or rabbit anti-GAL4 polyclonal Ab (1:100; Santa Cruz Biotechnology, Inc.) followed by secondary goat anti-rabbit (for polyclonal Ab) IgG HRP-conjugated Ab (1:1000; Dako A/S, Grostrup, Denmark). The signals were visualized by Immobilon Western Chemiluminescent HRP substrate (Millipore) and luminescent image analyzer LAS-3000mini (Fujifilm, Tokyo, Japan). The densitometric intensities were measured as means by using Multi Gauge version 3.0 (Fujifilm).
Protein Expression and Subcellular Fraction Assays
KvLQT1-WT, -delV595, and -P631fs/19 cDNA fragments were cloned into pCMV-Tag3B (Stratagene) to add a Myc tag at the N terminus of KvLQT1 (Nt-Myc-KvLQT1) or into pEGFP-C1 (Clontech) to add a green fluorescent protein (GFP) tag (GFP-KvLQT1). HEK293 cells (1.0 × 106) were seeded onto a poly-d-lysine-coated 60-mm dish (BD Biosciences). After 24 h, 1 μg of Nt-Myc-KvLQT1 and 1 μg of Myc-tagged C terminus KvLQT1 constructs were added to the dish with TransFectin lipid reagent (Bio-Rad). After 12, 24, 48, and 72 h of transfection, the cells were subjected to sonication in 2% SDS, 250 mm sucrose, 75 mm urea, 1 mm dithiothreitol, and 50 mm Tris-HCl, pH 7.5, and the protease inhibitor mixture (Sigma). After measuring the protein concentration by using BCA protein assay regent (Pierce), equal amounts of proteins were subjected to SDS-PAGE and experiments using Western blotting. The membrane was incubated with primary mouse anti-glyceraldehyde-3-phosphate dehydrogenase monoclonal Ab (1:100; Santa Cruz Biotechnology, Inc.) or rabbit anti-c-Myc polyclonal Ab (1:100; Santa Cruz Biotechnology, Inc.), followed by incubation with appropriate secondary Ab: rabbit anti-mouse IgG HRP-conjugated Ab (1:1000; Dako A/S) or goat anti-rabbit IgG HRP-conjugated Ab (1:1000; Dako A/S).
For the subcellular fraction assay, Nt-Myc-KvLQT1 or GFP-KvLQT1 constructs were transfected into HEK293 cells plated on a poly-d-lysine-coated 60-mm dish (BD Biosciences). After 48 h, cells were subjected to brief sonication in 150 mm NaCl, 1 mm EDTA, and 10 mm Tris-HCl, pH 7.6, containing 1% Triton X-100 and the protease inhibitor mixture. The cell lysates were centrifuged at 15,000 rpm for 15 min at 4 °C to remove cell debris, nuclei, and large particulates. The supernatant portion containing both the membrane proteins and cytosolic proteins was collected and further centrifuged at 40,000 rpm for 30 min at 4 °C to separate the membrane fraction from the cytosolic fraction as described by Wu et al. (24). The supernatant (cytosolic fraction) was removed after the centrifugation, and the pellet (membrane fraction) was subjected to the sonication in 2% SDS, 250 mm sucrose, 75 mm urea, 1 mm dithiothreitol, and 50 mm Tris-HCl, pH 7.5, and the protease inhibitor mixture. After measuring the protein concentration by using BCA reagent, equal amounts of proteins were subjected to the SDS-PAGE and Western blot analysis. The membrane was incubated with primary rabbit anti-c-Myc polyclonal Ab (1:100; Santa Cruz Biotechnology), mouse anti-GFP monoclonal Ab (1:1000; Clontech), or mouse anti-caveolin-1 monoclonal Ab (1:1000; BD Biosciences), followed by the incubation with the appropriate secondary Ab. Signals were visualized, and the densitometric intensities were measured as described earlier.
Immunofluorescence Microscopy
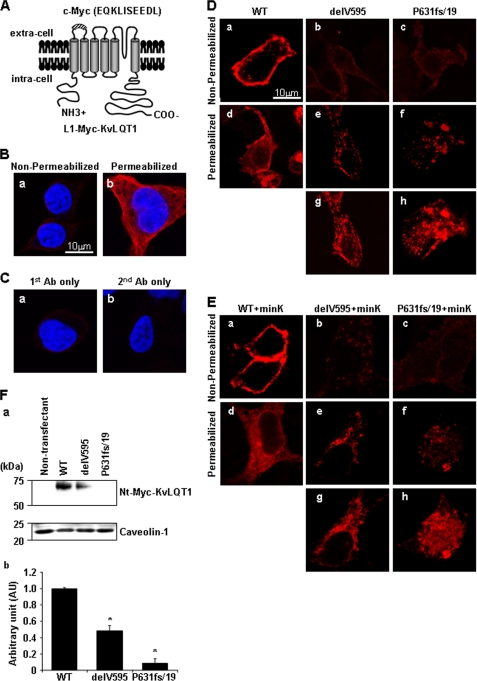
Mutant cDNA fragments containing substitution of AGA with TGA (P631stop mutation) or insertion of CC (P631fs2/34 mutation) were obtained by the primer-directed mutagenesis method from the KvLQT1-WT construct. We also constructed the P631fs/19-mutant carrying A633AA + R646LR, R633GR + A646AA, or A633AA + A646AA by primer-directed mutagenesis from the KvLQT1-P631fs/19 construct. All of the constructs were cloned into the pcDNA3.1(+) vector (Invitrogen). To introduce a c-Myc epitope (EQKLISEEDL) into the extracellular linker 1 (L1) between the S1 and S2 after the position of aa 146 of each KvLQT1 construct, we used a recombination PCR, and the PCR fragment was introduced into the pcDNA3.1(+)- based KvLQT1 constructs (L1-Myc-KvLQT1) (25).
HEK293 cells (7.0 × 104) were seeded onto poly-d-Lysine 8-well culture slides (BD Biosciences), and 24 h later, 0.3 μg of L1-Myc-KvLQT1 constructs or 0.15 μg of L1-Myc-KvLQT1 constructs plus 0.15 μg of MinK constructs were added into wells with 0.6 μl of TransFectin lipid reagent (Bio-Rad). After 48 h, the cells were washed with PBS, fixed in 4% paraformaldehyde and incubated with mouse anti-c-Myc monoclonal Ab (1:200; Santa Cruz Biotechnology, Inc.). After washing with PBS, Alexa Fluor 568 goat anti-mouse IgG1 Ab (1:400; Invitrogen) in 3% bovine serum albumin was applied. To detect the intracellular L1-Myc-KvLQT1 protein, HEK293 cells fixed in 4% paraformaldehyde were permeabilized with 0.15% Triton X-100 in PBS with 3% bovine serum albumin. The permeabilized cells transfected with L1-Myc-KvLQT1 constructs were incubated with the primary and secondary Abs. The permeabilized cells transfected with L1-Myc-KvLQT1 constructs were also incubated with rabbit anti-calnexin polyclonal Ab (1:100; Sigma), Alexa Fluor 568 goat anti-mouse IgG1 Ab, and sheep anti-rabbit IgG fluorescein isothiocyanate-conjugated Ab (1:400; Chemicon, Australia). To assess the efficacy of permeabilization, non-permeabilized and permeabilized cells were incubated with mouse anti-α-tubulin monoclonal Ab (1:1000; Sigma), and with Alexa Fluor 568 goat anti-mouse IgG1 Ab that detected the intracellular protein α-tubulin only in the permeabilized cells. The transfected HEK293 cells were mounted on cover glass using Mowiol 4-88 reagent (Calbiochem) with or without 4′,6-diamidino-2-phenylindole. Images were collected and analyzed with a laser confocal microscope (LSM510, Carl Zeiss (Jena, Germany)).
Luminometric Surface Expression Assay
In the luminometric surface expression assay, we tested L1-Myc-KvLQT1-WT, -delV595, -P631fs/19, -P631stop, -P631fs2/34, and -P631fs/19 derivatives; Nt-Myc-KvLQT1-WT; and Myc(−)-KvLQT1-WT. The assay was performed as previously described (26). Briefly, HEK293 cells were seeded at 2.0 × 105 in each well of a poly-d-lysine-coated multiwell plate (BD Biosciences). After 24 h, the cells were transfected with 0.6 μg of L1-Myc-KvLQT1 constructs, Nt-Myc-KvLQT1-WT, or Myc(−)-KvLQT1-WT along with 0.6 μg of MinK cDNA and 1.2 μl of TransFectin lipid reagent (Bio-Rad). After 48 h of transfection, the living cells were incubated with primary mouse anti-c-Myc monoclonal Ab (1:200; Santa Cruz Biotechnology) at 37 °C for 2 h, followed by fixation in 4% paraformaldehyde. After washing three times with PBS, the cells were incubated with the secondary rabbit anti-mouse IgG HRP-conjugated Ab (1:2000; Dako) in 3% bovine serum albumin for 40 min at room temperature. After washing with PBS, the bottoms of the wells were dipped with Immobilon Western Chemiluminescent HRP substrate (Millipore), and the plate was placed in the luminescent image analyzer LAS-3000mini (Fujifilm). The intensity of chemiluminescence from each well was measured with a densitometer using Multi Gauge version 3.0 (Fujifilm) and expressed as arbitrary units (AU).
Statistical Methods
Numerical data were expressed as means ± S.E. Statistical differences were analyzed using one-way analysis of variance and Student's t test for paired values. Means were compared by independent sample t tests without correction for multiple comparisons. A p value of <0.05 was considered statistically significant.
RESULTS
Identification of KCNQ1 Mutations in LQTS
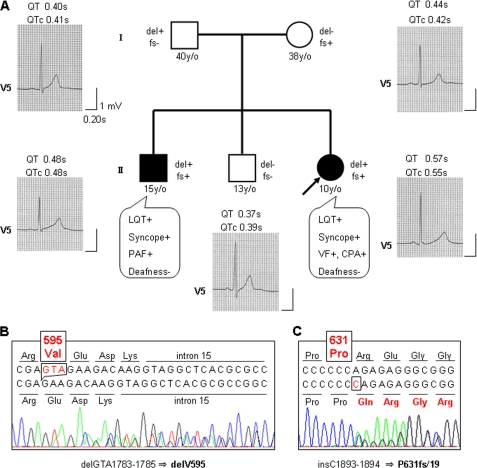
Mutational analysis of cardiac ion channel genes in the proband of LQTS family (Fig. 1A) identified two different KCNQ1 mutations, a deletion of three nucleotides at position 1783–1785 in exon 15, resulting in the predicted deletion of valine 595 (delV595) (Fig. 1B) and an insertion of cytosine between positions 1893 and 1894 in exon 16. This resulted in a frameshift mutation after the 631th residue (proline), which replaced the original 45 amino acids with 19 novel amino acids (P631fs/19) (Fig. 1C). Both mutations were not detected in 180 unrelated healthy Japanese individuals. The former mutation is newly described, whereas the latter mutation has been reported to be a cause of LQTS (27, 28). No mutation in the other LQT-causing genes, KCNH2, SCN5A, KCNE1, KCNE2, and KCNJ2, has been identified in the proband. A family study revealed that the affected eldest brother carried both mutations, whereas the unaffected second eldest brother carried no mutation. Each mutation was found in each parent: delV595 in the father and P631fs/19 in the mother (Fig. 1A). None of the family members suffered hearing loss.
FIGURE 1.
Pedigree of an LQTS family carrying KCNQ1 mutations. A, clinical finding, ECG finding, and KCNQ1 genotype of each member in the LQTS family. The filled square and filled circle indicate affected male and female, respectively. The open squares and open circle represent unaffected males and female, respectively. The arrow indicates the proband. The presence or absence of KCNQ1 mutations, delGTA1783–1785 (delV595) and insC1893–1894 (P631fs/19), is indicated as del+ or del− and fs+ or fs−, respectively. ECGs of V5 lead at rest and its QT and corrected QT (QTc) intervals are shown. The clinical characteristics of each member are also indicated. y/o, years old; LQT, QT prolongation; PAF, paroxysmal atrial fibrillation; VF, ventricular fibrillation; CPA, cardiopulmonary arrest. B and C, direct sequencing data from the proband. Deletion of GTA should result in deletion of valine at codon 595 (B), and insertion of C was predicted to cause a frameshift after proline at codon 631 (C).
Structure and Alignment of C Terminus of KvLQT1 around the Mutations
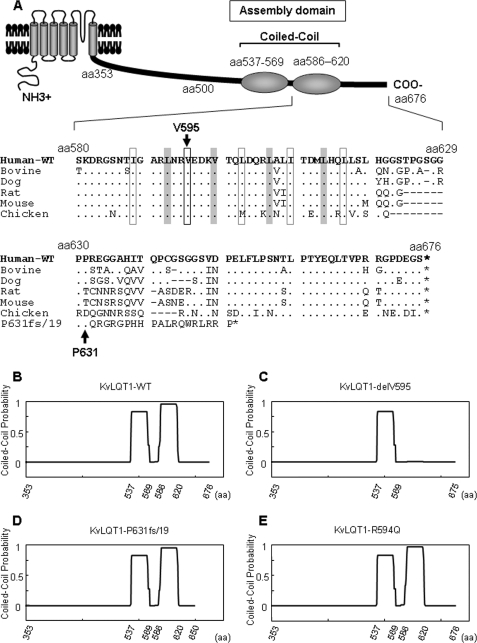
Valine 595 was located in four heptad repeats between aa 588 and 616, and evolutionarily conserved in the KvLQT1 channel among various species (Fig. 2A). Structures of WT, delV595, and P631fs/19 in this region were predicted by using the program COILS along with another LQTS-associated mutation R594Q (28). Two coiled-coil structures were predicted for WT, P631fs/19, and R594Q (Fig. 2, B, D, and E, respectively), whereas complete destruction of the second coiled-coil structure was predicted for delV595 (Fig. 2C).
FIGURE 2.
Alignment of amino acid sequences around the mutations and predicted coiled-coil structure of KvLQT1 cytoplasmic region. A (top), schematic representation of KvLQT1 α-subunit that has two coiled-coil structures (assembly domain) in the C-terminal region. Bottom, the alignment of amino acid sequences around the mutations. Amino acid sequences of human KvLQT1 (from aa 580 to 676) predicted from nucleotide sequences were aligned with those from bovine, dog, rat, mouse, and chicken. The shadowed boxes and black open squares indicate hydrophobic residues constituting heptad repeats. Additional amino acid sequences generated by the P631fs/19 mutation are also aligned. The asterisks, dots, and dashes indicate stop codons, identities to the human KvLQT1, and spaces introduced to obtain maximum homology, respectively. B–E, coiled-coil structure was predicted by using the COILS software for the KvLQT1 C-terminal cytoplasmic region, as shown for WT (B), delV595 (C), P631fs/19 (D), or R594Q (E). The probability at each position is shown as a line between 0 and 1.
Altered Electrophysiological Characters Caused by the Mutations
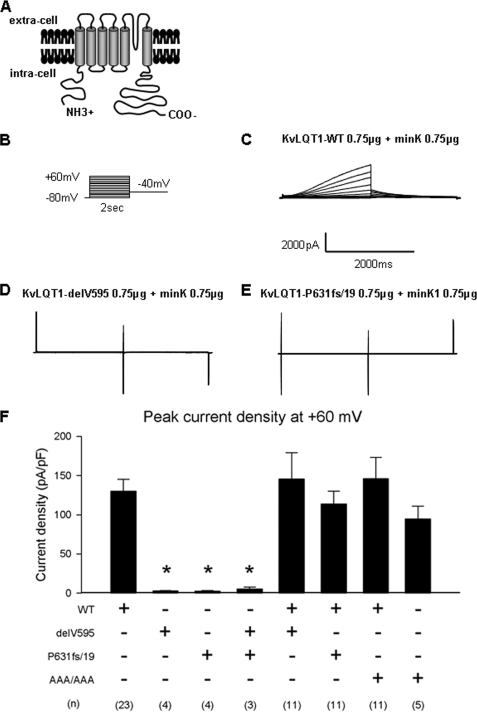
Whole-cell currents were recorded by a step pulse protocol from cells expressing EGFP and full-length KvLQT1-WT or -mutant in the presence of CD8-MinK (Fig. 3, A and B). KvLQT1-WT exhibited a slowly activating outward current similar to IKs (Fig. 3C). On the other hand, both KvLQT1-delV595 and KvLQT1-P631fs/19 produced little to no current (Fig. 3, D and E, respectively). Peak current densities measured at the test pulse of +60 mV were significantly smaller in cells expressing either delV595 (2.8 ± 0.6 pA/pF, n = 4; p < 0.001), P631fs/19 (2.6 ± 0.8 pA/pF, n = 4; p < 0.001), or delV595 + P631fs/19 (5.4 ± 2.2 pA/pF, n = 3; p < 0.001) than WT (130.0 ± 15.4 pA/pF, n = 23) (Fig. 3F). To investigate whether the mutant KvLQT1 channels would exhibit dominant negative suppression effects, KvLQT1-delV595 or KvLQT1-P631fs/19 was co-transfected with an equal amount of KvLQT1-WT in the presence of CD8-MinK. It was found that the co-expression of delV595 or P631fs/19 did not affect the peak current densities of KvLQT1-WT (WT + delV595: 145.8 ± 33.3 pA/pF, p = NS; WT + P631fs/19: 113.9 ± 16.4 pA/pF, p = NS), demonstrating that these mutations did not exert the dominant negative suppression (Fig. 3F). In addition, we examined the functional properties of KvLQT1-P631fs/19 carrying A633AA + A646AA (AAA/AAA) that lacked both ER retention signals of R633GR and R646LR (details are described below). The AAA/AAA mutant exhibited peak current density comparable with that of KvLQT1-WT, either alone (AAA/AAA: 94.6 ± 16.2 pA/pF, n = 5, p = NS) or in combination with KvLQT1-WT (WT + AAA/AAA: 146.1 ± 27.2 pA/pF, n = 11, p = NS) (Fig. 3F).
FIGURE 3.
Whole-cell patch clamp recording of the KvLQT1 channel. A, schematic representation of full-length KvLQT1 channel analyzed in the electrophysiological study. B, the pulse protocol of whole-cell patch clamp recording from CHO-K1 cells transiently transfected with the EGFP-KvLQT1 plasmid (full-length KvLQT1-WT or -mutant) in combination with the CD8-MinK plasmid. C–E, representative current traces for KvLQT1-WT, -delV595, and -P631fs/19, respectively. F, bar graph representing the levels of peak current densities measured at the end of the 2-s test pulse at +60 mV. Combinations of KvLQT1-WT plasmid with KvLQT1-WT or -mutant plasmid for transfection are indicated below. AAA/AAA, the ER retention signal-deficient mutant (P631fs/19 carrying A633AA + A646AA). Data are represented as means ± S.E. *, p < 0.0001 versus WT. The number of experiments for each combination is indicated in parentheses.
Altered KvLQT1 Subunit Binding Caused by the Mutation delV595
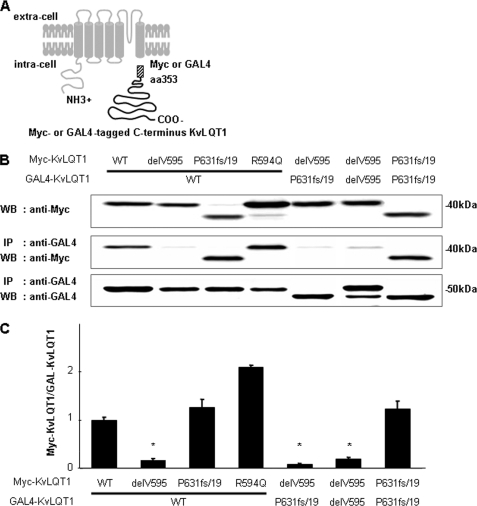
To investigate the functional alterations caused by the KCNQ1 mutations in the subunit assembly, we performed co-IP assays for KvLQT1-WT and -mutants. Myc-tagged C terminus KvLQT1-WT or -mutant (delV595, P631fs/19, or R594Q) constructs were co-transfected into COS-7 cells with GAL4-tagged C terminus KvLQT1-WT or -mutant construct (Fig. 4A). Western blot analyses of whole-cell supernatants demonstrated that these constructs expressed proteins at a similar level (Fig. 4B). To evaluate the binding of each subunit, we measured the amount of Myc-tagged proteins immunoprecipitated by anti-GAL4 monoclonal antibody (Fig. 4, B and C). The binding of C terminus KvLQT1-delV595 with C terminus KvLQT1-WT, -P631fs/19, or -delV595 significantly decreased when compared with the binding that occurred between two C terminus KvLQT1-WTs (0.17 ± 0.03, 0.07 ± 0.02, or 0.19 ± 0.04 AU, respectively, p < 0.001 in each case). Although the binding of C terminus KvLQT1-R594Q with C terminus KvLQT1-WT increased significantly (2.09 ± 0.05 AU, p < 0.001), the binding of C terminus KvLQT1-P631fs/19 with C terminus KvLQT1-WT or -P631fs/19 showed virtually no change. These data indicated that the delV595 mutation impaired the subunit assembly.
FIGURE 4.
Binding of C terminus KvLQT1 proteins. A, schematic representation of constructs used in this experiment. C-terminal cytoplasmic domain of KvLQT1 protein was tagged with Myc or GAL4. B (top), amounts of Myc-tagged C terminus KvLQT1 expressed in transfected COS-7 cells as measured by Western blot analysis of whole-cell supernatants. Middle, amount of Myc-tagged C terminus KvLQT1 after co-IP with GAL4-tagged C terminus KvLQT1. Bottom, amounts of GAL4-tagged C terminus KvLQT1 after the co-IP with GAL4-tagged C terminus KvLQT1. C, ratios of the amount of Myc-tagged C terminus KvLQT1 to that of GAL4-tagged C terminus KvLQT1 after the co-IP. Densitometric data for Myc-tagged C terminus KvLQT1-WT and GAL4-tagged C terminus KvLQT1-WT were arbitrarily defined as 1.0. Data are represented as means ± S.E. (n = 4 for each case). *, p < 0.001 versus WT.
Altered Localization of KvLQT1 Channel Caused by the Mutations
To investigate the functional consequence of the KCNQ1 mutations further, we examined cell surface expression and cytoplasmic distribution of mutant KvLQT1 proteins. HEK293 cells transfected with L1-Myc-KvLQT1-WT, -delV595, or -P631fs/19 constructs (Fig. 5A) were immunostained using anti-c-Myc Ab under non-permeabilized or permeabilized conditions. Under non-permeabilized conditions in which α-tubulin was not detected (Fig. 5B, a), L1-Myc-KvLQT1-WT was expressed at the cell surface (Fig. 5D, a), but L1-Myc-KvLQT1-delV595 showed reduced cell surface expression (Fig. 5D, b). Under the permeabilized condition in which α-tubulin was detected (Fig. 5B, b), L1-Myc-KvLQT1-delV595 represented an abnormal intracellular granular pattern with reduced fluorescence intensity (Fig. 5D, e and g), whereas a diffuse reticular pattern was found for L1-Myc-KvLQT1-WT (Fig. 5D, d). L1-Myc-KvLQT1-P631fs/19 showed abnormal localization similar to that of L1-Myc-KvLQT1-delV595 (Fig. 5D, c, f, and h). The abnormal localizations caused by the mutations were not suppressed by the presence of MinK, which had a critical chaperone-like function (Fig. 5E). To corroborate the reduced cell surface expression of KvLQT1-delV595 and -P631fs/19, Western blot analysis was performed for the membrane protein-enriched fraction from cells transfected with Nt-Myc-KvLQT1 constructs (schematic representation is shown in Fig. 6A). It was demonstrated that the expression of the KvLQT1 channel in the membrane fraction was significantly decreased by the delV595 and P631fs/19 mutations (0.49 ± 0.06 and 0.09 ± 0.06 AU, respectively, p < 0.001 in each case), although a similar amount of caveolin-1, a marker of the membrane proteins, was detected in the analyzed fractions (Fig. 5F). Because the reduced expression level of full-length KvLQT1 in the membrane fraction might be specific to the Myc-tagged KvLQT1, we examined the expression of mutant KvLQT1 proteins in cells transfected with the GFP-tagged full-length KvLQT1 constructs and found again the decreased expression of KvLQT1 carrying each mutation (data not shown).
FIGURE 5.
Distribution of transiently expressed Myc-tagged KvLQT1. A, schematic representation of the KvLQT1 channel containing the extracellular Myc epitope in the Linker 1 region between the membrane-spanning segments 1 and 2. B, HEK293 cells were fixed without (a; non-permeabilized condition) or with (b; permeabilized condition) 0.15% Triton X-100, respectively, and stained with anti-α-tubulin Ab (red) and 4′,6-diamidino-2-phenylindole (blue). C, HEK293 cells were fixed under the permeabilized condition and stained with only anti-c-Myc Ab without secondary Ab (a) or only secondary Ab without anti-c-Myc Ab (b) and 4′,6-diamidino-2-phenylindole (blue). D and E, HEK293 cells transfected with L1-Myc- KvLQT1-WT (a and d), -delV595 (b, e, and g), and -P631fs/19 (c, f, and h) in the absence (D) or presence (E) of MinK were fixed without (a–c; non-permeabilized condition) or with (d–f; permeabilized condition) 0.15% Triton X-100, respectively, and stained with anti-c-Myc Ab. g and h, highly sensitized images taken from e and f, respectively. Surface expression of each KvLQT1-mutant was impaired (b and c), and each mutant showed reduced fluorescence intensity (e and f) with abnormal granular distribution (g and h). Scale bar, 10 μm. F, expression of Myc-tagged KvLQT1 proteins in the membrane-enriched fraction obtained from non-transfectant, KvLQT1-WT-, KvLQT1-delV595-, and KvLQT1-P631fs/19-transfected HEK293 cells. a, upper lanes, detection of Nt-Myc-KvLQT1 proteins by anti-c-Myc Ab. Lower lanes, detection of caveolin-1 showing that the similar amounts of cell membrane protein were subjected to the Western blot analysis. b, densitometric data obtained from a are indicated as -fold expression relative to Nt-Myc-KvLQT1 normalized by caveolin-1 protein. The data for KvLQT1-WT were arbitrarily defined as 1.0. Data are represented as means ± S.E. (n = 4 for each case). *, p < 0.001 versus WT.
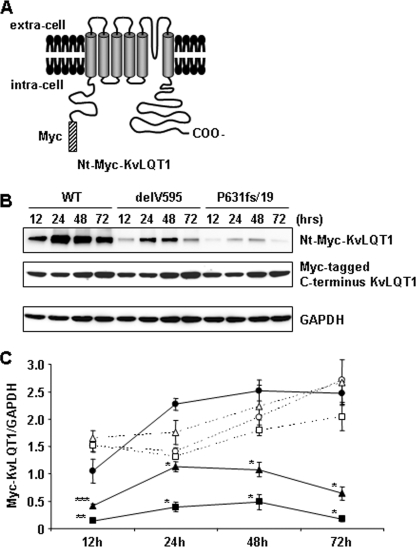
FIGURE 6.
Expression and stability of transiently expressed Myc-tagged KvLQT1. A, schematic representation of the KvLQT1 channel tagged with Myc at the N-terminal cytoplasmic region. B (top), expression of Nt-Myc-KvLQT1-WT, -delV595, and -P631fs/19. Middle, the expression of Myc-tagged C terminus KvLQT1-WT, -delV595, and -P631fs/19. Bottom, expression of glyceraldehyde-3-phosphate dehydrogenase. Whole cell lysates were extracted from HEK293 cells 12, 24, 48, and 72 h after the transfection with equal amounts of each construct and detected with anti-c-Myc Ab or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Ab, followed by secondary antibody. Western blot analysis showed lower expression and decreased stability of Nt-Myc-KvLQT1-delV595 and -P631fs/19 proteins as compared with Nt-Myc-KvLQT1-WT. C, data from representative experiments are shown. Data indicate the expression level of Myc-KvLQT1 proteins normalized by glyceraldehyde-3-phosphate dehydrogenase protein as compared with that of Nt-Myc-KvLQT1-WT protein, which was defined arbitrarily as 1.0. Densitometric data are expressed as AU and represented as means ± S.E. (n = 4–6 for each case). The closed circles, closed triangles, and closed squares represent Nt-Myc-KvLQT1-WT, -delV595, and -P631fs/19, respectively. The open circles, open triangles, and open squares represent Myc-tagged C terminus KvLQT1-WT, -delV595, and -P631fs/19, respectively. *, p < 0.001; **, p < 0.01; ***, p < 0.05 versus Nt-Myc-KvLQT1-WT at the same time point after the transfection.
Altered Stabilization and Subcellular Fraction of KvLQT1 Channel Caused by the Mutations
To investigate further the expressivity and stability of mutant KvLQT1 proteins, we examined the expression of Nt-Myc-KvLQT1-WT, -delV595, and -P631fs/19 proteins (Fig. 6A) in the cells co-transfected with Myc-tagged C terminus KvLQT1-WT, -delV595, and -P631fs/19 (Fig. 4A), respectively. Western blot analyses of total cellular proteins at 12, 24, 48, and 72 h after transfection in HEK293 cells demonstrated that both Nt-Myc-KvLQT1-delV595 and -P631fs/19 mutant proteins expressed statistically lower than the Nt-Myc-KvLQT1-WT protein, whereas the Myc-tagged C terminus KvLQT1 proteins expressed at similar levels (Fig. 6, B and C). In addition, the amount of mutant proteins decreased, especially at 72 h, suggesting that both mutations affected the stability of KvLQT1.
Abnormal Cytoplasmic Localization of Mutant KvLQT1
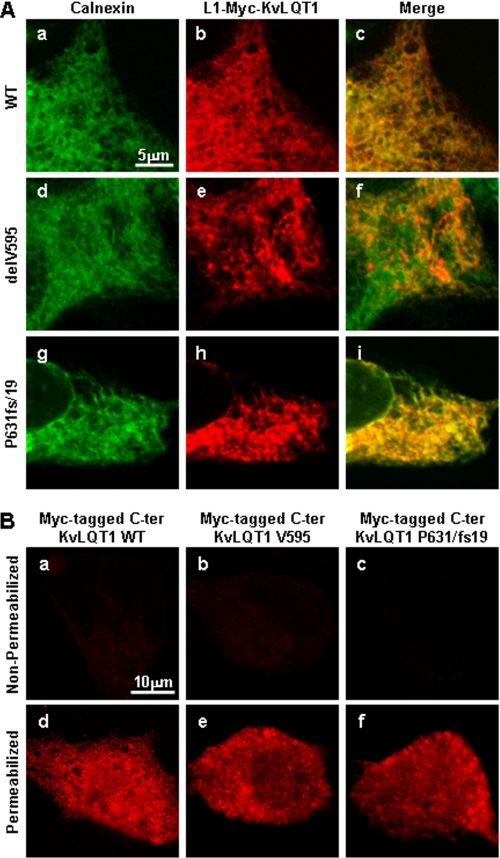
To examine the intracellular distribution of the mutant KvLQT1 proteins, cells transfected with L1-Myc-KvLQT1 constructs were co-immunostained for c-Myc and calnexin (a marker for ER) under the permeabilized conditions. Cytoplasmic reticular distribution of L1-Myc-KvLQT1-WT was found with calnexin (Fig. 7A, a–c). On the other hand, L1-Myc-KvLQT1-delV595 and -P631fs/ 19 showed reticular and abnormal intracellular granular patterns with aggregates, and these aggregates were at least in part co-localized with calnexin (Fig. 7A, d–f and g–i, respectively). These data suggested that both L1-Myc-KvLQT1-delV595 and -P631fs/19 were localized to the ER and adjacent organelle, presumably the Golgi apparatus. In addition, we examined the cellular distribution of Myc-tagged C terminus KvLQT1. It was found that both normal and mutant C terminus KvLQT1 proteins did not express on the cell surface and showed a diffuse intracellular distribution pattern (Fig. 7B).
FIGURE 7.
Intracellular localization of transiently expressed Myc-tagged KvLQT1. A, HEK293 cells transfected with L1-Myc-KvLQT1-WT (a–c), L1-Myc-KvLQT1-delV595 (d–f), and L1-Myc-KvLQT1-P631fs/19 (g–i) were fixed under the permeabilized condition and stained with anti-calnexin (a, d, and g) and anti-c-Myc (b, e, and h) Abs. Merged images are shown (c, f, and i). Myc-tagged mutant full-length KvLQT1 proteins, especially with the P631fs/19 mutation, showed an abnormal granular pattern, which is overlapped by the localization of calnexin. Scale bar, 5 μm. B, HEK293 cells transfected with Myc-tagged C terminus KvLQT1-WT (a and d), KvLQT1-delV595 (b and e), and KvLQT1-P631fs/19 (c and f) were fixed without (non-permeabilized conditions; a–c) or with (permeabilized conditions; d–f) 0.15% Triton X-100 and stained with anti-c-Myc Ab. C terminus KvLQT1 proteins did not express on the cell surface (a–c) and showed similar intracellular localizations (d–f). Scale bar, 10 μm.
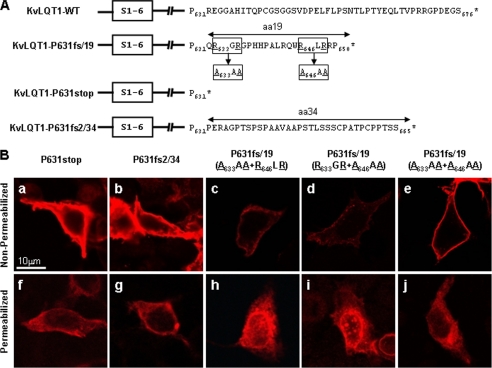
To investigate the cause of the trafficking defect due to the P631fs/19 mutation, we made two additional constructs (Fig. 8A). One was P631stop, in which the 19 amino acids generated by the frameshift were removed, and the other was an artificial frameshift derived from the insertion of two nucleotides, resulting in the addition of 34 amino acids (P631fs2/34). To our surprise, both L1-Myc-KvLQT1-P631stop and -P631fs2/34 were expressed well at the cell surface under the non-permeabilized condition (Fig. 8B, a and b, respectively). Furthermore, these KvLQT1 proteins showed cytoplasmic reticular distribution patterns under the permeabilized condition (Fig. 8B, f and g, respectively), indicating that the 19 residues in P631fs/19 caused the intracellular aggregation and trafficking defect. We hypothesized that there might be a retention signal to ER and/or Golgi in the 19 residues that caused the trafficking defect in the cytoplasm and found that there were R633GR and R646LR sequences (Fig. 8A). Because the RXR motif was acknowledged to be a retention signal to ER, we constructed three variants of L1- Myc-KvLQT1-P631fs/19 carrying A633AA + R646LR, R633GR + A646AA, or A633AA + A646AA (Fig. 8A) and tested for the trafficking. It was discovered that A633AA + R646LR and R633GR + A646AA did not suppress the defects (Fig. 8B, panels c and h and panels d and i), whereas A633AA + A646AA relieved the defects (Fig. 8B, e and j), suggesting that these RXR motifs were responsible for the trafficking defect.
FIGURE 8.
Distribution of transiently expressed Myc-tagged KvLQT1 with or without ER retention signal. A, amino acid sequences of KvLQT1 after codon 631 from WT, P631fs/19, P631stop, and P631fs2/34. Arginine residues in RGR and RLR sequences in KvLQT1-P631fs/19 mutant were substituted by alanine to investigate the role of arginines. S1–6, six membrane-spanning segments of KvLQT1. *, position of stop codon. B, HEK293 cells transfected with L1-Myc-KvLQT1-P631stop (a and f), Myc-KvLQT1-P631fs2/34 (b and g), and Myc-KvLQT1-P631fs/19 with A633AA + R646LR (c and h), R633GR + A646AA (d and i), or A633AA + A646AA (e and j) were fixed without (non-permeabilized conditions) or with (permeabilized conditions) 0.15% Triton X-100 and stained with anti-c-Myc Ab. L1-Myc-KvLQT1-P631stop, -P631fs2/34, and -P631fs/19 with A633AA + A646AA expressed well at the cell surface (a, b, and e) without abnormal granular pattern in the cytoplasm (f, g, and j), respectively. Scale bar, 10 μm.
Quantitative Analysis of Cell Surface Expression of KvLQT1 Channel Protein
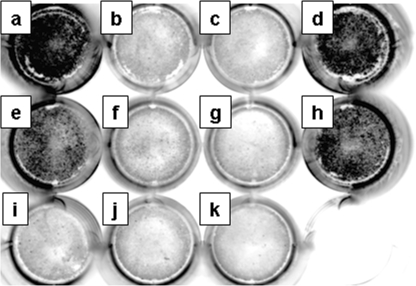
To quantitatively investigate the cell surface expression of the KvLQT1 channel, we developed a luminometric analysis of the L1-Myc-tagged KvLQT1 (Fig. 9). As shown in Table 2, L1-Myc-KvLQT1-delV595, L1-Myc-KvLQT1-P631fs/19, and L1-Myc-KvLQT1-P631fs/19 carrying A633AA + R646LR, and -P631fs/19 carrying R633GR + A646AA, Nt-Myc-KvLQT1-WT, and Myc(−)-KvLQT1-WT showed significantly less expression on the cell surface than the L1-Myc-KvLQT1-WT (p < 0.001 in each case) On the other hand, L1-Myc-KvLQT1-P631stop, -P631fs2/34, and -P631fs/19 carrying A633AA + A646AA were expressed at a similar level as L1-Myc-KvLQT1-WT.
FIGURE 9.
Luminometric assay of surface expression for KvLQT1. Cell surface expression of KvLQT1 from HEK293 transfectants (Figs. 5 and 8) was quantitatively analyzed by a luminometric assay. Representative images of the 12-well cell culture plate with confluent cells transfected with L1-Myc-KvLQT1-WT (a), L1-Myc-KvLQT1-delV595 (b), L1-Myc-KvLQT1-P631fs/19 (c), L1-Myc-KvLQT1-P631stop (d), L1-Myc-KvLQT1-P631fs2/34 (e), L1-Myc-KvLQT1-P631fs/19 carrying A633AA + R646LR (f), L1-Myc-KvLQT1-P631fs/19 carrying R633GR + A646AA (g), L1-Myc-KvLQT1-P631fs/19 carrying A633AA + A646AA (h), Nt-Myc-KvLQT1-WT (i), Myc(−)-KvLQT1-WT (j), or non-transfectant (k) are shown.
TABLE 2.
Quantitative analysis of cell surface expression of L1-Myc-KvLQT1 protein
Data are represented as means ± S.E., n = 9 in each assay.
| Transfected construct | Chemiluminescence intensity | Relative intensity |
|---|---|---|
| AU | ||
| L1-Myc-KvLQT1-WT | 592,820 ± 4,836 | 1.000 ± 0.008 |
| L1-Myc-KvLQT1-delV595 | 102,387 ± 13,760a | 0.173 ± 0.023a |
| L1-Myc-KvLQT1-P631fs/19 | 117,828 ± 11,248a | 0.199 ± 0.019a |
| L1-Myc-KvLQT1-P631stop | 560,114 ± 58,401 | 0.945 ± 0.099 |
| L1-Myc-KvLQT1-P631fs2/34 | 537,019 ± 35,679 | 0.906 ± 0.060 |
| L1-Myc-KvLQT1-P631fs/19(A633AA + R646LR) | 213,523 ± 36,101a | 0.360 ± 0.061a |
| L1-Myc-KvLQT1-P631fs/19(R633GR + A646AA) | 121,504 ± 13,719a | 0.205 ± 0.023a |
| L1-Myc-KvLQT1-P631fs/19(A633AA + A646AA) | 519,126 ± 23,794 | 0.876 ± 0.040 |
| Nt-Myc-KvLQT1-WT | 119,797 ± 7,856a | 0.202 ± 0.013a |
| Myc(−)-KvLQT1-WT | 57,936 ± 11,481a | 0.098 ± 0.019a |
a p < 0.001 versus chemiluminescence intensity from cells transfected with L1-Myc-KvLQT1-WT.
DISCUSSION
In the present study, we found two KCNQ1 mutations, delV595 and P631fs/19, which caused LQTS without hearing loss in two compound heterozygotes (proband and affected brother). No significant QT elongation was observed in either heterozygote (father and mother), implying that both mutations lacked the dominant negative effect. As shown in Fig. 3, both mutants were non-functional by themselves, and the mutant channels lacked the dominant negative suppression properties, consistent with the clinical observations that the heterozygous carriers of neither delV595 nor P631fs/19 mutations showed the LQT phenotype. The most important finding in this study was that both mutations caused intracellular trafficking abnormality due to novel mechanisms, impaired complex formation (delV595), and newly generated ER retention signal (P631fs/19).
Schmitt et al. (11) reported that functional KvLQT1 channel complex is composed of four α-subunits, and the assembly domain in the C terminus of the α-subunit was required for the interaction of each subunit. However, all of the LQTS-associated mutations so far reported within this domain were missense mutations, N576D, T587M, G589D, A590T, R591H, R594Q, D611Y, and L619M, and the impairment of subunit binding was not demonstrated for these mutations. We also predicted the structural changes due to these mutations by using COILS and found that they would not disrupt the coiled-coil structure (data not shown), as was the case with R594Q. Because we showed that R594Q did not affect the subunit binding, it was suggested that these missense mutations might not impair the subunit assembly. On the other hand, the delV595 mutation that was predicted to disrupt the coiled-coil structure was found to impair the subunit binding in this study. In addition, we showed that the delV595 mutation reduced the intracellular expression and affected the intracellular trafficking of the KvLQT1 channel, which, in turn, abolished the IKs current. These observations implied that the subunit assembly might be a prerequisite for the membrane trafficking of the KvLQT1 channel. It remains to be resolved why the impairment of subunit binding caused the intracellular trafficking abnormality, but the subunit assembly domain appeared to act as a module for proper subunit assembly and scaffolding for interaction with other proteins that are required for membrane trafficking and/or regulation of channel processing (10, 25, 29).
On the other hand, although the P631fs/19 mutation was not predicted to disrupt the coiled-coil structures, it caused the trafficking defect and complete loss of electrophysiological function of the KvLQT1 channel. Because RGR and RLR sequences similar to the ER retention signal (RXR motif) were found in the 19 residues generated by the frameshift, it was speculated that these two motifs caused the trafficking defect via increased retention to cytoplasmic organelle (30). Indeed, the trafficking defect of P631fs/19 was suppressed by substitutions of arginine by alanine. Reports have indicated that the RXR motif was used as a retention signal to ER in some channel proteins, including HERG and ATP-sensitive potassium (KATP) channels (31, 32). Although the trafficking defects were reported to cause LQTS, particularly for the HERG channel and in part for the KvLQT1 channel (17, 33, 34), the molecular mechanisms causing the defect were completely different from that of the P631fs/19 mutation found in this study. Interestingly, it was revealed that each ER retention motif was sufficient to cause the trafficking defect, because the disruption of only one motif did not suppress the retention (Fig. 8B).
We found that the KvLQT-P631fs/19 formed the heteromultimer with the KvLQT1-WT subunit (Fig. 4B), and hence it might modify the trafficking of the KvLQT1 channel containing a subunit with the P631fs/19 mutation. However, our data showed that the co-expression of KvLQT-P631fs/19 did not exert significant dominant negative effects on the current density of the KvLQT1-WT channel, indicating that the P631fs/19 mutation was a recessive mutation. In other words, the KvLQT-P631fs/19 subunit could not efficiently retain the KvLQT1-WT subunit in the cytoplasm, although these KvLQT1 subunits could form the heteromultimer. On the other hand, we found that the P631fs/19 mutation severely affected the expression and stability of full-length KvLQT1 protein in the transfectants. Therefore, the number of mutant KvLQT1 subunits was fewer than the number of normal KvLQT1 subunits in the ER, where the subunit assembly occurred. This may be the reason for the P631fs/19 mutation not exerting the dominant negative effects.
Consistent with our data, it has been reported that the individuals carrying the P631fs/19 mutation in the homozygous state but not in the heterozygous state, showed the LQTS phenotype (35). On the other hand, the P631fs/19 mutation has been reported to exhibit the Romano Ward syndrome phenotype (autosomal-dominant LQTS) (27). Why this mutation developed the LQTS phenotype in the heterozygous state in the latter report remains to be solved, but the P631fs/19 mutation might exert its abnormality in the presence of additional factors, facilitating the stability and/or heteromultimer formation in ER.
We found that the expression of full-length KvLQT1 protein was reduced by the delV595 and P631fs/19 mutations, whereas the expression of C terminus KvLQT1 was not affected by the mutations (Fig. 6). In addition, the reduced expression was more prominent with the P631fs/19 mutation than the delV595 mutation. The decreased expression of mutant KvLQT1 proteins might be due to the fact that these mutants showed trafficking abnormality and retention in the ER/Golgi apparatus, because there is a quality control of protein in ER, by which non-native and unassembled subunits of multimeric proteins are degraded by the ubiquitin-proteasome machinery (36, 37).
There are several limitations in this study. First, we used different cell lines (CHO-K1, COS-7, and HEK293) in the electrophysiological study, pull-down experiments, and cell biological experiments, respectively, leaving a possibility that we might be seeing a cell-specific effect. Second, most of the cellular experiments were done in the transiently transfected cells. Therefore, a possibility remains that the data represent the results of overexpression. Third, the direct links among the impaired subunit assembly, retention in ER, and reduced expression/stability of the delV595-KvLQT1 channel were not demonstrated at the molecular level.
In summary, we investigated functional alterations caused by KCNQ1 mutations, delV595 and P631fs/19. These mutations were found to cause trafficking defects via different mechanisms, impaired complex formation and retention to ER, respectively. These observations provide novel insights into the molecular pathogenesis of LQTS.
Acknowledgments
We thank Dr. C. Antzelevitch and Dr. Y. Wu Masonic Medical Research Laboratory) for helpful discussions and comments on the electrophysiological analysis. We also thank Dr. M. Yanokura, M. Emura, A. Nishimura (Tokyo Medical and Dental University), and M. Fukuoka (Hokkaido University) for technical assistance.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, grants from the Japan-Korea Collaboration Program and from the Japan-France Collaboration Program from the Japan Society for Promotion of Science, and research grants from the Ministry of Health, Labor, and Welfare, Japan.
- LQTS
- long QT syndrome
- ECG
- electrocardiogram
- IKs
- slow component of delayed rectifier potassium current
- ER
- endoplasmic reticulum
- IKr
- rapid component of the delayed rectifier potassium current
- QTc
- corrected QT
- WT
- wild-type
- aa
- amino acid(s)
- EGFP
- enhanced green fluorescent protein
- co-IP
- co-immunoprecipitation
- Ab
- antibody
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- AU
- arbitrary units
- pF
- picofarad
- KATP
- ATP-sensitive potassium.
REFERENCES
- 1.Lehnart S. E., Ackerman M. J., Benson D. W., Jr., Brugada R., Clancy C. E., Donahue J. K., George A. L., Jr., Grant A. O., Groft S. C., January C. T., Lathrop D. A., Lederer W. J., Makielski J. C., Mohler P. J., Moss A., Nerbonne J. M., Olson T. M., Przywara D. A., Towbin J. A., Wang L. H., Marks A. R. (2007) Circulation 116, 2325–2345 [DOI] [PubMed] [Google Scholar]
- 2.Neyroud N., Tesson F., Denjoy I., Leibovici M., Donger C., Barhanin J., Fauré S., Gary F., Coumel P., Petit C., Schwartz K., Guicheney P. (1997) Nat. Genet. 15, 186–189 [DOI] [PubMed] [Google Scholar]
- 3.Goldenberg I., Moss A. J. (2008) J. Am. Coll. Cardiol. 51, 2291–2300 [DOI] [PubMed] [Google Scholar]
- 4.Wang Q., Curran M. E., Splawski I., Burn T. C., Millholland J. M., VanRaay T. J., Shen J., Timothy K. W., Vincent G. M., de Jager T., Schwartz P. J., Toubin J. A., Moss A. J., Atkinson D. L., Landes G. M., Connors T. D., Keating M. T. (1996) Nat. Genet. 12, 17–23 [DOI] [PubMed] [Google Scholar]
- 5.Curran M. E., Splawski I., Timothy K. W., Vincent G. M., Green E. D., Keating M. T. (1995) Cell 80, 795–803 [DOI] [PubMed] [Google Scholar]
- 6.Napolitano C., Priori S. G., Schwartz P. J., Bloise R., Ronchetti E., Nastoli J., Bottelli G., Cerrone M., Leonardi S. (2005) JAMA 294, 2975–2980 [DOI] [PubMed] [Google Scholar]
- 7.Yang W. P., Levesque P. C., Little W. A., Conder M. L., Shalaby F. Y., Blanar M. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4017–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 9.Schmitt N., Calloe K., Nielsen N. H., Buschmann M., Speckmann E. J., Schulze-Bahr E., Schwarz M. (2007) Biochem. Biophys. Res. Commun. 358, 304–310 [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Marquardt M. L., Tester D. J., Sampson K. J., Ackerman M. J., Kass R. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20990–20995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt N., Schwarz M., Peretz A., Abitbol I., Attali B., Pongs O. (2000) EMBO J. 19, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jespersen T., Membrez M., Nicolas C. S., Pitard B., Staub O., Olesen S. P., Baró I., Abriel H. (2007) Cardiovasc. Res. 74, 64–74 [DOI] [PubMed] [Google Scholar]
- 13.Li X., Xu J., Li M. (1997) J. Biol. Chem. 272, 705–708 [DOI] [PubMed] [Google Scholar]
- 14.Schulteis C. T., Nagaya N., Papazian D. M. (1998) J. Biol. Chem. 273, 26210–26217 [DOI] [PubMed] [Google Scholar]
- 15.Phartiyal P., Jones E. M., Robertson G. A. (2007) J. Biol. Chem. 282, 9874–9882 [DOI] [PubMed] [Google Scholar]
- 16.Gong Q., Anderson C. L., January C. T., Zhou Z. (2002) Am. J. Physiol. Heart Circ. Physiol. 283, H77–H84 [DOI] [PubMed] [Google Scholar]
- 17.Anderson C. L., Delisle B. P., Anson B. D., Kilby J. A., Will M. L., Tester D. J., Gong Q., Zhou Z., Ackerman M. J., January C. T. (2006) Circulation 113, 365–373 [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z., Gong Q., Epstein M. L., January C. T. (1998) J. Biol. Chem. 273, 21061–21066 [DOI] [PubMed] [Google Scholar]
- 19.Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 20.Lupas A., Van Dyke M., Stock J. (1991) Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 21.Aizawa Y., Ueda K., Scornik F., Cordeiro J. M., Wu Y., Desai M., Guerchicoff A., Nagata Y., Iesaka Y., Kimura A., Hiraoka M., Antzelevitch C. (2007) J. Cardiovasc. Electrophysiol. 18, 972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inagaki N., Hayashi T., Arimura T., Koga Y., Takahashi M., Shibata H., Teraoka K., Chikamori T., Yamashina A., Kimura A. (2006) Biochem. Biophys. Res. Commun. 342, 379–386 [DOI] [PubMed] [Google Scholar]
- 23.Arimura T., Hayashi T., Matsumoto Y., Shibata H., Hiroi S., Nakamura T., Inagaki N., Hinohara K., Takahashi M., Manatsu S. I., Sasaoka T., Izumi T., Bonne G., Schwartz K., Kimura A. (2007) Biochem. Biophys. Res. Commun. 357, 162–167 [DOI] [PubMed] [Google Scholar]
- 24.Wu L., Yong S. L., Fan C., Ni Y., Yoo S., Zhang T., Zhang X., Obejero-Paz C. A., Rho H. J., Ke T., Szafranski P., Jones S. W., Chen Q., Wang Q. K. (2008) J. Biol. Chem. 283, 6968–6978 [DOI] [PubMed] [Google Scholar]
- 25.Kanki H., Kupershmidt S., Yang T., Wells S., Roden D. M. (2004) J. Biol. Chem. 279, 33976–33983 [DOI] [PubMed] [Google Scholar]
- 26.Margeta-Mitrovic M. (2002) Methods 27, 311–317 [DOI] [PubMed] [Google Scholar]
- 27.Neyroud N., Richard P., Vignier N., Donger C., Denjoy I., Demay L., Shkolnikova M., Pesce R., Chevalier P., Hainque B., Coumel P., Schwartz K., Guicheney P. (1999) Circ. Res. 84, 290–297 [DOI] [PubMed] [Google Scholar]
- 28.Splawski I., Shen J., Timothy K. W., Lehmann M. H., Priori S., Robinson J. L., Moss A. J., Schwartz P. J., Towbin J. A., Vincent G. M., Keating M. T. (2000) Circulation 102, 1178–1185 [DOI] [PubMed] [Google Scholar]
- 29.Robinson J. M., Deutsch C. (2005) Neuron 45, 223–232 [DOI] [PubMed] [Google Scholar]
- 30.Michelsen K., Yuan H., Schwappach B. (2005) EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phartiyal P., Sale H., Jones E. M., Robertson G. A. (2008) J. Biol. Chem. 283, 3702–3707 [DOI] [PubMed] [Google Scholar]
- 32.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 33.Delisle B. P., Anson B. D., Rajamani S., January C. T. (2004) Circ. Res. 94, 1418–1428 [DOI] [PubMed] [Google Scholar]
- 34.Wilson A. J., Quinn K. V., Graves F. M., Bitner-Glindzicz M., Tinker A. (2005) Cardiovasc. Res. 67, 476–486 [DOI] [PubMed] [Google Scholar]
- 35.Novotny T., Kadlecova J., Janousek J., Gaillyova R., Bittnerova A., Florianova A., Sisakova M., Toman O., Chroust K., Papousek I., Spinar J. (2006) Pacing Clin. Electrophysiol. 29, 1013–1015 [DOI] [PubMed] [Google Scholar]
- 36.Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]