Abstract
miR-210 is a key player of cell response to hypoxia, modulating cell survival, VEGF-driven endothelial cell migration, and the ability of endothelial cells to form capillary-like structures. A crucial step in understanding microRNA (miRNA) function is the identification of their targets. However, only few miR-210 targets have been identified to date. Here, we describe an integrated strategy for large-scale identification of new miR-210 targets by combining transcriptomics and proteomics with bioinformatic approaches. To experimentally validate candidate targets, the RNA-induced silencing complex (RISC) loaded with miR-210 was purified by immunoprecipitation along with its mRNA targets. The complex was significantly enriched in mRNAs of 31 candidate targets, such as BDNF, GPD1L, ISCU, NCAM, and the non-coding RNA Xist. A subset of the newly identified targets was further confirmed by 3′-untranslated region (UTR) reporter assays, and hypoxia induced down-modulation of their expression was rescued blocking miR-210, providing support for the approach validity. In the case of 9 targets, such as PTPN1 and P4HB, miR-210 seed-pairing sequences localized in the coding sequence or in the 5′-UTR, in line with recent data extending miRNA targeting beyond the “classic” 3′-UTR recognition. Finally, Gene Ontology analysis of the targets highlights known miR-210 impact on cell cycle regulation and differentiation, and predicts a new role of this miRNA in RNA processing, DNA binding, development, membrane trafficking, and amino acid catabolism. Given the complexity of miRNA actions, we view such a multiprong approach as useful to adequately describe the multiple pathways regulated by miR-210 during physiopathological processes.
Introduction
miRNAs are 21–23-nucleotide non-protein coding RNA molecules that regulate the stability and/or the translational efficiency of target messenger RNAs (1–3). Mature miRNAs are loaded into the RNA-induced silencing complex (RISC)3 and mediate the translational inhibition of target mRNA, albeit a few opposing examples have been described as well (4–6). The rules that guide miRNA-mRNA interaction are very complex and still under investigation. However, the current paradigm states that a Watson-Crick pairing between the mRNA and the 5′-region of the miRNA centered on nucleotides 2–7, termed “seed sequence,” is required for miRNA-mediated inhibition (7). RISC-miRNA complexes can move the mRNAs they bind to the P-bodies, which are specialized cytoplasmic compartments where translational repression and mRNA turnover is thought to occur (8). Because P-bodies contain many enzymes involved in mRNA exonucleolitic degradation, miRNAs may also have a secondary quantitative inhibitory effect on mRNAs. A role for miRNAs in mRNA destabilization is also suggested by studies reporting robust correlations between the levels of miRNAs and the message of multiple predicted or validated targets (9–11).
miR-210 is currently regarded as “master miRNA” of hypoxic response, because it was found up-regulated by hypoxia in all the cell types tested to date (12). Previously, we characterized miR-210 regulation and its functional relevance in endothelial cell (EC) response to hypoxia (13). We found that miR-210 increases EC tubulogenesis and migration, whereas miR-210 blockade in the presence of hypoxia inhibits these processes and induces apoptosis. These effects are mediated, at least in part, by the direct inhibition of the receptor-tyrosine kinase ligand Ephrin-A3 (EFNA3). Furthermore, other miR-210 targets have been recently identified: E2F3, NPTX1, RAD52, ACVR1B, MNT, CASP8AP2, FGFRL1, and HOXA-1 and -9 (14–20), suggesting multiple roles of this miRNA in the cellular adaptation to low oxygen. Considering that the function of each miRNA is mediated by the modulation of a subset-specific mRNA targets and that each miRNA is thought to regulate hundreds of mRNAs (21), the identification of new miR-210 targets represents an important step in elucidating its function. Several tools for predicting miRNA targets have been developed. However, the overlap between their output is rather limited (7), raising the distinct possibility of false positive predictions. On the other hand, genuine targets could be excluded by a cut-off that depends on base pairing type, as well as by conservation or 3′-UTR localization requirements. Indeed, many reports have recently demonstrated that certain miRNAs can inhibit mRNAs subsequent to targeting of their coding regions or of the 5′-UTR (5, 6, 22–24). To overcome these limitations of bioinformatic predictions, several biochemical approaches for target identification have been developed (25–27).
In this study, candidate miR-210 targets have been selected using a combination of bioinformatic, proteomic, and transcriptomic approaches, using ECs with manipulated levels of miR-210. These potential targets were validated based on their association to RISC complexes enriched for miR-210, leading to the identification of a panel of novel targets.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human umbilical vein EC (HUVEC; Clonetics) were grown in EGM-2 (Lonza) containing 2% FBS. HEK-293, and MCF7 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Hypoxic conditions were maintained in an InVivo200 hypoxia work station (Biotrace International) with oxygen maintained at 0.2%.
miRNA Blockade and Overexpression
Locked nucleic acid (LNA) oligonucleotides against miR-210 or a control scramble sequence (Exiqon) were transfected by siRNA Transfection Reagent (Santa Cruz Biotechnology), following manufacturer instructions, in 40% confluent HUVEC or HEK-293 (4 × 103 cells/cm2) at the final concentration of 40 nm. After 16 h, cells were re-fed with fresh medium, and experiments were performed 24 h later. miR-210 mimic or a control scramble sequence (Ambion) were transfected in MCF7 using siPORT NeoFX transfection agent (Ambion) at the final concentration of 20 nm. Transient miR-210 overexpression was also obtained by transfection of pSUPER-pre-miR-210 using Fugene6 (Roche Applied Science). To obtain stable miR-210-overexpressing cells, HUVEC were infected by retroviral vectors bearing a pre-miR-210 sequence or a control scramble sequence, as previously described (13).
Transcriptomics
Gene expression profiles were measured using Affymetrix HG-U133A-plus GeneChips and were detailed under supplemental materials. The complete dataset of our study is available on the NCBI Gene Expression Omnibus data base, GEO series GSE16962.
Gene Ontology and Pathway Analysis
Differentially expressed Gene Ontology classes and BioCarta pathways were evaluated using the Babelomics suite (28).
miRNAs Target Prediction
Bioinformatic prediction of target genes and miRNA binding sites was performed using: PicTar (version 2006), TargetScan (version 4.2), microRNA.org (September 2008 release), MirBase Targets (version 5), DIANA-microT (v3.0 and v4.0), and EIMMo (7). Hypothetical proteins were not considered for further analysis.
Immunoprecipitation of c-Myc-Ago2-containing RISC
Transfected cells were harvested in 1 ml/p15 dish of cold lysis buffer (150 mm KCl, 25 mm Tris-HCl, pH 7.4, 5 mm EDTA, 0.5% Nonidet P-40) supplemented with 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, protease inhibitors mixture tablets (Roche Applied Science), and 100 units/ml of RNasin Plus (Promega). After 30 min at 4 °C, samples were precleared for 10 min with 75 μl/plate of A/G-agarose beads (Santa Cruz Biotechnology) and spun at 4 °C for 30 min at 20,000 × g in a microcentrifuge. Next, lysates were incubated at 4 °C with 2.5 μg/plate of anti-c-Myc antibody (9E10, Santa Cruz Biotechnology) for 3 h and then 50 μl/plate of A/G-agarose beads were added to each sample. After 1 h, immunocomplexes were washed two times with lysis buffer and resuspended in 200 μl of TRIzol (Invitrogen). RNA was purified and specific mRNAs were measured by quantitative real-time PCR (qPCR). Average values of 4 genes that preliminary experiments showed to be RISC-associated but not miR-210 targets (B2M, GAPDH, GUSB, and RPL13) were used for normalization.
mRNA Conservation
The conservation of miR-210 seed sites was evaluated using annotated human genes and their orthologs, as defined by UCSC whole-genome alignments (29).
Statistical Analysis
Variables were analyzed by both Student's t test and one way analysis of variance, and a p ≤ 0.05 was deemed statistically significant, unless differently specified. Values are expressed as ± S.E.
Additional Information
See supplemental materials for information on plasmids, transcriptomics, proteomics (two-dimensional electrophoresis, and mass spectrometry), Western blotting, luciferase assays, miRNA, and mRNA quantification.
RESULTS
Proteomic Profile of miR-210-overexpressing Cells
To identify the biological consequences of miR-210 induction, we first analyzed the proteome of miR-210-overexpressing cells. HUVEC overexpressing pre-miR-210 were generated by retroviral infection, yielding a selected population that expressed mature miR-210 levels comparable with those observed in hypoxic cells (13). HUVEC overexpressing miR-210 or a scramble sequence as control were analyzed by two-dimensional electrophoresis (2-DE). Comparison of the 2-DE maps revealed statistically significant differences between the two groups in the expression of 28 proteins (supplemental Fig. SF1), which were excised from the 2-DE gels and identified by mass spectrometry (supplemental Table ST1). Results are summarized in Table 1. Whereas the 17 up-regulated proteins were most likely indirect targets, the 11 down-modulated proteins may be inhibited by miR-210 either directly or indirectly.
TABLE 1.
Proteins modulated by miR-210
| Gene | Description | Spot | MW kDa/pIa | Fold increaseb | S.E. | p |
|---|---|---|---|---|---|---|
| Up-regulated | ||||||
| CNN2 | Calponin-2 | 34 | 33.7/6.94 | 2.15 | 0.12 | 0.002 |
| EHD1 | EH-domain-containing protein 1 | 62 | 60.6/6.35 | 1.75 | 0.07 | 0.002 |
| EIF5A | Eukaryotic translation initiation factor 5A | 6 | 16.8/5.07 | 3.29 | 0.34 | 0.008 |
| GIPC1 | RGS19-interacting protein 1 | 75 | 36.0/5.90 | 1.64 | 0.11 | 0.037 |
| HNRPL | Heterogeneous nuclear ribonucleoprotein L | 91 | 64.1/8.46 | 1.52 | 0.07 | 0.035 |
| HSPD1 | 60 kDa heat shock protein | 70 | 61.1/5.70 | 1.69 | 0.15 | 0.030 |
| KHSRP | Far upstream element-binding protein 2 | 15 | 73.1/6.84 | 2.69 | 0.30 | 0.009 |
| KHSRP | Far upstream element-binding protein 2 | 81 | 73.1/6.84 | 1.59 | 0.05 | 0.041 |
| LMNA | Lamin A/C | 1 | 74.1/6.57 | 5.14 | 0.60 | 0.007 |
| LMNA | Lamin A/C | 39 | 74.1/6.57 | 2.04 | 0.21 | 0.026 |
| LOC403161 | Nicotinate phosphoribosyltransferase-like protein | 58 | 57.5/5.51 | 1.80 | 0.12 | 0.007 |
| MCCC2 | Methylcrotonoyl-CoA carboxylase β-chain | 53 | 61.3/7.58 | 1.85 | 0.24 | 0.046 |
| MTHFD1 | C-1-tetrahydrofolate synthase | 21 | 101.5/6.89 | 2.59 | 0.27 | 0.012 |
| NUP54 | Nucleoporin 54 kDa | 87 | 54/6.53 | 1.54 | 0.10 | 0.030 |
| PCBP1 | Poly(rC)-binding protein 1 | 92 | 37.5/6.66 | 1.51 | 0.03 | 0.002 |
| PDHA1 | Pyruvate dehydrogenase E1 component α-subunit | 100 | 43.3/8.35 | 1.43 | 0.11 | 0.036 |
| SARS | Seryl-tRNA synthetase | 68 | 58.2/8.35 | 1.71 | 0.14 | 0.020 |
| TUBA3 | Tubulin α-3 chain | 87 | 49.9/4.96 | 1.54 | 0.10 | 0.030 |
| UCHL1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 79 | 24.8/5.33 | 1.61 | 0.16 | 0.043 |
| Down-regulated | ||||||
| BCAS2 | Breast carcinoma amplified sequence 2 | 41 | 26.1/5.48 | 0.50 | 0.06 | 0.023 |
| CBX1 | Chromobox protein homolog 1 | 28 | 21.4/4.85 | 0.44 | 0.06 | 0.020 |
| CTSB | Cathepsin B precursor | 60 | 37.8/5.88 | 0.57 | 0.06 | 0.029 |
| CTSB | Cathepsin B precursor | 69 | 37.8/5.88 | 0.59 | 0.05 | 0.024 |
| EIF1AY | Eukaryotic translation initiation factor 1A. Y-chromosomal | 97 | 16.5/5.07 | 0.68 | 0.05 | 0.031 |
| NDUFV2 | NADH-ubiquinone oxidoreductase 24 kDa subunit | 80 | 27.4/8.22 | 0.62 | 0.05 | 0.040 |
| NES | Nestin | 36 | 177.4/4.35 | 0.47 | 0.06 | 0.028 |
| P4HB | Protein disulfide-isomerase precursor | 17 | 57.1/4.76 | 0.38 | 0.07 | 0.030 |
| PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 | 72 | 49.9/5.88 | 0.60 | 0.06 | 0.035 |
| SEC13L1 | SEC13-related protein | 29 | 35.5/5.22 | 0.45 | 0.07 | 0.038 |
| TCEB2 | Transcription elongation factor B polypeptide 2 | 78 | 13.3/4.73 | 0.62 | 0.04 | 0.033 |
| VIM | Vimentin | 10 | 53.6/5.06 | 0.33 | 0.07 | 0.029 |
| VIM | Vimentin | 2 | 53.6/5.06 | 0.21 | 0.01 | 0.000 |
| VIM | Vimentin | 12 | 53.6/5.06 | 0.37 | 0.06 | 0.045 |
a kDa/isoelectric point.
b Ratio of normalized spot density from cells overexpressing miR-210 versus control cells.
Transcriptomic Profile Changes Induced by miR-210 Overexpression or Blockade
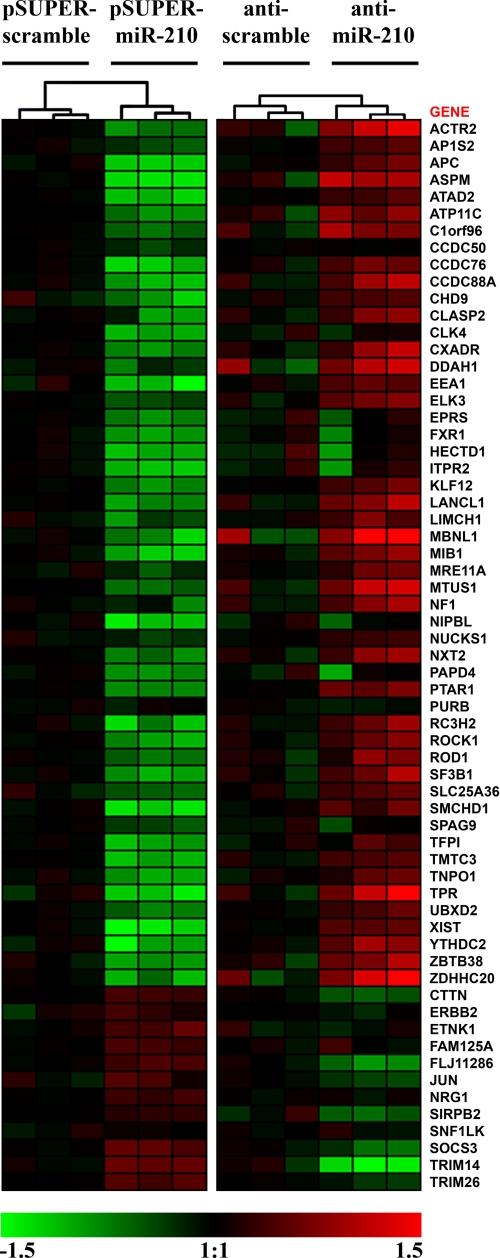
To further investigate the effects of miR-210 expression, we analyzed gene expression modifications induced by miR-210 modulation using Affymetrix technology. To test the effect of miR-210 increase, total RNAs derived from HUVEC overexpressing miR-210 or a scramble sequence under the control of a retroviral vector were analyzed. Using a reciprocal approach, to determine the effect of miR-210 inhibition, HUVEC in which miR-210 activity was blocked by a complementary LNA oligonucleotide, that binds with high affinity and specificity to miR-210, were analyzed. A scrambled LNA was used as control. To limit the number of false positive, only transcripts displaying inverse modulation following miR-210 overexpression or miR-210 blockade were further analyzed. For instance, a gene repressed by miR-210 up-regulation was considered only if it was induced upon miR-210 inhibition as well. When the complementary lists derived from miR-210 up- and down-modulated datasets (supplemental Tables ST2 and ST3) were intersected, we identified a subset of 63 genes (inversely-modulated genes, Fig. 1). The principal effect of a miRNA is the inhibition of the translation of the target mRNA. However, it was demonstrated that the repression of many miRNA targets is frequently associated with their destabilization (3). Among the inversely modulated genes, 51 were both down-modulated by miR-210 overexpression and up-regulated by miR-210 blockade, representing potential direct targets.
FIGURE 1.
Genes modulated by miR-210. Heat map representing mRNAs modulated by miR-210 overexpression (pSUPER-miR-210) that displayed an inverse modulation following miR-210 inhibition (anti-miR-210) (n = 3). Gene: official gene symbol. Green, down-modulation; Red, up-regulation. All differences are statistically significant (p < 0.005).
Over-represented Gene Ontology Categories among miR-210-modulated Genes
We next evaluated the cellular processes affected by miR-210, both directly and indirectly, according to Gene Ontology (GO) (30). We analyzed the 63 inversely modulated genes identified by transcriptomics and the 28 modulated proteins identified by proteomics, searching for enriched GO categories. Results in supplemental Table ST4 show that miR-210-modulated genes were involved in RNA processing, in DNA binding, in differentiation and development, in the transport through cytoplasmic membranes and through the nucleus, and in the amino acids catabolism. We also analyzed gene interactions within pathways (supplemental Table ST4), and we found that “Hypoxia-Inducible Factor in the Cardiovascular System” was the pathway that displayed the highest enrichment. Other pathways modulated by miR-210 were: ATM pathway, involved in DNA repair; FAS and TNFR1 apoptotic pathways; integrin and agrin pathways, that both regulate proliferation, motility, and cellular shape, by modulating cytoskeleton organization and actin polymerization.
Potential Targets Identified by Proteomics and Transcriptomics Are Enriched for miR-210 Seed-pairing Sites
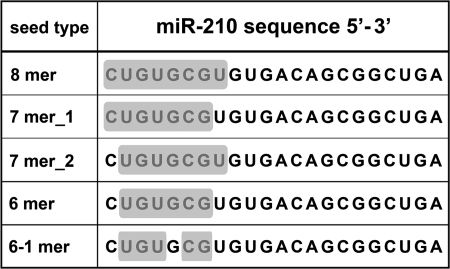
To discriminate between direct and indirect miR-210 targets, we took advantage of the many available target-prediction software. Surprisingly, none of the 62 candidates (51 from transcriptomic and 11 from proteomic analyses) was recognized as a direct target by PicTar, TargetScan, and Diana-microT algorithms. Other prediction softwares recognized few candidates as direct miR-210 targets, but their number remained low (BCAS2, SF3B1, and SMCHD1 by Sanger miRBase; AP1S2, KLF12, and ROD1 by microRNA.org; KLF12 by EIMMo). It is worth noting that these software limit the search of miRNA-mRNA interactions to the 3′-UTR regions of mRNA. Considering that many reports have demonstrated the presence of functional miRNA pairing sites both in the coding sequence and in the 5′-UTR of mRNAs (5, 6, 22–24, 27), we performed a low-stringency miR-210 seed-pairing search in the whole sequence of miR-210 potential targets and, as control, in a group of randomly picked genes that were not modulated after miR-210 overexpression (non-target genes, supplemental Table ST2). In keeping with previous observations (7), different kinds of seed-target associations were considered (Fig. 2). Results showed that miR-210 potential targets were significantly enriched for miR-210 putative binding sites, compared with the non-target list (1.33, p < 0.04). As a negative control, we screened both target and non-target gene lists for potential binding sites to miR-126, an endothelial-specific miRNA that is abundantly expressed in HUVEC (13), and no significant differences were found (data not shown). As summarized in supplemental Table ST5, 42 of 62 potential targets contained one or more miR-210 seed-pairing sites, 29 genes displayed one or more pairing motifs in the coding sequence, and seed-pairing sequences were also found in the 5′-UTR of 5 potential targets. (Sequence alignments are schematized in supplemental Fig. SF2.)
FIGURE 2.
miR-210 seed-pairing. The figure shows highlighted with a gray background the different types of miR-210 seed sequences that were used for a low-stringency search of miR-210 seed-pairing among the differentially expressed genes identified by transcriptomics and proteomics.
Confirmation of Potential Targets by Immunoprecipitation of miR-210-enriched RISC
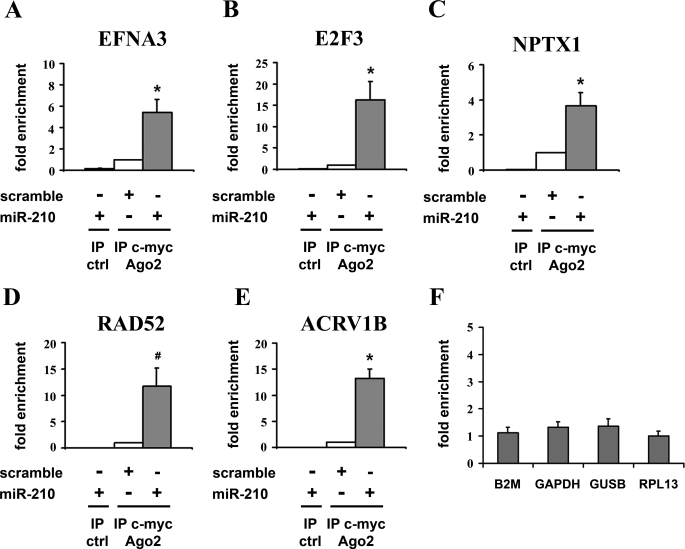
To validate experimentally the potential targets identified with the low-stringency miR-210 seed-pairing search, we set up a biochemical assay based on the immunoprecipitation of RISC complexes enriched for miR-210 and its targets. To this aim, we used a c-Myc-tagged allele of Ago2, a core component of the RISC complex, associating both with miRNAs and their mRNA targets (31, 32). Easily transfectable HEK-293 cells were co-transfected with expression vectors for miR-210 and c-Myc-Ago2, yielding cells enriched of miR-210/c-Myc-Ago2-containing RISC complexes. Consequently, we expected miR-210 endogenous targets to be over-represented in the RISC of transfected cells. Then, we immunoprecipitated c-Myc-Ago2 and evaluated the levels of co-immunoprecipitated mRNAs by qPCR. Background controls were represented by c-Myc immunoprecipitates derived from cells transfected with miR-210 but not c-Myc-Ago2. To validate this approach, five well established miR-210 target genes were assayed (13–17). Fig. 3 shows that EFNA3, E2F3, NPTX1, RAD52, and ACVR1B were significantly enriched in immunoprecipitates of miR-210-overexpressing cells compared with cells transfected with a scramble sequence. Conversely, B2M, GAPDH, GUSB, and RPL13, that according to transcriptomic and target-prediction analysis were not miR-210 targets, did not display any enrichment, and thus were used for normalization (Fig. 3F). We next performed the same assay evaluating both the 42 seed-pairing potential targets and the whole list of predicted miR-210 target genes according to Pictar and TargetScan. A selection of cardiovascular-related genes identified as predicted targets by microRNA.org was assayed as well (see supplemental Table ST6 for the complete list of tested genes). Only genes enriched significantly (p < 0.05) more than 2-fold were considered as positives. Furthermore, to minimize the number of false positives, each gene was re-assayed with an independent primer couple. It is worth noting that, with few exceptions such as NCAM and EIF1AY, all the tested genes were easily detectable and similarly expressed in both HEK-293 and HUVEC (median Delta Ct 0.4 ± 0.3, supplemental Table ST7) and that the background control immunoprecipitations displayed low-to-undetectable signals for all the assayed genes (data not shown). As expected, target prediction algorithms displayed a >50% validation rate (46% for Pictar and 53% for TargetScan). MiR-210 seed-pairing potential targets identified by transcriptomics and proteomics displayed a lower, but robust validation rate (16/42, 38%). Intriguingly, for most of them (68%), miR-210 seed-pairing sites were in the coding sequence, and CBX1 and TNPO1 even contained miR-210 seed-pairing sites in the 5′-UTR. Table 2 shows only genes enriched in miR-210-loaded RISCs, while the complete dataset is shown in supplemental Table ST6. miR-210 seed-pairing sites of the validated targets displayed a poor degree of conservation: whereas all but 2 were conserved in chimp, only 4 were conserved also in mouse and/or rat (supplemental Fig. SF3). The GO analysis of the cellular processes affected by the 31 newly identified targets, along with 5 identified previously, showed a significant enrichment of cell cycle-related GO classes, further demonstrating a role for miR-210 in growth control (supplemental Table ST8).
FIGURE 3.
miR-210 known targets were enriched in miR-210-containing RISC. HEK-293 cells were co-transfected with expression vectors for either miR-210 or a scramble sequence and c-Myc-Ago2. Then, c-Myc or control antibody were used to immunoprecipitate the miR-210/c-Myc-Ago2-containing complexes. EFNA3 (A), E2F3 (B), NPTX1 (C), RAD52 (D), and ACRVB1 (E) were enriched in the immunoprecipitates of the mir-210 loaded RISC, whereas B2M, GAPDH, GUSB, and RPL13 (F) did not display any significant enrichment and thus were used for normalization (n = 3–11; *, p < 0.005; #, p < 0.05).
TABLE 2.
Genes enriched in miR-210 containing RISC
| Gene | Description | Fold enrichment | S.E. | p |
|---|---|---|---|---|
| Genes identified by proteomics | ||||
| CBX1 | Chromobox protein homolog 1 | 2.20 | 1.26 | 0.009 |
| P4HB | Protein-disulfide isomerase precursor | 6.52 | 2.19 | 0.054 |
| PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 | 2.67 | 1.38 | 0.021 |
| Genes identified by transcriptomics | ||||
| APC | Adenomatosis polyposis coli | 5.97 | 1.40 | 0.002 |
| ATP11C | ATPase, Class VI, type 11C | 5.84 | 1.73 | 0.018 |
| CHD9 | Chromodomain helicase DNA-binding protein 9 | 6.07 | 1.32 | 0.001 |
| CLASP2 | Cytoplasmic linker-associated protein 2 | 4.67 | 1.88 | 0.051 |
| DDAH1 | Dimethylarginine dimethylaminohydrolase 1 | 15.64 | 1.34 | 0.001 |
| ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | 4.10 | 1.57 | 0.020 |
| HECTD1 | HECT domain-containing 1 | 3.99 | 1.30 | 0.001 |
| MIB1 | Mindbomb homolog 1 (Drosophila) | 6.31 | 1.72 | 0.014 |
| NIPBL | Nipped-B homolog (Drosophila) | 3.64 | 1.52 | 0.014 |
| PTAR1 | Protein prenyltransferase α-subunit repeat containing 1 | 6.37 | 1.63 | 0.009 |
| SMCHD1 | Structural maintenance of chromosomes flexible hinge domain containing 1 | 5.08 | 1.28 | 0.001 |
| TNPO1 | Transportin 1 | 4.29 | 1.71 | 0.035 |
| XIST | X (inactive)-specific transcript | 6.88 | 1.35 | 0.001 |
| Genes identified by target prediction | ||||
| ABCB9 | ATP-binding cassette, subfamily B (MDR/TAP) member 9 (ABCB9), transcript variant 3 | 5.26 | 2.03 | 0.047 |
| BDNF | Brain-derived neurotrophic factor | 4.86 | 1.17 | 0.001 |
| CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | 8.50 | 1.33 | 0.001 |
| CPEB2 | Cytoplasmic polyadenylation element-binding protein 2 | 3.23 | 1.66 | 0.050 |
| FAM116A | Family with sequence similarity 116, member A | 2.01 | 1.33 | 0.040 |
| GPD1L | Glycerol-3-phosphate dehydrogenase 1-like | 3.12 | 1.39 | 0.009 |
| HOXA3 | Homeobox A3 | 7.87 | 1.92 | 0.014 |
| ISCU | Iron-sulfur cluster scaffold enrichment homolog (E. coli) | 5.16 | 1.47 | 0.003 |
| KIAA1161 | KIAA1161 | 26.48 | 3.19 | 0.024 |
| MDGA1 | MAM domain cont. glycosylphosphatidylinositol anchor 1 | 2.95 | 1.35 | 0.007 |
| MID1IP1 | MID1-interacting protein 1 (gastrulation-specific G12 homolog (zebrafish)) | 36.36 | 1.82 | 0.001 |
| NCAM1 | Neural cell adhesion molecule 1 | 2.33 | 1.42 | 0.043 |
| SEH1L | SEH1-like (S. cerevisiae) | 2.72 | 1.38 | 0.014 |
| SERTAD2 | SERTA domain-containing 2 | 4.88 | 1.79 | 0.026 |
| UBQLN1 | Ubiquilin 1 | 3.52 | 1.33 | 0.003 |
Further Validation of the Identified miR-210 Targets
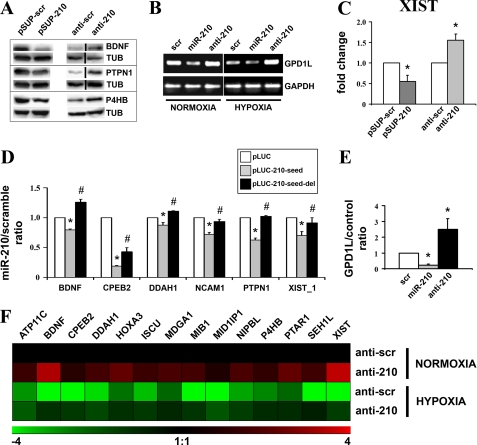
To further validate our results, protein levels of a selection of identified targets were evaluated after miR-210 modulation. As shown in Fig. 4A, BDNF, PTPN1, and P4HB were down-modulated following miR-210 overexpression and induced when miR-210 was inhibited (see supplemental Fig. SF4 for Western blotting quantification). A slightly different experimental approach was used to validate the GPD1L target. We were unable to generate an antibody that discriminated GPD1L from GPD1. However, we found that miR-210 overexpression down-modulated and miR-210 blockade increased GPD1L mRNA (Fig. 4B, see supplemental Fig. SF4 for quantification). These regulations were even more dramatic when hypoxic cells were analyzed: GPD1L mRNA levels were down-modulated by hypoxia per se, miR-210 overexpression further down-modulated GPD1L; in contrast, miR-210 inhibition completely prevented GPD1L mRNA decrease. MiR-210 ability to modulate RNA levels was further demonstrated when Xist was measured (Fig. 4C). We also confirmed that miR-210 directly regulated the expression of a selection of identified targets. To this aim, miR-210 seed-pairing sites and the immediately surrounding sequences contained in BDNF, CPEB2, DDAH1, NCAM1, PTPN1, and XIST (supplemental Fig. SF2) were cloned downstream of a luciferase open reading frame. The luciferase activity of these constructs was evaluated following the overexpression of either miR-210 or a scramble sequence. Fig. 4D shows that miR-210 inhibited the reporter constructs containing an intact miR-210 binding site (pLUC-210-seed), whereas this effect was prevented by the deletion of the seed complementary nucleotides (pLUC-210-seed del). These data confirm that miR-210 directly inhibits the tested targets. It is worth noting that Xist contains two potential miR-210 seed-paring sites, and only the one in position 6197–6202 (XIST_1) was modulated by miR-210 (Fig. 4D), whereas the one in position 11332–11337 was not functional in the assayed conditions (not shown). GPD1L was confirmed to be a direct target as well. GPD1L-3′-UTR was cloned downstream of a luciferase open reading frame, and the luciferase activity was evaluated in cells overexpressing or down-modulating miR-210. A significant negative effect was observed following miR-210 overexpression (Fig. 4E), whereas miR-210 inhibition increased the luciferase activity, indicating that GPD1L-3′-UTR was also inhibited by basal levels of miR-210.
FIGURE 4.
Confirmation of a panel of identified targets. A, HEK-293 cells were transfected with plasmids encoding either miR-210 (pSUP-210) or a scramble sequence (pSUP-scr). Alternatively, cells were transfected with either scramble (anti-scr) or anti-miR-210 (anti-210) LNA oligonucleotides. Then, 40 h later, cell extracts were derived, and the protein levels of the indicated proteins were assayed. Representative Western blots are shown (n = 3–5). Black lines indicate where irrelevant lanes were removed. B, MCF7 were transfected with scramble, miR-210, or anti-miR-210 oligonucleotides and cultured in normoxic or hypoxic conditions. Then, 40 h later, total RNA was extracted, and the indicated mRNAs were measured. The figure shows semi-quantitative RT-PCR of GPD1L mRNA (n = 3). GAPDH mRNA levels were used for normalization control. C, HEK-293 cells were transfected as indicated in panel A. Then, total RNA was extracted and Xist levels were assayed by qPCR. GAPDH mRNA levels were used for normalization purposes (n = 4; *, p < 0.05). D, for each indicated gene, HEK-293 were transfected with vector alone (pLUC) or firefly luciferase constructs that contain either an intact miR-210 binding site (pLUC-210-seed), or a mutated miR-210 binding site (pLUC-210-seed-del). Each pLUC plasmid was co-transfected with a plasmid encoding Renilla luciferase along with a plasmid encoding either miR-210 or a scramble sequence. Firefly luciferase values were normalized according to Renilla luciferase activity, and the ratio of luciferase activity of each construct in the presence or in the absence of exogenous miR-210 was calculated (n = 4–6; *, pLUC versus pLUC-210-seed p < 0.05; #, pLUC-seed versus pLUC-210-seed-del p < 0.05). E, MCF7 were transfected with pGL3 (control) or pGL3-GPD1L-3′-UTR (GPD1L) along with scramble, miR-210 or anti-miR-210 oligonucleotides. The GPD1L/control ratio of luciferase activity upon miR-210 overexpression or blockade of was calculated (n = 3; *, p < 0.05). F, HUVEC were transfected with anti-scramble or anti-210 LNA oligonucleotides, and, 18 h later, cells were exposed to hypoxia for a further 24 h. Then, total RNA was extracted, and the indicated mRNAs were measured. The heat map represents the average modulations in different groups, compared with the anti-scramble/normoxic control (n = 3, p < 0.04). Green and red colors indicate down- and up-regulation, respectively.
Newly Identified miR-210 Targets Are Modulated by miR-210 upon Hypoxia
We wanted to determine whether newly identified miR-210 targets were modulated by hypoxia. To this aim, HUVEC were exposed to hypoxia for 24 or for 48 h and their mRNA levels were measured. Supplemental Table ST9 shows that all of them were down-modulated by hypoxia, and the difference was statistically significant at one or both time points. The only exception was represented by ABCB9 that was down-modulated at both timepoints, but differences did not reach statistical significance. Then, we assayed whether miR-210 blockade prevented hypoxia-induced decrease of a subset of 14 targets. We found that miR-210-inhibition, at least in part, rescued target gene down-modulation induced by hypoxia (Fig. 4F and supplemental Table ST10). We conclude that miR-210 play an instrumental role in the inhibition of the identified genes induced by hypoxia in HUVEC.
DISCUSSION
In this study, we took a molecular approach encompassing proteomics, transcriptomics, bioinformatics, and biochemistry to investigate miR-210 function. Specifically, we studied how perturbations of miR-210 levels or activity influenced gene expression, both directly and indirectly, affecting specific molecular pathways and cellular functions. Furthermore, we identified, among the modulated genes, 31 new direct miR-210 targets, allowing to better delineate the molecular pathways underpinning miR-210 action.
Our approach to study gene expression modulation by miR-210 was 2-fold, proteomic and transcriptomic. Whereas proteomics is the technique of choice to analyze post-transcriptional regulation, it is also limited by its low sensitivity. Thus, we also measured gene expression modifications induced by both miR-210 overexpression and blockade. Transcriptomic experiments are highly sensitive; however, often yield extensive lists of modulated genes, and it is challenging to evaluate the directly affected genes, or the genes of relevance for a given process. To address this issue, we limited our analysis to those genes that exhibited inverse patterns when miR-210 was overexpressed or down-regulated. A similar approach was recently used to study miR-140 function and targets (33) and to demonstrate that miR-210 modulates the c-Myc antagonist MNT (18).
GO analysis confirmed that miR-210 activity is involved in differentiation and cell cycle regulation (13, 14, 17, 18, 34, 35) and highlighted new functions in RNA processing, DNA binding, membrane trafficking, amino acid catabolism, and development. Of note, one of the most enriched GO classes, “heart development,” was consistent with miR-210 up-regulation observed in cardiac hypertrophy and heart failure (36, 37), events characterized by the re-expression of fetal genes. Furthermore, in keeping with HIF1α-dependent regulation of miR-210 in hypoxic conditions (12), the HIF1α pathway was found as the most enriched miR-210-modulated pathway, suggesting a potential bi-directional relationship between miR-210 and the HIF pathway. Other enriched pathways, such as ATM, FAS, and TNFR1, correlated with the anti-apoptotic role of miR-210 (13, 16, 38). Likewise, integrin and agrin pathway modulations may be instrumental for miR-210-dependent regulation of EC migration and in vitro differentiation previously described by our group (13).
Potential miR-210 direct targets were then assessed for direct association with miR-210 within RISC complexes. To this end, we immunoprecipitated RISC complexes enriched for miR-210 targets. Our analysis included candidate genes, identified both by proteomics and transcriptomics, containing miR-210 seed-pairing sequences. Moreover, given the reported specificity of Pictar and Targetscan (26), predicted targets identified by these algorithm were assayed as well. One important aspect of the adopted assay is that, despite of miR-210 overexpression, non-targets were not squelched out of the RISC complex. Furthermore, it was previously demonstrated that the overexpression of the RISC component Ago2 in HEK-293 cells does not substantially alter the global gene expression program (26). It has been shown that the immunoprecipitation of non-crosslinked RNA-protein complexes, including Ago-miRNAs, may co-purify mRNAs that are not specific targets (39, 40). To minimize this potential artifact source, miR-210-containing RISC complexes were compared with RISC complexes derived from cells transfected with a scramble sequence: Thus, most artifactual targets were presumably excluded by the background subtraction. Results show that 31 of 76 tested genes were targeted by miR-210 directly. However, we cannot exclude that, among non-enriched genes, bona fide miR-210 targets exist. Indeed, our approach was not intended to provide a comprehensive compilation of miR-210 targets, but it was aimed at the identification of a sizable number of targets, allowing to highlight certain nodal elements of the molecular pathways underpinning miR-210 function.
To this aim, very stringent selection parameters were adopted to minimize the number of false positives. Furthermore, it is worth noting that the transcriptomic and the proteomic experiments were performed in HUVEC cells, whereas the IP-RISC experiments were, for technical reasons, performed in the highly transfectable HEK-293 cells. Whereas tested genes were expressed in both cell types, one should consider that alternative splicing and polyadenylation may eliminate miRNA target sites, as demonstrated for highly proliferating cells that express shortened 3′-UTRs (7, 41, 42). Results obtained were not limited to the HEK-293 cell system. Indeed, miR-210 was necessary for the down-modulation of the identified targets in hypoxic HUVEC.
The validity of the RISC immunoprecipitation approach was also confirmed by a recent study that identified a number of miR-210 targets with an experimental approach similar to the one we adopted (20). It is worth noting that the targets identified by Huang et al. display very little overlap with the ones we demonstrated. Indeed, relevant differences in the experimental procedures and in the cell system are present, and none of the studies is saturating, leaving avenues to further investigations in the field.
In our low-stringency research of miR-210 seed-pairing among the differentially expressed genes identified by transcriptomics and proteomics, we kept limitations to a minimum. Thus, our search was not confined to the gene 3′-UTR, but included the coding sequence and the 5′-UTR, both recently demonstrated as involved in miRNA-mRNA binding (5, 6, 22–24, 27). It was found that miR-210 seed-pairing sequences were more frequent among the putative targets compared with controls. Furthermore, we did not impose any conservation bias. Indeed, whereas many miRNAs are evolutionarily conserved, others are species-specific, including human miRNAs not conserved in chimpanzee (43), consistent with miRNA roles ranging from generating cellular to organismal diversity. Thus, it is not surprising that a certain gene may be targeted by miR-210 in humans but not in other vertebrates.
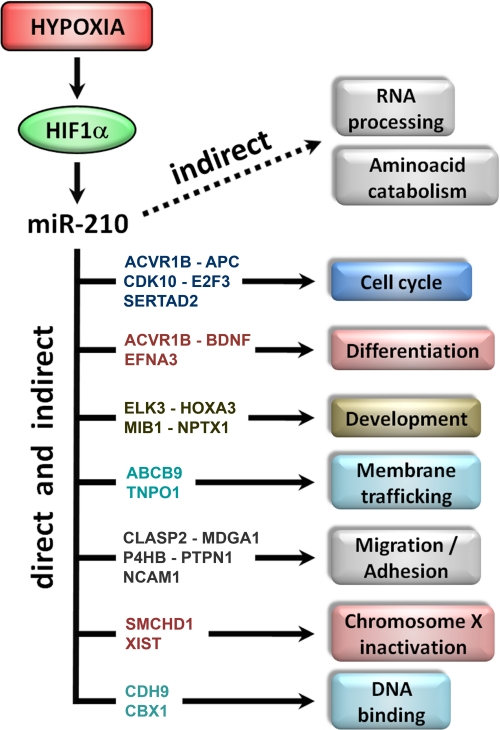
Fig. 5 summarizes the identified cellular processes affected by miR-210, both directly and indirectly, and how some of the new validated targets could be involved in the direct regulation of these events. CLASP2, MDGA1, NCAM1, and P4HB are involved in cellular migration and adhesion and could, at least in part, mediate miR-210 regulation of tubulogenesis and migration observed in hypoxic ECs (13). Furthermore, it has been shown that, following hindlimb ischemia, PTPN1 negatively regulates VEGFR2 and modulates cell-cell adhesions (44). Moreover, in keeping with our findings, previous studies found that CPEB2 and the ternary complex factor ELK-3 proteins are modulated by hypoxia (45, 46). Many identified targets, such as the tumor suppressor APC, ACVR1B, CDK10, SERTAD2, as much the previously identified E2F3 and MNT (14, 18), are involved in cell cycle regulation. These results are in keeping with miR-210 overexpression observed in breast cancer, where miR-210 levels correlate with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer (18, 47, 48). Indeed, it has been shown that CDK10 expression is reduced in breast cancer (49), suggesting a role for miR-210 in this regulation. Furthermore, miR-210 is overexpressed in many other solid cancer types (50), it was detected in serum of patients affected by diffuse large B-cell lymphoma (51) and by breast, colon, lung, and ovarian cancer (52). It was also found that elevated expression of miR-210 in melanoma and in pancreatic tumors associates with poorer survival (18, 53). Another novel target is GPD1L, a gene recently linked to sudden infant cardiac death and the Brugada syndrome, a disease that induces syncope, ventricular arrhythmias, and sudden death (54–56). Interestingly, acute myocardial ischemia involving the right-ventricular outflow tract is known to induce Brugada-like electrophysiological and electrocardiogram alterations (57–59). Whereas miR-210 and GPD1L levels were not measured in these pathological conditions, it is worth noting that hypoxia is both a crucial component of ischemia and a potent miR-210 inducer (12). To the best of our knowledge, to date a single report shows that miRNAs could target non-coding RNAs (27). Thus, an unexpected target is Xist (60), a long non-protein coding RNA involved in mammalian X-chromosome inactivation during early female embryogenesis. Although Xist is confined in the nucleus, recent data demonstrate that miRNAs can be imported to the nucleus (61). In keeping with our data, Xist is down-modulated by ischemia (62), and recently was demonstrated an inappropriate Xist expression/localization on active X-chromosome in breast cancer (63). Given high miR-210 levels observed in breast cancer (47, 48), one can hypothesize that miR-210 may play a role in this deregulation. Furthermore, another identified miR-210 target, SMCHD1, localizes to the inactive X-chromosome specifically, and has a role in the maintenance of its inactivation (64). Another interesting finding is that the miR-210 seed-pairing sequence of Xist in position 6197–6202 represents the 3′-end of a perfect palindrome within Xist (see supplemental Fig. SF2 for details), suggesting several possible speculations: miR-210 access to Xist palindrome sequence could be modulated in different physiological situations and, in turn, Xist could be regulated by both miR-210 and miR-210* (the complementary mature strand). Moreover, miR-210 may also regulate Tsix, the Xist antisense partner that blocks X-chromosome silencing (60). Further studies to functionally evaluate miR-210/Xist interaction are ongoing.
FIGURE 5.
Cellular processes modulated by miR-210. miR-210 is induced by hypoxia through HIF1α. The diagram summarizes the cellular processes affected by miR-210, both directly and indirectly, according to GO analysis. Newly identified targets possibly involved in each event are shown.
We can conclude that this study establishes a new frame for further studies determining the relevance of each of the newly identified or predicted molecular pathways in mediating miR-210-dependent response to hypoxia. Our integrative approach may also serve as a model that can be applied for the functional dissection of additional miRNAs.
Acknowledgment
We thank Yuri D'Alessandra (Centro Cardiologico Monzino-IRCCS) for skillful help in proteomic analysis.
This work was supported in part by Ministero della Salute Grants RF06-conv.29/07-1, RF07-conv-85.1, RBLA035a4X-001, RF05-conv.66-1, RF07-55.1, and RF07-strat.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables ST1–11, Figs. SF1–4, and “Materials.”
- RISC
- RNA-induced silencing complex
- miRNA
- microRNA
- EC
- endothelial cell
- GO
- gene ontology
- HUVEC
- human umbilical vein endothelial cells
- LNA
- locked nucleic acid
- UTR
- untranslated region.
REFERENCES
- 1.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2.Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 4.Vasudevan S., Tong Y., Steitz J. A. (2007) Science 318, 1931–1934 [DOI] [PubMed] [Google Scholar]
- 5.Ørom U. A., Nielsen F. C., Lund A. H. (2008) Mol. Cell 30, 460–471 [DOI] [PubMed] [Google Scholar]
- 6.Tsai N. P., Lin Y. L., Wei L. N. (2009) Biochem. J., in press [DOI] [PubMed] [Google Scholar]
- 7.Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker R., Sheth U. (2007) Mol. Cell 25, 635–646 [DOI] [PubMed] [Google Scholar]
- 9.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., Bhanot S., Monia B. P. (2006) Cell Metab. 3, 87–98 [DOI] [PubMed] [Google Scholar]
- 10.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 11.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 12.Ivan M., Harris A. L., Martelli F., Kulshreshtha R. (2008) J. Cell Mol. Med. 12, 1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasanaro P., D'Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M. C., Martelli F. (2008) J. Biol. Chem. 283, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannakakis A., Sandaltzopoulos R., Greshock J., Liang S., Huang J., Hasegawa K., Li C., O'Brien-Jenkins A., Katsaros D., Weber B. L., Simon C., Coukos G., Zhang L. (2008) Cancer Biol. Ther. 7, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulkkinen K., Malm T., Turunen M., Koistinaho J., Ylä-Herttuala S. (2008) FEBS Lett. 582, 2397–2401 [DOI] [PubMed] [Google Scholar]
- 16.Crosby M. E., Kulshreshtha R., Ivan M., Glazer P. M. (2009) Cancer Res. 69, 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno Y., Tokuzawa Y., Ninomiya Y., Yagi K., Yatsuka-Kanesaki Y., Suda T., Fukuda T., Katagiri T., Kondoh Y., Amemiya T., Tashiro H., Okazaki Y. (2009) FEBS Lett. 583, 2263–2268 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., Sun H., Dai H., Walsh R. M., Imakura M., Schelter J., Burchard J., Dai X., Chang A. N., Diaz R. L., Marszalek J. R., Bartz S. R., Carleton M., Cleary M. A., Linsley P. S., Grandori C. (2009) Cell Cycle 8, 2756–2768 [DOI] [PubMed] [Google Scholar]
- 19.Kim H. W., Haider H. K., Jiang S., Ashraf M. (2009) J. Biol. Chem. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X., Ding L., Bennewith K. L., Tong R. T., Welford S. M., Ang K. K., Story M., Le Q. T., Giaccia A. J. (2009) Mol. Cell 35, 856–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhdanov V. P. (2009) Mol. Biosyst. 5, 638–643 [DOI] [PubMed] [Google Scholar]
- 22.Rigoutsos I. (2009) Cancer Res. 69, 3245–3248 [DOI] [PubMed] [Google Scholar]
- 23.Lee I., Ajay S. S., Yook J. I., Kim H. S., Hong S. H., Kim N. H., Dhanasekaran S. M., Chinnaiyan A., Athey B. D. (2009) Genome Res. 19, 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X., Duan X., Qian J., Li F. (2009) Genetica 137, 159–164 [DOI] [PubMed] [Google Scholar]
- 25.Karginov F. V., Conaco C., Xuan Z., Schmidt B. H., Parker J. S., Mandel G., Hannon G. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19291–19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendrickson D. G., Hogan D. J., Herschlag D., Ferrell J. E., Brown P. O. (2008) PLoS ONE 3, e2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009) Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Shahrour F., Carbonell J., Minguez P., Goetz S., Conesa A., Tárraga J., Medina I., Alloza E., Montaner D., Dopazo J. (2008) Nucleic Acids Res. 36, W341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004) Mol. Cell 15, 185–197 [DOI] [PubMed] [Google Scholar]
- 32.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. (2004) Science 305, 1437–1441 [DOI] [PubMed] [Google Scholar]
- 33.Nicolas F. E., Pais H., Schwach F., Lindow M., Kauppinen S., Moulton V., Dalmay T. (2008) Rna 14, 2513–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchiya S., Oku M., Imanaka Y., Kunimoto R., Okuno Y., Terasawa K., Sato F., Tsujimoto G., Shimizu K. (2009) Nucleic Acids Res. 37, 3821–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bianchi N., Zuccato C., Lampronti I., Borgatti M., Gambari R. (2009) BMB Rep. 42, 493–499 [DOI] [PubMed] [Google Scholar]
- 36.van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thum T., Galuppo P., Wolf C., Fiedler J., Kneitz S., van Laake L. W., Doevendans P. A., Mummery C. L., Borlak J., Haverich A., Gross C., Engelhardt S., Ertl G., Bauersachs J. (2007) Circulation 116, 258–267 [DOI] [PubMed] [Google Scholar]
- 38.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005) Nucleic Acids Res. 33, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mili S., Steitz J. A. (2004) Rna 10, 1692–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirino Y., Mourelatos Z. (2008) Rna 14, 2254–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008) Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji Z., Lee J. Y., Pan Z., Jiang B., Tian B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berezikov E., Thuemmler F., van Laake L. W., Kondova I., Bontrop R., Cuppen E., Plasterk R. H. (2006) Nat. Genet. 38, 1375–1377 [DOI] [PubMed] [Google Scholar]
- 44.Nakamura Y., Patrushev N., Inomata H., Mehta D., Urao N., Kim H. W., Razvi M., Kini V., Mahadev K., Goldstein B. J., McKinney R., Fukai T., Ushio-Fukai M. (2008) Circ. Res. 102, 1182–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hägele S., Kühn U., Böning M., Katschinski D. M. (2009) Biochem. J. 417, 235–246 [DOI] [PubMed] [Google Scholar]
- 46.Gross C., Dubois-Pot H., Wasylyk B. (2008) Oncogene 27, 1333–1341 [DOI] [PubMed] [Google Scholar]
- 47.Foekens J. A., Sieuwerts A. M., Smid M., Look M. P., de Weerd V., Boersma A. W., Klijn J. G., Wiemer E. A., Martens J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13021–13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camps C., Buffa F. M., Colella S., Moore J., Sotiriou C., Sheldon H., Harris A. L., Gleadle J. M., Ragoussis J. (2008) Clin. Cancer Res. 14, 1340–1348 [DOI] [PubMed] [Google Scholar]
- 49.Iorns E., Turner N. C., Elliott R., Syed N., Garrone O., Gasco M., Tutt A. N., Crook T., Lord C. J., Ashworth A. (2008) Cancer Cell 13, 91–104 [DOI] [PubMed] [Google Scholar]
- 50.Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrie C. H., Gal S., Dunlop H. M., Pushkaran B., Liggins A. P., Pulford K., Banham A. H., Pezzella F., Boultwood J., Wainscoat J. S., Hatton C. S., Harris A. L. (2008) Br. J. Haematol. 141, 672–675 [DOI] [PubMed] [Google Scholar]
- 52.Lodes M. J., Caraballo M., Suciu D., Munro S., Kumar A., Anderson B. (2009) PLoS One 4, e6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greither T., Grochola L., Udelnow A., Lautenschlager C., Wurl P., Taubert H. (2009) Int. J. Cancer, in press [DOI] [PubMed] [Google Scholar]
- 54.Benito B., Brugada R., Brugada J., Brugada P. (2008) Prog. Cardiovasc. Dis. 51, 1–22 [DOI] [PubMed] [Google Scholar]
- 55.London B., Michalec M., Mehdi H., Zhu X., Kerchner L., Sanyal S., Viswanathan P. C., Pfahnl A. E., Shang L. L., Madhusudanan M., Baty C. J., Lagana S., Aleong R., Gutmann R., Ackerman M. J., McNamara D. M., Weiss R., Dudley S. C., Jr. (2007) Circulation 116, 2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Norstrand D. W., Valdivia C. R., Tester D. J., Ueda K., London B., Makielski J. C., Ackerman M. J. (2007) Circulation 116, 2253–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omichi C., Fujimoto N., Kawasaki A., Kasai A. (2009) Int. J. Cardiol., in press [DOI] [PubMed] [Google Scholar]
- 58.Di Diego J. M., Fish J. M., Antzelevitch C. (2005) J. Electrocardiol. 38, 14–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kataoka H. (2000) Am. J. Cardiol. 86, 1056. [DOI] [PubMed] [Google Scholar]
- 60.Ng K., Pullirsch D., Leeb M., Wutz A. (2007) EMBO Rep. 8, 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hwang H. W., Wentzel E. A., Mendell J. T. (2007) Science 315, 97–100 [DOI] [PubMed] [Google Scholar]
- 62.Yoshida T., Kurella M., Beato F., Min H., Ingelfinger J. R., Stears R. L., Swinford R. D., Gullans S. R., Tang S. S. (2002) Kidney Int. 61, 1646–1654 [DOI] [PubMed] [Google Scholar]
- 63.Sirchia S. M., Tabano S., Monti L., Recalcati M. P., Gariboldi M., Grati F. R., Porta G., Finelli P., Radice P., Miozzo M. (2009) PLoS ONE 4, e5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blewitt M. E., Gendrel A. V., Pang Z., Sparrow D. B., Whitelaw N., Craig J. M., Apedaile A., Hilton D. J., Dunwoodie S. L., Brockdorff N., Kay G. F., Whitelaw E. (2008) Nat. Genet. 40, 663–669 [DOI] [PubMed] [Google Scholar]