Abstract
The human DNA polymerase κ homolog Sulfolobus solfataricus DNA polymerase IV (Dpo4) produces “−1” frameshift deletions while copying unmodified DNA and, more frequently, when bypassing DNA adducts. As judged by steady-state kinetics and mass spectrometry, bypass of purine template bases to produce these deletions occurred rarely but with 10-fold higher frequency than with pyrimidines. The DNA adduct 1,N2-etheno-2′-deoxyguanosine, with a larger stacking surface than canonical purines, showed the highest frequency of formation of −1 frameshift deletions. Dpo4 T239W, a mutant we had previously shown to produce fluorescence changes attributed to conformational change following dNTP binding opposite cognate bases (Beckman, J. W., Wang, Q., and Guengerich, F. P. (2008) J. Biol. Chem. 283, 36711–36723), reported similar conformational changes when the incoming dNTP complemented the base following a templating purine base or bulky adduct (i.e. the “+1” base). However, in all mispairing cases, phosphodiester bond formation was inefficient. The frequency of −1 frameshift events and the associated conformational changes were not dependent on the context of the remainder of the sequence. Collectively, our results support a mechanism for −1 frameshift deletions by Dpo4 that involves formation of active complexes via a favorable conformational change that skips the templating base, without causing slippage or flipping out of the base, to incorporate a complementary residue opposite the +1 base, in a mechanism previously termed “dNTP-stabilized incorporation.” The driving force is attributed to be the stacking potential between the templating base and the incoming dNTP base.
Introduction
The integrity of the genetic code is maintained by DNA polymerases, but mutations do occur (1). Mutations can be advantageous in enabling adaptation (e.g. in microbial populations or the immune system), but the loss of genetic integrity can lead to many diseases, including cancer, atherosclerosis, and neurodegenerative diseases (1–3), as well as teratology and aging (4).
In the past decade, the number of recognized DNA polymerases has expanded considerably. Of these, several have been shown to be so-called translesion DNA polymerases, DNA polymerases specializing in bypass of templating adducts that normally stall the more efficient and processive replicative DNA polymerases (1, 5, 6). However, translesion DNA polymerases have relatively low fidelity when copying undamaged DNA and are prone to cause frameshift mutations, particularly deletions that reduce the nucleotide count by 1 in newly formed DNA strands, i.e. “−1” frameshifts. Frameshifts occur in the presence of many DNA adducts and are the basis of one major type of damage in the classic “Ames test” for bacterial genotoxicity, i.e. Salmonella typhimurium TA1538 and TA98 (7). Frameshift deletions also occur with unmodified DNA, particularly while copying long runs of purines or pyrimidines (8). For example, Solfataricus acidocaldarius DNA polymerase Dbh was found to make −1 deletion frameshift mutations with a frequency up to 50% (9, 10), and human pol κ was found to generate −1 deletion frameshifts at a frequency of 2 × 10−3 (11, 12).
The model translesion DNA polymerase Sulfolobus solfataricus DNA polymerase IV (Dpo4),4 a homolog of human pol κ, has also been found to produce frameshifts (13–18). Dpo4 bypass of bulky templating adducts, e.g. an N2-benzo[a]pyrene diol epoxide-G adduct, 1,N2-ϵ-G, M1G, and 1,N2-propano-2′-deoxyguanosine, all produced −1 frameshifts at varying frequencies and even +1 (and −1) frameshifts when Dpo4 bypassed a template AP site (14, 16–18). Dpo4 can also produce −1 frameshifts with unmodified DNA (19).
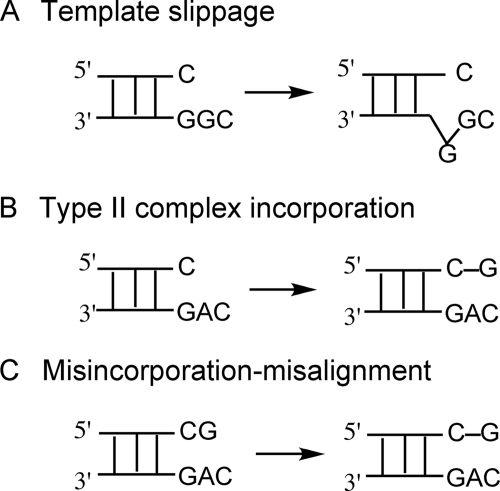
Three basic models have been proposed to explain how −1 deletion frameshifts occur (20) (Fig. 1). (i) The primer and template DNA strands misalign prior to dNTP addition opposite the following templating base, i.e. the “+1” base (the Streisinger slippage model) (8), which requires a repetitive sequence (Fig. 1A). (ii) The incoming dNTP “skips” over the templating base (often described as the formation of a “type II complex” (21) as well as a “dNTP-stabilized misalignment mechanism”) (Fig. 1B) (22–24). (iii) The dNTP is first misincorporated and then misaligns with a complementary template base (termed the “misincorporation-misalignment mechanism”) (20, 25) (Fig. 1C). Although much has been published on these mechanisms and their roles in particular situations, the low frequency of DNA polymerase-mediated frameshifts has made physical studies difficult, aside from NMR studies with mispaired oligonucleotides (in the absence of polymerases) and some x-ray structures of DNA polymerases (see below).
FIGURE 1.
Three mechanisms for the formation of −1 deletion frameshifts. A, template slippage mechanism. The repeatable template base G is slipped from the +1 site to the insertion site, prior to incorporation. B, type II complex (21) incorporation mechanism (also termed “dNTP-stabilized misincorporation” mechanism). dGTP incorporation occurs opposite the +1 C (type II complex) followed directly by phosphodiester bond formation. C, misincorporation-misalignment mechanism. dGTP is first misincorporated opposite A and extended, followed by repositioning opposite the next base (C).
To help establish which of these models, or an alternative one, is utilized by Dpo4, we employed a Dpo4 mutant containing a single Trp, T239W, with which we recently reported fluorescence changes attributed to conformational changes associated with correct pairing (26). Dpo4 T239W produced fluorescence changes upon binding of a complementary dNTP, a rapid increase in fluorescence upon formation of an active ternary complex followed by a slower decrease following phosphodiester bond formation, with the latter attributed to relaxation of the conformation. However, we also observed rapid fluorescence increases that corresponded to noncomplementary dNTP binding but without fluorescence decreases (i.e. phosphodiester bond formation was relatively inefficient). In this study, we investigated these events associated with −1 frameshifts (because they only occurred when the incoming dNTP complemented the +1 base) by examining the fluorescence changes of Dpo4 T239W and comparing the data with quantitative analyses of product formation using steady-state kinetics and LC/MS-MS. We observed that formation of −1 frameshift products was facilitated by the presence of a templating purine or an adduct with relatively high stacking potential (1,N2-ϵ-G), and we conclude that such events involve a rapid and favorable conformational change despite very slow phosphodiester bond formation.
EXPERIMENTAL PROCEDURES
Materials
Unlabeled dNTPs were purchased from New England Biolabs (Beverly, MA), and [γ-32P]ATP (specific activity 3 × 103 Ci mmol−1) was from PerkinElmer Life Sciences. Bio-Spin columns were obtained from Bio-Rad. The Dpo4 single Trp mutant (Dpo4 T239W) was expressed in Escherichia coli and purified to electrophoretic homogeneity as described previously (14, 26), with the 80 °C heat step omitted in the case of the mutant because of the possibility of decreased mutant stability. Purified Dpo4 T239W was stored in small aliquots at −80 °C in 50 mm Tris-HCl buffer (pH 7.7 measured at 22 °C) containing 50 mm NaCl, 1 mm dithiothreitol, and 50% glycerol (v/v). All other reagents were of the highest quality commercially available.
Oligonucleotides
Unmodified 13-mers (primers) and 18- mers and adducted 18-mer (templates) oligonucleotides containing O6-MeG, 8-oxoG, or a stabilized AP site (i.e. 3′,5′-disubstituted tetrahydrofuran) (Table 1) were synthesized and purified using high pressure liquid chromatography by Midland Certified Reagent Co. (Midland, TX). For analysis of fluorescence changes in the absence of phosphodiester bond formation, the 3′ end of the primer strand was rendered incapable of phosphoryl transfer by deletion of the 3′-OH group and is termed “dd” in the text. The extinction coefficients for the oligonucleotides were estimated by the Borer method (27).
TABLE 1.
Oligodeoxynucleotides used in analyses
X indicates A, C, G, or T; Xdd indicates a terminal 2′,3′-dideoxy-X (incapable of phosphodiester bond formation); YYY indicates GGC, GGT, AAC, CAG, GAG, GAC, TTC, GTC, TCG, CCT, GCT, GCA, G(O6-MeG)C, G(8-oxoG)C, G(1,N2-ϵ-G)C, G(1,N2-ϵ-G)T, or G(AP)C. The primer terminal residue X of the 13-mer primer was paired with the first (3′) Y in the YYY sequence of the template strand. For the LC-MS experiments, the 5′-most T of the primer was replaced with 2′-deoxyuridine (see Table 2).
| 13-mer | 5′-GGGGGAAGGATTX-3′ |
| 13dd-mer | 5′-GGGGGAAGGATTXdd-3′ |
| 18-mer | 3′-CCCCCTTCCTAAYYYACT-5′ |
The 5′ end of each 13-mer primer was labeled with [γ-32P]ATP using T4 polynucleotide kinase at 37 °C for 30 min. After removal of excess [γ-32P]ATP using a Bio-Spin 6 column (Bio-Rad), the labeled primer and (unmodified or modified) template (molar ratio 1:1) were heated at 95 °C for 5 min and then slowly cooled to room temperature to form the 13-mer/18-mer duplexes, which were used for all steady-state and pre-steady-state kinetic experiments.
Reaction Conditions for Dpo4 T239W Assays and Product Analysis Methods
Unless stated otherwise, DNA polymerization reactions with Dpo4 T239W were carried out in 50 mm Tris-HCl buffer (pH 7.5 at 25 °C) containing 50 mm NaCl, 5 mm dithiothreitol, and 5% (v/v) glycerol) at 37 °C (26). Although bovine serum albumin is often used in DNA polymerase assays, it was not added here because it contains two Trp residues that could interfere with the fluorescence of the Dpo4 mutant (26); removal of albumin was previously found not have an effect on the rate of DNA polymerization. All reactions were initiated by mixing a dNTP/MgCl2 solution (final MgCl2 concentration of 5 mm) with preincubated enzyme/DNA mixtures. After the reaction, 5-μl aliquots were quenched by the addition of EDTA/formamide solution (50 μl of 20 mm EDTA in 95% formamide (v/v) with 0.5% bromphenol blue (w/v) and 0.05% xylene cyanol (w/v)). Products were resolved using 20% polyacrylamide (w/v), 8 m urea denaturing gel electrophoresis and visualized and quantitated by phosphorimaging analysis using a Bio-Rad Molecular Imager FX instrument and Quantity One software.
Single dNTP Incorporation and Extension
Reactions were initiated by mixing 200 nm Dpo4 T239W and 100 nm DNA mixtures with 1 mm dNTP and 5 mm MgCl2 complex at 37 °C. Each 30 s an aliquot was quenched by the addition of EDTA (to 4 min). The zero time point value was determined without the addition of dNTP. The products were visualized and quantitated by phosphorimaging analysis using a Bio-Rad Molecular Imager FX instrument and Quantity One software.
LC-MS/MS Analysis of Primer Extension Products
The template base at the insertion position was varied to characterize all possible misincorporation and deletion products after addition of only three dNTPs, in the absence of the dNTP that would be correctly incorporated opposite the insertion base. The primer was extended by incubating Dpo4 T239W (5 μm), DNA (10 μm), a mixture of three dNTPs (1 mm each, with the normally pairing dNTP omitted), and MgCl2 (5 mm) in Tris-HCl buffer (pH 7.5, final volume 100 μl) at 37 °C for 12 h. In the case of the modified G base used here (i.e. 1,N2-ϵ-G), all four dNTPs were added. Each reaction was terminated by extraction of the remaining dNTPs using a Bio-Spin 6 chromatography column, and concentrated Tris-HCl, dithiothreitol, and EDTA solutions were added to restore the final concentrations to 50, 5, and 1 mm, respectively. E. coli uracil DNA glycosylase (20 units, Sigma) was then added; the solution was incubated at 37 °C for 6 h to remove the uracil residues on the extended primer and then heated at 95 °C for 1 h in the presence of 0.25 m piperidine, followed by removal of the solvent by in vacuo centrifugation (14). The dried sample was resuspended in 100 μl of H2O for spectrometry analysis.
LC-MS/MS analysis was performed using an Acquity UPLC system (Waters) connected to a Finnigan LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operating in the electrospray ionization negative ion mode. An Acquity UPLC BEH octadecylsilane (C18) column (1.7 μm, 1.0 × 100 mm) was used with the following LC conditions (all at 50 °C) with Buffer A (10 mm NH4CH3CO2 plus 2% CH3CN (v/v)) and Buffer B (10 mm NH4CH3CO2 plus 95% CH3CN (v/v)). The conditions used were similar to those reported previously (14, 28, 29). The calculations of the collision-induced dissociation fragmentations of oligonucleotide sequences were done using a program linked to the Mass Spectrometry Group (Medicinal Chemistry) of the University of Utah.
Steady-state Kinetic Assays
Steady-state single-base incorporation experiments were performed by adding a single dNTP at various concentrations (12 concentrations for each assay) to a preincubated Dpo4 T239W/primer-template duplex (molar ratio of DNA polymerase to primer-template <10%). Primer conversion to product was kept to <20% by adjusting the polymerase concentration and incorporation time (30). Graphs of kobs versus dNTP concentration were fit to a hyperbolic equation to yield kcat and Km values using nonlinear regression in GraphPad Prism version 3.0 (GraphPad, San Diego). The corresponding errors were also obtained to indicate the goodness of fit.
Stopped-flow Fluorescence Measurements
An OLIS RSM-1000 spectrofluorimeter (On-line Instrument Systems, Bogart, GA) with a 4 × 4 mm observation cell was used in the measurement of transient fluorescent assays. For optimal signal-to-noise ratios for observing changes in Trp fluorescence, 3.16-mm slits (yielding 20 nm bandwidth) were employed along with both 335-nm long pass (CVI Laser Corp., Albuquerque, NM) and 355-nm bandpass filters (Newport, Irvine, CA) attached to the sample photomultiplier tube in tandem. MgCl2 (5 mm) was included in both syringes. In typical experiments, one syringe contained Dpo4 T239W plus oligonucleotide complex in 50 mm Tris-HCl buffer (pH 7.5 at 25 °C) containing 50 mm NaCl, 5 mm dithiothreitol, and 5% (v/v) glycerol, and the second syringe contained various concentrations of the complementary dNTP (either paired with the “0” or +1 templating base) in the same Tris buffer (with 5 mm MgCl2). After rapid mixing, the final concentration of Dpo4-oligonucleotide complex was 1 μm. In all cases, standard assays were performed, including all components except the reagent producing a change (e.g. dNTP in the case of fluorescence changes versus dNTP concentrations). The signal-to-noise ratio was decreased somewhat due to an inner filter effect; this phenomenon was least problematic in the situations where dATP was mixed with oligonucleotides. The fluorescence plots presented here are generally averages of eight individual shots.
Analysis of Stopped-flow Fluorescence Data
The rates of conformational change at different dNTP concentrations can be obtained by nonlinear regression fitting of the fluorescence increase signal against time using Equation 1,
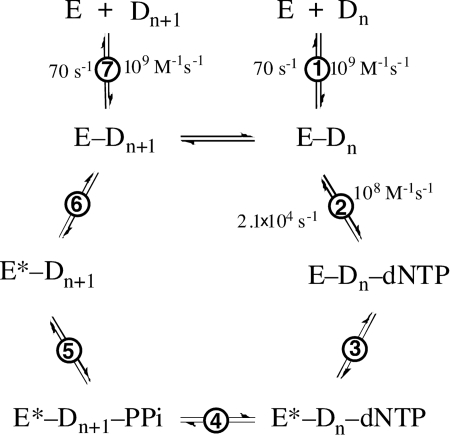
where y is the fluorescence signal produced over time; A is the amplitude of the signal; kobs is the observed rate constant, and t is time. The kobs apparent values were then plotted against dNTP concentrations (Equation 2) to obtain the ground state dissociation constant of dNTP (Kd,dNTP), maximal forward conformational change rate (k3), and reverse conformational change rate (k−3) (Fig. 2) (26),
FIGURE 2.
General DNA polymerization mechanism. Note that the pyrophosphate release step precedes the relaxation of conformation in this scheme (26). The rate constants shown are from Ref. 26.
All nonlinear regression analyses used the OLIS RSM-1000 stopped-flow spectrofluorimeter software and GraphPad Prism version 3.0. The corresponding errors were obtained to indicate the goodness of fit.
RESULTS
Analysis of Full-length Primer Extension Products as a Function of Template Sequence
LC-MS/MS analysis of the full-length extension products of primer was used to distinguish −1 deletion frameshifts from other miscoding events (Table 2). In each case, the template base at the insertion position was different from any bases in the 5′-template sequence, and only three dNTPs were added, excluding the dNTP that would correctly pair opposite the templating base (0 base). In this experimental design, the DNA polymerase has to mispair or frameshift because of the inability to proceed with normal base pairing at the insertion site (Fig. 1) (if all four dNTPs are present, the primer is prone to rapid incorporation and extension in an accurate mode, not revealing potential products resulting from slower misincorporations and frameshifts (29, 31, 32)). For full-length extension bypass of 1,N2-ϵ-G, dCTP was also added because all four dNTPs were found to be incorporated slowly opposite this lesion by wild-type Dpo4 (14). All possible extension products were considered, including misincorporation products, −1 deletion frameshift products, products with blunt-end additions, and partial extension products. Each product in the oligonucleotide product mixtures could be readily identified and quantified by LC-MS/MS (two examples are shown in detail in supplemental Figs. S1–S5 and supplemental Tables S1–S9).
TABLE 2.
LC-MS/MS analysis of full-length extension products using Dpo4 T239W and partial dNTP mixtures
The reaction conditions are as follows: 5 μm Dpo4, 10 μm DNA primer-template complex, 1 mm dNTP (without the correct dNTP corresponding to the base at the incorporation position), 5 mm MgCl2, 12-h incubation time; MS, charge −3; peak area calculated from m/z limit, ±0.5 atomic mass units. Sequences 5′-GGGGGAAGGAU and 3′-CCCCCTTCCTA are completed below (where underlined U indicates 2′-deoxyuridine).
| Additional primer and template sequence | Extension products | % | Frameshift productsa |
|---|---|---|---|
| % | |||
| -TC | TCCGCGA | 91 | |
| -AGACGCT | TC GCGAC | 9 | 9 |
| -TC | TCCCGAC | 89 | |
| -AGAGCTG | TC CGACC | 11 | 11 |
| -TC | TCTGTGA | 64 | |
| -AGGCACT | TCAGTGA | 24 | |
| TC GTGA | 12 | 12 | |
| TC | TCATACT | 18 | |
| -AGCATGA | TCATACTA | 3 | |
| TCCTACT | 4 | ||
| TCTTACT | 68 | ||
| TCTTACTT | 3 | ||
| TC TACT | 2 | 2 | |
| -TA | TACCTAC | 53 | |
| -ATCGATG | TACCTACC | 5 | |
| TACCTACT | 4 | ||
| TAACTAC | 15 | ||
| TATCTAC | 21 | ||
| TA CTAC | 2 | 2 | |
| -TC | TCCGCGT | 43 | |
| -AGTCGCA | TCCGCGT G | 4 | |
| TCGGCGT C | 25 | ||
| TCTGCG T | 25 | ||
| TCTGCG | 2 | ||
| TC GCGT | 1 | 1 | |
| -TC | TCCGTG | 16 | |
| -AG(1,N2-ϵ-G)CACTa | TCCGTGA | 6 | |
| TCAGTC | 22 | ||
| TCAGTGA | 10 | ||
| TCGGTGA | 21 | ||
| TC GTGA | 25 | 25 | |
| -TC | TCCATGA | 9 | |
| -AG(1,N2-ϵ-G)TACTa | TCAATG | 16 | |
| TCAATGA | 6 | ||
| TCGATGA | 8 | ||
| TCTATG | 11 | ||
| TC ATG | 29 | ||
| TC ATGA | 21 | 50 | |
a With the two templates containing 1,N2-ϵ-G, all four dNTPs were added.
Collectively, when A or G was in the 0 templating position, Dpo4 T239W bypass yielded 9–12% of the −1 frameshift products but only 0–2% when T or C was in the 0 templating position. All other products were the result of other miscoding events (misinsertion). Changing the template sequence to include either repeated or nonrepeated template sequences had little effect on the production of −1 deletion frameshifts. As with wild-type Dpo4, Dpo4 T239W incorporated all four dNTPs opposite a 0 templating 1,N2-ϵ-G with significant percentages of −1 frameshifts (25–50%) (14). All −1 frameshift products arose from complementary incorporation opposite the +1 base.
Fluorescence Changes in Dpo4 T239W-Oligonucleotide Complexes Observed following Mixing with dNTPs
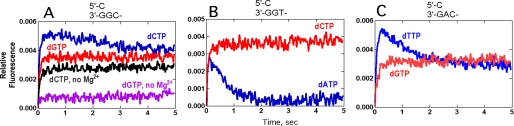
Stopped-flow mixing of dCTP opposite a (5′)- C/(3′)-GGC- pair (i.e. dCTP positioned opposite a G and with a free 3′-OH to permit phosphodiester bond formation) showed a fast fluorescence increase, followed by a slow relaxation phase (following rapid phosphodiester bond formation), in agreement with our previous results (Fig. 3A) (26). Mixing of dGTP with this primer-template complex showed only the fluorescence increase without a detectable relaxation phase, suggesting a much slower phosphodiester bond formation step, relaxation step, or both. Additions of dNTPs to other (primer-) template sequences produced similar results (Fig. 3, B and C). These experiments generally included 5 mm Mg2+; if Mg2+ was omitted (using EDTA to trap residual metal ions), no fluorescence increases were observed (Fig. 3A).
FIGURE 3.
Fluorescence changes of Dpo4 T239W after addition of dNTPs to primer-template complexes. The relevant portions of the oligonucleotide sequences (within the region of dNTP binding) and the dNTP are shown above the graphs. The reactions included 1.0 μm Dpo4 T239W, 1.0 μm 13-/18-mer primer-template complex (Table 1), 0.5 mm dNTP, and 5 mm Mg2+ at 37 °C. All assays contained Mg2+ except for the two traces with EDTA (0.3 mm) (A).
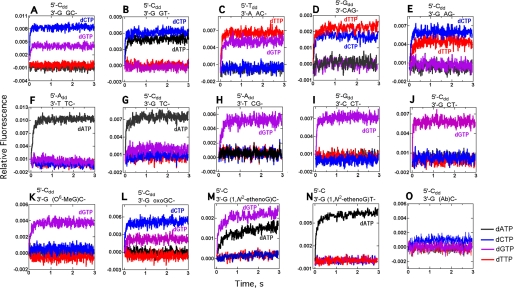
Addition of each of the four dNTPs to a (5′)-C primer (3′-dd) annealed to the templates (3′)-GGC-, (3′)-GGT- (0 templating base in boldface), or a (5′)-T primer (3′-dd) annealed to the template (3′)-AAC- (Fig. 4, A–C) produced two fluorescence increases with each primer-template complex, one from binding of the dNTP complementary to the 0 templating base and the other from binding of the dNTP complementary to the following base (5′ or +1 base). Because these template sequences contained repeated sequences (either AA or GG), the 0 template base could have slipped to the −1 position to complement the 3′-primer base, thereby pulling the +1 base into the 0 templating position (Fig. 1A) (10, 26). The expectation was that fluorescence changes would be observed only if the complementary base was in the 0 position (21). However, adding dNTPs opposite nonrepeated template sequences gave comparable results, with rapid fluorescence increases upon addition of dNTPs complementary to 0 and +1 bases (Fig. 4, D and E). Interestingly, a fluorescence change was observed only upon binding of the dNTP complementary to a 0 templating pyrimidine, regardless of whether repeating or nonrepeating base template sequences were used (Fig. 4, F–J).
FIGURE 4.
Fluorescence changes for Dpo4 T239W following mixing of each of four dNTPs opposite a 13/18-mer primer (3′-dideoxy terminated)-template pair. The relevant portion of the DNA sequence within the region of dNTP binding is shown above each graph (see Table 1 for remainder). The final concentrations of the Dpo4 T239W-oligonucleotide complex and dNTP were 1.0 and 0.5 mm, respectively. dATP (black), dTTP (red), dCTP (blue), and dGTP (purple).
Several DNA adducts were included as 0 templates in these fluorescence experiments (Fig. 4, K–O). Only the addition of dGTP opposite a +1 C following O6-MeG produced a fluorescence increase (Fig. 4K). The fluorescence changes observed upon addition of all four dNTPs opposite 8-oxoG are comparable with those observed opposite G (Fig. 4, A and L). Increased fluorescence was observed upon complementary pairing with the +1 C or T following 1,N2-ϵ-G (Fig. 4, m and N), consonant with a complex observed in crystal structures with wild-type Dpo4 (14). dATP pairing opposite 1,N2-ϵ-G also gave a fluorescence increase, in agreement with a previous finding that dATP was relatively efficiently incorporated opposite this lesion (as shown by LC-MS/MS analysis of full-length primer extension products by wild-type Dpo4 (14)). Dpo4 T239W gave comparable results (Table 2). None of the four dNTPs produced a fluorescence increase when added opposite an abasic site (Fig. 4O).
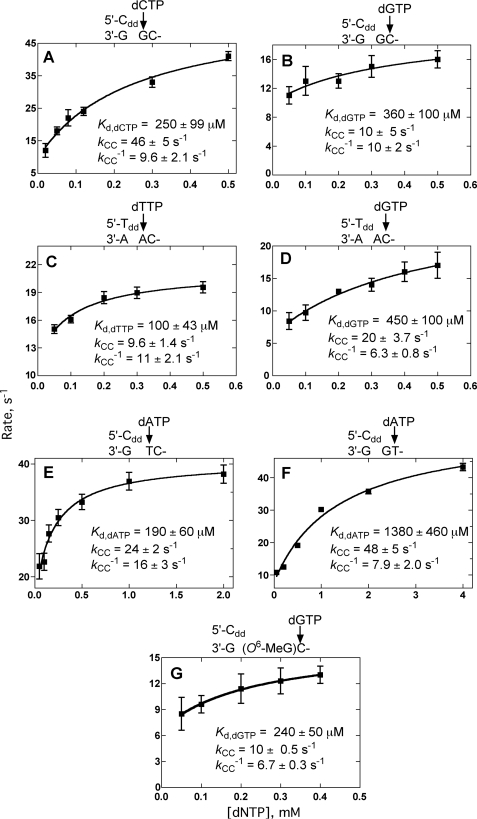
dNTP Binding Affinity and Conformational Change Rates Determined by Stopped-flow Fluorescence Measurements
In the absence of phosphodiester bond formation (using 3′-dd terminated primer-templates), pre-steady-state parameters were measured by adding varying concentrations of dNTPs opposite the 0 or +1 base (supplemental Fig. S6 and Fig. 5, A–G) by fitting plots of the increased fluorescence signal versus time to single exponential equations (Equation 1). Plotting kobs versus [dNTP] to a hyperbola (Equation 2) yielded the ground state dissociation constant, Kd,dNTP, and the forward and reverse conformational change rates (k3 and k−3, respectively, see Fig. 2) (Fig. 5) (26). Surprisingly, these kinetic parameters were similar whether the added dNTP was complementary to the 0 or +1 base.
FIGURE 5.
Estimation of ground state dNTP binding affinity Kd,dNTP and conformational change rate constants (k3 and k−3) (Fig. 2) by fitting the observed rates of fluorescence changes as a function of dNTP concentration. See supplemental Fig. S6 for data traces. The relevant portion of each oligonucleotide sequence and the dNTP are shown above each graph, and the calculated parameters are shown.
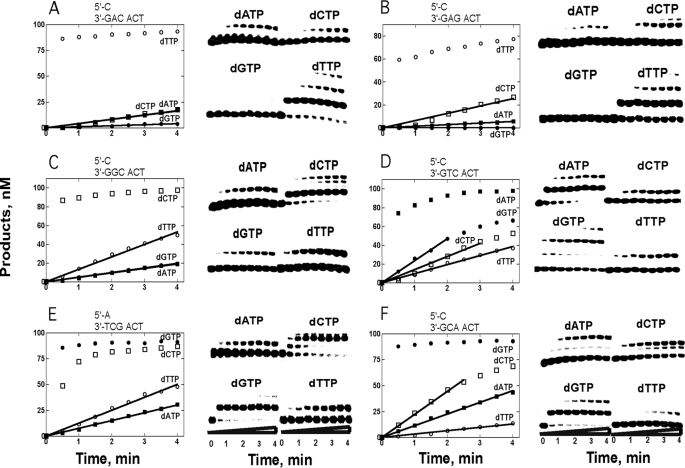
Steady-state Kinetic Analysis of Incorporation/Extension by Dpo4 T239W with Individual dNTPs
Incorporation of dNTPs complementary to a 0 templating base always showed a burst phase (Fig. 6, A–F), which was not quantified here but has been described previously with wild-type Dpo4 (32) and Dpo4 T239W (26). None of the miscoding events, including those leading to −1 frameshift mutations, showed a detectable burst phase and thus are produced inefficiently (kcat ≤0.012 s−1). These slow rates were reflected by a lack of a detectable decrease in fluorescence following phosphodiester bond formation in these cases.
FIGURE 6.
Single dNTP primer incorporation/extension opposite 18-mer templates catalyzed by Dpo4 T239W. The reactions contained 200 nm Dpo4 T239W, 100 nm DNA, 1 mm dNTP, and 5 mm Mg2+ at 37 °C for 4 min. The graph in each part shows product concentration plotted against reaction time.
Steady-state kinetic analysis (Table 3) yielded quantitative information about the various miscoding events, but these data cannot distinguish between misincorporation opposite the 0 template base or incorporation opposite the +1 base, yielding a frameshift. These data (Fig. 6) confirm that rapid, favorable conformational changes related to +1 complementary dNTP binding did not lead to high efficiency phosphodiester bond formation.
TABLE 3.
Steady-state kinetic parameters for the single dNTP incorporation by Dpo4 T239W
The extent of conversion of primer to the product was kept to <20% by adjustment of the enzyme concentration and reaction time.
| Primer-templatea | dNTP | kcat ×103 | Km,dNTP | kcat/Km × 106 | Misincorporation ratio |
|---|---|---|---|---|---|
| s−1 | μm | μm−1s−1 | |||
| 5′-13-mer–C | T | 47 ± 1 | 48 ± 4 | 980 | |
| 3′-18-mer–GAC- | A | 4.2 ± 0.2 | 2000 ± 180 | 2.1 | 2.1 × 10−3 |
| C | 0.25 ± 0.01 | 280 ± 30 | 0.89 | 8.9 × 10−4 | |
| G | 0.36 ± 0.07 | 2600 ± 800 | 0.14 | 1.4 × 10−4 | |
| 5′-13-mer–C | T | 33.6 ± 3.1 | 36 ± 2 | 930 | |
| 3′-18-mer–GAG- | A | 1.4 ± 0.1 | 770 ± 140 | 1.8 | 1.9 × 10−3 |
| C | 0.16 ± 0.01 | 67 ± 7 | 2.4 | 2.6 × 10−3 | |
| G | 0.063 ± 0.014 | 2600 ± 800 | 0.024 | 2.9 × 10−5 | |
| 5′-13-mer–A | G | 120 ± 3 | 11 ± 1 | 11,000 | |
| 3′-18-mer–TCG- | A | 1.7 ± 0.2 | 2700 ± 470 | 0.63 | 5.8 × 10−5 |
| C | 12 ± 1 | 480 ± 90 | 25 | 2.3 × 10−3 | |
| T | 2.4 ± 0.2 | 430 ± 100 | 5.6 | 5.1 × 10−4 | |
| 5′-13-mer–C | G | 150 ± 20 | 78 ± 22 | 1900 | |
| 3′-18-mer–GCA- | A | 1.3 ± 0.1 | 700 ± 150 | 1.9 | 1.0 × 10−3 |
| C | 0.11 ± 0.01 | 2300 ± 450 | 0.048 | 2.5 × 10−5 | |
| T | 1.2 ± 0.11 | 860 ± 190 | 1.4 | 7.4 × 10−4 | |
| 5′-13-mer–C | C | 34.6 ± 1.7 | 17 ± 3 | 2000 | |
| 3′-18-mer–GGC- | A | 0.25 ± 0.01 | 270 ± 20 | 0.93 | 4.7 × 10−4 |
| G | 0.23 ± 0.01 | 150 ± 20 | 1.5 | 7.5 × 10−4 | |
| T | 2.7 ± 0.2 | 900 ± 150 | 3.0 | 1.5 × 10−3 | |
| 5′-13-mer–C | A | 26.1 ± 1.6 | 37 ± 6 | 710 | |
| 3′-18-mer–GTC- | C | 0.49 ± 0.02 | 320 ± 30 | 1.5 | 2.1 × 10−3 |
| G | 1.5 ± 0.05 | 690 ± 50 | 2.2 | 3.1 × 10−3 | |
| T | 0.51 ± 0.03 | 660 ± 90 | 0.77 | 1.1 × 10−3 | |
| 5′-13-mer–C | C | 2.2 ± 0.1 | 216 ± 43 | 10 | |
| 3′-18-mer–G(O6-MeG)C- | A | 0.44 ± 0.02 | 113 ± 18 | 3.9 | 0.38 |
| G | 0.15 ± 0.01 | 91 ± 8 | 1.6 | 0.16 | |
| T | 2.6 ± 0.1 | 219 ± 27 | 12 | 1.2 | |
a See Table 1 for the rest of the oligonucleotide sequences.
DISCUSSION
Frameshift mutations are considered very deleterious, because adding or subtracting a nucleotide produces non-sense protein products. Translesion DNA polymerases appear to be prone to frameshift mutations (9–11, 19). The reason for this behavior may be that these polymerases form relatively few protein contacts with the incoming dNTP and primer-template complex to help anchor the substrates at one site, leading to formation of alternative active site configurations that lead to different products. The ability to effectively bind two template residues within the active site is also apparently unique to Y family polymerases and probably contributes to their propensity to generate frameshift mutations.
Crystal structures of Dpo4 often have purines or bulky adducts as 0 template bases with the dNTP base bound via hydrogen bonds with the +1 base. These structures were originally termed type II structures (21), as opposed to the typical type I structures that simply have the incoming base paired with the 0 templating base. Type II structures provide insight as to how a −1 frameshift event occurs; the dNTP binds opposite the +1 base, followed by phosphodiester bond formation, followed by additional polymerization events to “seal” the bulge formed by the excluded nucleotide. This simple set of events may at first seem obvious when the templating 0 base is a DNA adduct, which is often large in relation to natural bases and therefore the incoming dNTP would need to find an alternative location to bind. A change in the polymerase structure might be anticipated when comparing type I and type II pairing modes, but this is not the case. Instead, the incoming dNTP binds in the same site and orientation relative to the polymerase. In type II complexes, it is the primer terminus that is not translocated toward the active site residues of Dpo4.
Other models used to explain the phenomenon of frameshift mutations (Fig. 1) rely on placement of the +1 base to occupy the 0 templating position, which allows for at least two fates of the 0 base, either it is flipped out or it takes the place of the −1 base at the primer terminus via a “slippage” mechanism. However, regarding Dpo4, these latter models are largely unsupported structurally.
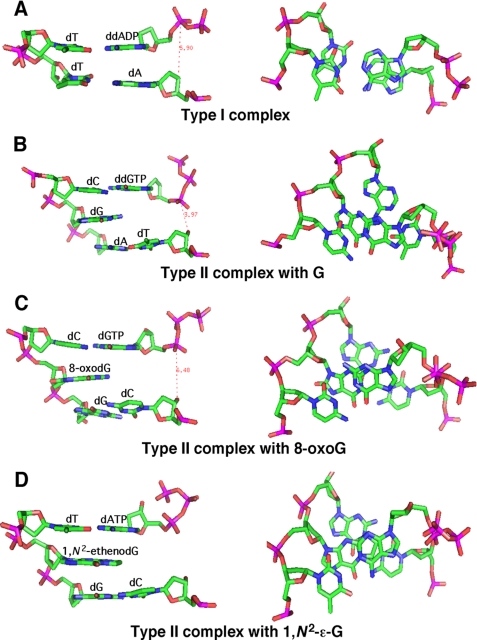
Our results indicate that the stacking potential of the 0 templating base is a crucial factor facilitating formation of −1 frameshift mutations (Fig. 7, A–D). Bulky 0 templating adducts are prone to −1 frameshifts. The LC-MS/MS results (Table 2) indicate that 1,N2-ϵ-G caused −1 frameshift deletions during ∼25–50% of all adduct bypass events, a result similar to that found with wild-type Dpo4 (14). Of the unmodified 0 templating bases, purines were ∼10-fold more likely to cause −1 frameshift deletions than the relatively small pyrimidines (Table 2), although these events were less frequent relative to 1,N2-ϵ-G (9–12% for purines and 0–2% for pyrimidines). Dpo4 type II crystal structures either contain a 0 templating purine or DNA adduct at least the size of a purine, and in each case there is stacking overlap between this base and the incoming base (Fig. 7). Additionally, the fact that there has not been a reported type II structure containing a 0 templating natural pyrimidine can arguably be considered evidence, by default, that the −1 frameshift mutation is unfavored by Dpo4 in these cases. Stacking potential and not necessarily a larger relative stacking surface may also lead to increased −1 frameshift mutations, as was inferred by the recently solved type II crystal structure of a 0 templating hydrophobic thymidine isostere 2′-deoxy-2,4-difluorotoluyl with an incoming dA:dGTP pair at the +1 position (34). Overall, the data suggest that the energy supplied by the increased stacking surface and/or stacking potential of the 0 template base and the accompanying hydrogen bonds formed between the incoming base and +1 base combine to provide sufficient stability to counter the large contortions of the active site that occur when the dNTP is positioned opposite the +1 base.
FIGURE 7.
H-bond and stacking interactions for type I and type II complexes containing G, 8-oxoG, or 1,N2-ϵ-G at the insertion position. The ternary type I complexes (Protein Data Bank code 2ATL (15)) and type II complexes containing G (1JXL (21)), 8-oxoG (2C22 (33)), or 1,N2-ϵ-G (2bqr (14)) all form H-bonds between the incoming d(d)NTP and the opposite template base. The type I complex shows stacking interactions between ddADP and the primer terminus base A. Type II complexes show stacking interactions of d(d)NTP with the −1 unpaired template base (G, 8-oxoG, or 1,N2-ϵ-G). The reaction distance between the 3′-OH of the primer and the α-phosphorus atom of dNTP is shortened due to the tilted configuration of the primer terminus and dNTP.
The single Trp Dpo4 mutant T239W detects what we interpret to be conformational changes specific for incorporation of a complementary dNTP during formation of both type I and type II ternary complexes (26). Because of the proximity of Trp-239 to the duplex, and its location within a flexible “linker” region, Trp-239 is likely sensitive to the position of the DNA duplex relative to nearby subdomains of Dpo4. We suggest that the thumb (and possibly the little finger) domain is translocating the primer-template into an orientation that places the 3′-OH group in a position for phosphoryl transfer. Intuitively, if this happens with a type II complex, then the conformations sampled by the thumb/little finger domains will have to move further out of a thermodynamically favored free energy well (i.e. the thumb/little finger has to “stretch” further to facilitate phosphodiester bond formation), and this may result in a slower rate of product formation (Table 3). However, we found that in the cases when a type II complex forms, conformational changes upon dNTP binding, as reported by the rapid increase in fluorescence signal, were favorable, comparable with formation of type I complexes, even when accompanied by an extremely slow rate of phosphodiester bond formation. This result cannot be explained by a longer “stretching” of the thumb/little fingers domains, because the higher energy cost would likely result in an unfavorable forward conformational change. A more reasonable assessment is that Dpo4 recognizes a complementary +1 template:dNTP pair as it would a complementary 0 template:dNTP pair and responds by making similar movements (as long as the energy requirements are met, e.g. the 0 templating base stacks favorably with the incoming base, see above).
The observation that Dpo4 exhibits favorable fluorescent changes upon formation of ternary complexes that do not lead to catalysis was unexpected. Tsai and Johnson (35) showed that fluorescence changes within the fingers region of the replicative bacteriophage pol T7− report the binding and incorporation of mispaired dNTPs, obviously events that cause relatively high distortion within the active center, but the reported conformational changes upon binding of incorrect dNTPs were unfavorable relative to correct pairing events. However, pol T7− is a high fidelity polymerase, and Dpo4 is not.
An important point is that the propensity of Dpo4 to form type II complexes does not appear to be dependent on sequence context, aside from the purine selectivity. Mutational frequency analysis of Dpo4 by Kokoska et al. (19) showed that a coding sequence with a 0 templating C flanked by a +1 G (i.e. 3′-CG-) gave the highest rate of formation of −1 frameshifts. Our fluorescence and LC-MS/MS experiments show that sequences having pyrimidines at the 0 position are the least likely to form −1 frameshift deletions. The reason for the apparent discrepancy is not obvious. Although the 3′-CG- sequence often led to −1 frameshift deletions in the lacZ template sequence used in the Kokoska et al. study (19), there were other relative “hot spots” with completely unrelated sequences. In some locations, the 3′-CG- sequence did not lead to −1 frameshift mutations, suggesting that hot spots may best be defined by a larger number of nucleotides. However, our results indicate that a “hot spot” may indeed be only 1 base, the identity of the 0 templating base determining the reaction course. Some of the local sequences in the lacZ template, perhaps defined by several bases, may discourage bypassing templates even when the polymerase rapidly forms type II complexes. Studies involving more global changes in primer-template context are required.
In summary, the biochemical approaches used in this study provide strong evidence that the type II complex structure often exhibited in ternary crystal structures of Dpo4 with “0-position” templating adduct or purine bases is the complex on the pathway to produce −1 frameshift deletions by this DNA polymerase (Fig. 1B), and that a driving force is base stacking between the incoming dNTP base and the 0 templating base. Because type II complex formation, as reported by Dpo4 T239W fluorescence, did not depend on sequence context, we rule out the model of template slippage as a driving mechanism to form these deletions (Fig. 1A). This conclusion is further supported by the LC-MS/MS data. We are aware that other DNA polymerases are highly sensitive to sequence context, e.g. family X and Y DNA polymerases make common frameshift errors while copying repeated sequences. We do not rule out the “misincorporation-misalignment” mechanism (Fig. 1C) or the possibility that 0 templating bases will “flip” out with some other DNA adducts, but because these scenarios are not seen in ternary crystal structures except with an AP site or a very bulky lesion (13, 36), the type II complex (Fig. 1B) remains our favored model for unmodified DNA and several adducts.
Acknowledgments
We thank I. D. Kozekoff for synthesis of the oligonucleotide containing 1,N2-ϵ-G, R. L. Eoff for comments on drafts of the manuscript, and K. Trisler for technical assistance in preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES010375 and P30 ES000267 (to F. P. G.) from the USPHS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S9.
- Dpo4
- S. solfataricus P2 DNA polymerase IV
- AP
- apurinic (site)
- 1,N2-ϵ-G
- 1,N2-etheno-2′-deoxyguanosine
- LC
- liquid chromatography
- MS/MS
- mass spectrometry; (tandem)
- M1G
- 3(2′-deoxyribosyl)pyrimido[1,2-α]purin-10(3H)-one
- 8-oxoG
- 7,8-dihydro-8- oxo-2′-deoxyguanosine.
REFERENCES
- 1.Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (eds) (2006) DNA Repair and Mutagenesis, 2nd Ed., American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Loeb L. A., Christians F. C. (1996) Mutat. Res. 350, 279–286 [DOI] [PubMed] [Google Scholar]
- 3.Bond J. A., Gown A. M., Yang H. L., Benditt E. P., Juchau M. R. (1981) J. Toxicol. Environ. Health 7, 327–335 [DOI] [PubMed] [Google Scholar]
- 4.Shigenaga M. K., Hagen T. M., Ames B. N. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10771–10778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodgate R. (1999) Genes Dev. 13, 2191–2195 [DOI] [PubMed] [Google Scholar]
- 6.Goodman M. F. (2002) Annu. Rev. Biochem. 71, 17–50 [DOI] [PubMed] [Google Scholar]
- 7.McCann J., Spingarn N. E., Kobori J., Ames B. N. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. (1966) Cold Spring Harbor Symp. Quant. Biol. 31, 77–84 [DOI] [PubMed] [Google Scholar]
- 9.Potapova O., Grindley N. D., Joyce C. M. (2002) J. Biol. Chem. 277, 28157–28166 [DOI] [PubMed] [Google Scholar]
- 10.Wilson R. C., Pata J. D. (2008) Mol. Cell 29, 767–779 [DOI] [PubMed] [Google Scholar]
- 11.Ohashi E., Bebenek K., Matsuda T., Feaver W. J., Gerlach V. L., Friedberg E. C., Ohmori H., Kunkel T. A. (2000) J. Biol. Chem. 275, 39678–39684 [DOI] [PubMed] [Google Scholar]
- 12.Wolfle W. T., Washington M. T., Prakash L., Prakash S. (2003) Genes Dev. 17, 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling H., Boudsocq F., Woodgate R., Yang W. (2004) Mol. Cell 13, 751–762 [DOI] [PubMed] [Google Scholar]
- 14.Zang H., Goodenough A. K., Choi J. Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., Guengerich F. P. (2005) J. Biol. Chem. 280, 29750–29764 [DOI] [PubMed] [Google Scholar]
- 15.Rechkoblit O., Malinina L., Cheng Y., Kuryavyi V., Broyde S., Geacintov N. E., Patel D. J. (2006) PLoS Biol. 4, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer J., Xing G., Yagi H., Sayer J. M., Jerina D. M., Ling H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14905–14910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eoff R. L., Stafford J. B., Szekely J., Rizzo C. J., Egli M., Guengerich F. P., Marnett L. J. (2009) Biochemistry 48, 7079–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Musser S. K., Saleh S., Marnett L. J., Egli M., Stone M. P. (2008) Biochemistry 47, 7322–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokoska R. J., Bebenek K., Boudsocq F., Woodgate R., Kunkel T. A. (2002) J. Biol. Chem. 277, 19633–19638 [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K., Kunkel T. A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4946–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling H., Boudsocq F., Woodgate R., Yang W. (2001) Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 22.Bloom L. B., Chen X., Fygenson D. K., Turner J., O'Donnell M., Goodman M. F. (1997) J. Biol. Chem. 272, 27919–27930 [DOI] [PubMed] [Google Scholar]
- 23.Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1997) J. Biol. Chem. 272, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 24.Kunkel T. A. (1986) J. Biol. Chem. 261, 13581–13587 [PubMed] [Google Scholar]
- 25.Kunkel T. A., Soni A. (1988) J. Biol. Chem. 263, 14784–14789 [PubMed] [Google Scholar]
- 26.Beckman J. W., Wang Q., Guengerich F. P. (2008) J. Biol. Chem. 283, 36711–36723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borer P. N. (1975), in Handbook of Biochemistry and Molecular Biology, (Fasman G. D. ed) 3rd Ed., pp. 589–590, CRC Press, Cleveland, OH [Google Scholar]
- 28.Choi J. Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., Guengerich F. P. (2006) J. Biol. Chem. 281, 38244–38256 [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Eoff R. L., Kozekov I. D., Rizzo C. J., Egli M., Guengerich F. P. (2009) J. Biol. Chem. 284, 17687–17699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman M. F., Creighton S., Bloom L. B., Petruska J. (1993) Crit. Rev. Biochem. Mol. Biol. 28, 83–126 [DOI] [PubMed] [Google Scholar]
- 31.Zhang H., Eoff R. L., Kozekov I. D., Rizzo C. J., Egli M., Guengerich F. P. (2009) J. Biol. Chem. 284, 3563–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H., Bren U., Kozekov I. D., Rizzo C. J., Stec D. F., Guengerich F. P. (2009) J. Mol. Biol. 392, 251–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zang H., Irimia A., Choi J. Y., Angel K. C., Loukachevitch L. V., Egli M., Guengerich F. P. (2006) J. Biol. Chem. 281, 2358–2372 [DOI] [PubMed] [Google Scholar]
- 34.Irimia A., Eoff R. L., Pallan P. S., Guengerich F. P., Egli M. (2007) J. Biol. Chem. 282, 36421–36433 [DOI] [PubMed] [Google Scholar]
- 35.Tsai Y. C., Johnson K. A. (2006) Biochemistry 45, 9675–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling H., Sayer J. M., Plosky B. S., Yagi H., Boudsocq F., Woodgate R., Jerina D. M., Yang W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]