Abstract
Heparin and heparan sulfate glycosaminoglycans (HSGAGs) comprise a chemically heterogeneous class of sulfated polysaccharides. The development of structure-activity relationships for this class of polysaccharides requires the identification and characterization of degrading enzymes with defined substrate specificity and enzymatic activity. Toward this end, we report here the molecular cloning and extensive structure-function analysis of a 6-O-sulfatase from the Gram-negative bacterium Flavobacterium heparinum. In addition, we report the recombinant expression of this enzyme in Escherichia coli in a soluble, active form and identify it as a specific HSGAG sulfatase. We further define the mechanism of action of the enzyme through biochemical and structural studies. Through the use of defined substrates, we investigate the kinetic properties of the enzyme. This analysis was complemented by homology-based molecular modeling studies that sought to rationalize the substrate specificity of the enzyme and mode of action through an analysis of the active-site topology of the enzyme including identifying key enzyme-substrate interactions and assigning key amino acids within the active site of the enzyme. Taken together, our structural and biochemical studies indicate that 6-O-sulfatase is a predominantly exolytic enzyme that specifically acts on N-sulfated or N-acetylated 6-O-sulfated glucosamines present at the non-reducing end of HSGAG oligosaccharide substrates. This requirement for the N-acetyl or N-sulfo groups on the glucosamine substrate can be explained through eliciting favorable interactions with key residues within the active site of the enzyme. These findings provide a framework that enables the use of 6-O-sulfatase as a tool for HSGAG structure-activity studies as well as expand our biochemical and structural understanding of this important class of enzymes.
Introduction
Heparan sulfate glycosaminoglycans (HSGAGs)3 comprise an important polysaccharide constituent of many proteoglycans (see Ref. 1, for a review). These glycans are linear polymers based on the variably repeating disaccharide unit (uronic acid 1→4 glucosamine)n, where n represents a variably repeating number (typically 10–200). As present in nature, these sugars possess an extensive chemical heterogeneity that is largely attributed to the mosaic arrangement of O- and N-linked sulfates present at different positions along each sugar chain (2, 3). Additional structural variations include the presence of N-linked acetates at the glucosamine C2 position as well as epimerization of the uronic acid C5 carboxylate to form either β-d-glucuronic acid or α-l-iduronic acid. Fundamental to understanding HSGAG structure-activity relationships is the appreciation that polydispersity of the glycan fine structure is not random. Instead, it is the end product of a complex and concerted biosynthetic pathway involving numerous modifying enzymes, whose relative expression levels and specific activities are regulated in a cell- and tissue-specific fashion. This programmed diversity of HSGAG structure (4) ultimately plays out at a functional level, namely through the dynamic regulation of numerous biochemical signaling pathways (2) relating to such processes as cell growth and differentiation (5), cell death (6, 7), intercellular communication, adhesion and tissue morphogenesis (8). HSGAGs are present as structurally defined binding epitopes on the cell surface and hence also play an important role in microbial pathogenesis (9, 10).
In contrast to the complex enzymatic process by which these polysaccharides are made, it appears that their catabolism is considerably more straightforward, both in the scope of its purpose and the means by which it is carried out at the biochemical level. Sequential HSGAG degradation has been demonstrated in several microorganisms (11–13), which depend on these sulfated polysaccharides not only as a carbon source but often as a means of scavenging inorganic sulfate (14). The Gram-negative soil bacterium Flavobacterium heparinum (a.k.a. Pedobacter heparinus) is an excellent example of this process. As such this organism has proven to be a rich biological source for the isolation and molecular cloning of several GAG degrading enzymes (15, 16). Like the enzymes of the lysosomal pathway, many of the flavobacterial enzymes possess well defined substrate specificity. Given the utility of other HSGAG-degrading enzymes derived from F. heparinum, including the heparinases as well as the Δ4,5-glycuronidase (17) and the 2-O-sulfatase (18), we reasoned that cloning and characterization of additional sulfatases would enable the development of important tools for investigating HSGAG structure. This is especially relevant given extensive experimental evidence that points to the regulated expression of endolytic HSGAG desulfating enzymes (especially 6-O-sulfatase) and their secretion into the extracellular matrix as a mechanism of modulating GAG structures critical for protein interactions (19, 20). Therefore, the use of HSGAG degrading enzymes as analytical tools is central to unlocking the structural basis of HSGAG function and their potential use in generating structure-specific, bioactive glycans for therapeutic applications (21). To utilize these enzymes correctly and efficiently requires not only a detailed understanding of the biochemistry of these enzymes; it also requires their ample availability for in vitro use. Both of these criteria already have been satisfied in structure-function studies of several HSGAG-related degrading enzymes, such as heparinase I (22), heparinase II (23), a unique unsaturated glycuronidase (17), and the 2-O-sulfatase (24). We have been able to clone and identify two additional sulfatases downstream to these enzymes, namely the 6-O-sulfatase and N-sulfamidase.
In this study, we report the molecular cloning and recombinant expression in Escherichia coli of the F. heparinum sulfohydrolase, namely the glucosamine 6-O-sulfatase, and in the accompanying manuscript (39) we report detailed studies on the N-sulfamidase as well as a model for how the 2-O- and 6-O-sulfatases, and N-sulfamidase work in concert. In both studies, we also present detailed molecular structural modeling and biochemical analyses of the recombinant enzyme as it relates to substrate specificity, optimal reaction conditions, and the role of divalent metal ions in the action of the enzyme. Taken together with our previous studies regarding other members of the heparinase-sulfatase family of enzymes, we have now unraveled the complete framework for HSGAG degradation in F. heparinum.
EXPERIMENTAL PROCEDURES
Reagents
Fluorescent glucopyranoside substrates 4-methyllumbelliferyl-α/β-d-glucopyranoside (4-MU-α-d-Glc and 4-MU-β-d-Glc) were purchased from EMD Biosciences, Inc. (San Diego, CA). 6-O-Sulfated fluorogenic glycopyranoside derivatives were obtained through Toronto Research Chemicals (Toronto, Canada). Glucosamine and galactosamine monosaccharides, and arylsulfate substrates 4-catechol-sulfate and 4-methyllumbelliferyl-sulfate were purchased from Sigma. Exo-glucosidases were purchased from MP Biomedicals (Irvine, CA). Materials for genomic library construction and screening were obtained from Stratagene (La Jolla, CA). Reagents for site-directed mutagenesis were also obtained from Stratagene. PCR enzymes, TOP10 chemically competent cells, and oligonucleotide primers were obtained from Invitrogen. Additional molecular cloning reagents were purchased from New England Biolabs (Beverly, MA) or the manufacturers listed.
Molecular Cloning of Flavobacterial 6-O-Sulfatase
The flavobacterial sulfatase gene was cloned by PCR from a λZAPII flavobacterial genomic library originally screened using DNA hybridization probes specific to the 2-O-sulfatase (18). Library construction, hybridization screening, and phage excision were as described. An overlapping clone was expanded by chromosomal walking and restriction mapping using the Lambda DASH II genomic cloning kit (Stratagene) for the ligation of size-fractionated genomic DNA (generated by partial Sau3AI digestion). 2-O-Sulfatase-positive clones from an amplified library were plaque purified through three successive rounds, and the DNA were purified from a high titer lysate using standard techniques. For DNA sequencing, recombinant phage DNA was subcloned into pBluescript SK±. The coding sequence of the putative sulfatase gene (described throughout this paper as orfb) was identified by the canonical PFAM (25) sulfatase family identifier (CXPXRXXXXS/TG) and subsequently PCR amplified using the following primer set for orfb (6-O-sulfatase), 5′-GAA TTC ATA TGG GTA AAT TGA AAT TAA TTT TA-3′ (forward) and 5′-GGA TCC TCG AGT TAT AAA GCT TCA GTT GGA TTC GT-3′ (reverse). The amplified gene was subcloned into the T7-based bacterial expression vector pET28a (Novagen) as an NdeI-XhoI cassette (restriction sites underlined). Cloning as such allowed the gene to be expressed as an NH2-terminal His6 fusion with an intervening thrombin cleavage site for facile removal of this tag following protein purification.
Bacterial Expression and Protein Purification
Recombinant protein expression in E. coli strain BL21(DE3) and one-step affinity purification by nickel chelation chromatography were as described for the 2-O-sulfatase (18). The prediction of the NH2-terminal signal sequence and putative cleavage site for the protein was made by the computational method of von Heijne (26). Engineering and expression of the truncated protein (minus signal sequence) was as described above for the full-length gene with the exception of substituting the 5′ primer that was used in the original PCR amplification step. The internal primer included 5′-TCT AGA CAT ATG TCT TGC CAG CAG CCT AAA C-3′ for orfb with the NdeI site underlined. As such, the orfb gene sequence begins at Met-18. Removal of the His6 tag was achieved by site-specific protease cleavage using the thrombin cleavage capture kit (Novagen). Proteolysis conditions were generally as described for other recombinantly expressed flavobacterial heparin-degrading enzymes (17, 18). Following concentration of orfb by ultrafiltration, the cleaved protein was dialyzed against 4 liters of 50 mm Tris, pH 7.5, and 0.1 m NaCl, 4 °C, overnight using a 3-ml slide-a-lyzer cassette with a 10,000 molecular weight cutoff (Pierce).
Final protein concentration was determined calorimetrically using the Bradford assay (Bio-Rad) and confirmed by UV absorption spectroscopy using theoretical molar extinction coefficients (ϵ280) of 94,730 m−1 (61,572 Da) for the NH2-terminal truncated orfb (6-O-sulfatase). This value was calculated for the thrombin-cleaved protein lacking a His6 purification tag. The enzyme was stored at 4 °C at a concentration of ∼10 mg/ml and full enzyme activity was retained for several months under these conditions.
Site-directed Mutagenesis of Putative 6-O-Sulfatase Active Site Residues
Based on homology modeling of the 6-O-sulfatase (described below), three residues were initially chosen for mutagenesis: Cys-80, Asp-186, and Asp-374. All three residues were mutated to alanines. Site-directed mutagenesis was completed by thermal cycling using the QuikChangeTM method (Stratagene) generally as recommended by the manufacturer. The same pET28 (His6) vector used for bacterial expression of the wild type 6-O-sulfatase was likewise used as the template for linear amplification. The following oligonucleotide primer pairs (forward and reverse) were used: 5′-CTG TTT TGT AAC CAA TGC AGT Tgc CGG GCC ATC CAG GGC TAC-3′, 5′-GTA GCC CTG GAT GGC CCG gcA ACT GCA TTG GTT ACA AAA CAG-3′ (C80A); 5′-C CTT GAA AAA AGG GAC CAT GcT AAA CCC TTT CTG ATG ATT TAC-3′, 5′-G TAA ATC ATC AGA AAG GGT TTA gCA TGG TCC CTT TTT TCA AGG-3′ (D186A); 5′-CC ATT ATT GTC TAT ACT TCC GcT CAG GGC TTT TAT TTG GGT G-3′, 5′-CA CCC AAA TAA AAG CCC TGA gCG GAA GTA TAG ACA ATA ATG G-3′. Base pair changes are noted in lowercase, boldface type. Parental DNA was restricted using DpnI. Mutated DNA was transformed into XL Blue chemically competent cells. Targeted mutations were confirmed by direct DNA sequencing of both strands.
Arylsulfatase Assay
Arylsulfatase activity was measured independently using two chromogenic substrates, 4-catechol sulfate and 4-methylumbelliferyl sulfate. The catechol substrate assay was conducted generally as described (27). Briefly, 10 mm substrate was incubated with ∼30 μm recombinant enzyme overnight (12–15 h) at 37 °C in a 100-μl reaction volume that included 50 mm MES, pH 7.0, and 2 mm CaCl2. Reactions were quenched by the addition of 5 μl of 5 m NaOH, and colorimetric activity was determined spectroscopically at 515 nm. The fluorimetric arylsulfatase assay using 4-methylumbelliferyl sulfate was as described (28) with some modifications. Reaction conditions included 10 μm enzyme, 2 mm 4-methylumbelliferyl sulfate, 50 mm sodium acetate, pH 6.0, and 5 mm CaCl2 in a 20-μl reaction volume. Enzyme incubation temperature was 30 °C. Activity was measured as a function of time ranging from 3 to 24 h; the reactions were quenched by the addition of 200 μl of 0.5 m Na2CO3, pH 10.7. Detection of fluorescent methylumbelliferone was measured at this alkaline pH using a SpectraMax microtiter plate reader (GE Healthcare) set at excitation and emission wavelengths of 360 and 440 nm, respectively. Fluorescence intensity was corrected against background (minus enzyme control). In both assays, 0.5 units of arylsulfatase from Aerobacter aerogenes (Sigma) were used as a positive control.
Pilot 6-O-Sulfatase Assay
Initial assessment of substrate specificity and pH optima was made using a capillary electrophoresis-based assay for the detection of desulfated products. Preliminary enzyme activity was measured against the following series of fluorescently derivatized, monosulfated gluco- and galactopyranosides: 4-MU-GlcNAc,6S, 4-MU-GlcNS, 4-MU-GalNAc,6S, and 4-MU-Gal6S (where Glc refers to glucosamine and Gal refers to galactose). Standard reactions included 1 mm substrate, 1–10 μm enzyme, 50 mm sodium acetate, pH 5.5–6.5, and 5 mm CaCl2 in a 20-μl reaction volume. For the pilot experiment, exhaustive reactions involved overnight incubations at 30 °C. Enzyme was inactivated by heat denaturation at 95 °C for 10 min, followed by a 10-fold dilution into water. Reaction products were resolved by capillary electrophoresis using a 25-cm long, 75-μm inner diameter fused silica capillary (Agilent Technologies). Electrophoresis was carried out under negative polarity by applying a voltage of −15 kV (∼1.2 watts) for 10 min. Substrate desulfation was measured as a percentage of substrate depletion relative to a minus enzyme control as monitored by the loss of UV absorbance at 315 nm detected at ∼4 min. A standard capillary electrophoresis buffer included 50 mm Tris and 10 μm dextran sulfate (average molecular mass of 10,000 Da) adjusted to pH 2.0 with phosphoric acid.
The effect of pH was likewise measured by capillary electrophoresis using the following three sets of buffers with overlapping pH ranging from 4.5 to 8.0: 50 mm sodium citrate at 4.5, 5.0, and 5.5; 50 mm MES at 5.5, 6.0, 6.5, and 7.0; and 50 mm MOPS at 6.5, 7.0, 7.5, and 8.0. Reactions included 1 μm enzyme, 2 mm 4-MU-GlcNAc,6S, 50 mm buffer, and 5 mm CaCl2 in a 20-μl reaction volume. Assay was initiated by the addition of 2 μl of a 10× enzyme stock to 18 μl of preheated reaction mixture. Reactions were carried out at 30 °C for 60 min and quenched by heat and dilution as described. The ability of the enzyme to desulfate unsaturated heparin and chondroitin disaccharides was assessed essentially as described for CE-based compositional analyses of enzymatically generated glycosaminoglycan di- and tetrasaccharides (29). For these studies, the following disaccharide substrates were tested: ΔUGlcNAc,6S, ΔU2SGlcNAc,6S, ΔUGlcNS, ΔUGlcNS,6S, ΔU2SGlcNS,6S, and ΔUGalNAc,6S. Reactions included 500 μm substrate, 10 μm enzyme, 50 mm sodium acetate, pH 6.5, and ±2 mm CaCl2 in a 20-μl reaction volume.
Coupled Enzyme Assay for the Determination of Biochemical Reaction Conditions and Steady-state Kinetics
Indirect measurement of enzyme activity was also made using a fluorimetrically based plate assay in which the prerequisite desulfation of the appropriate glucopyranoside 1→4-methlybelliferone substrate by the 6-O-sulfatase was coupled to the glucosidase-mediated hydrolysis of the stereo-specific 1→4-glycosidic linkage between the pyranose ring and the adjoining fluorophore. Release of the free fluorophore (4-MU) was monitored spectroscopically as described above for the arylsulfatase assay using 4-MU sulfate. Hydrolysis of the substrate 4-MU-β-d-GlcNAc,6S at the 6-OH position was coupled to β-glucosidase purified from sweet almonds (MP Biomedicals, catalogue number 195197). The efficacy of the coupled assay was contingent on the intrinsically poor ability that the glucosidase possesses for hydrolyzing the glycosidic bond when the adjoining glucosamine is modified by a sulfate. The presumption of the sulfatase activity being the rate-limiting step was established experimentally. Reaction conditions were optimized to satisfy three criteria: 1) linear readout of the fluorescent signal that was directly proportional to sulfatase activity; 2) quantitative release of 4-MU by glucosidase activity under the biochemical conditions examined; and 3) negligible fluorescent quenching of free chromophore.
The standard reaction conditions for the assay included 2 μm recombinant enzyme, 50 mm sodium acetate buffer, pH 5.5, and 5 mm CaCl2 in a 20-μl reaction volume. The 4-MU-GlcNAc,6S substrate concentration was varied from 0.1 to 2 mm. Each well of a microtiter plate (prechilled on ice) was treated with 2 μl of enzyme, followed by gentle vortexing of the plate and spin down of the well contents for 1 min at 500 × g and 4 °C. The assay was initiated by transferring the 96-well plate to a heating block preequilibrated at 30 °C. The incubation of the 6-O-sulfatase enzyme was carried out at 30 °C for 20 min, after which the enzyme activity was inactivated by heat denaturation (95 °C, 10 min). For the glucosidase enzyme, the microtiter plate was once again chilled on ice and 40 units of β-glucosidase was added to each well. These were followed by vortex of the plate for mixing and spin down at 500 × g for 1 min at 4 °C. The contents were then transferred to a heating block preequilibrated at 37 °C. Incubation proceeded for 60 min prior to being quenched with 200 μl of 0.5 m Na2CO3, pH 10.7. Reactions were transferred to a black 96-well, flat-bottom fluoroimmunoassay plate and fluorescence measured as described above for the detection of free 4-MU. Fluorescent signal was adjusted to background (minus sulfatase control). For β-glucosidase, this background hydrolysis was somewhat dependent on the initial 4-MU-GlcNAc,6S concentration, but was typically less than 10%. Molar conversion of product was extrapolated from a standard curve generated from various concentrations of 4-MU from 0 to 300 μm. Michaelis-Menten kinetics was extrapolated from Vo versus substrate concentration plots fit by non-linear regression to pseudo-first order kinetics and all obtained data represent the mean of three experimental trials.
The coupled enzyme assay was also used for the assessment of enzyme activity for select site-directed mutants. Enzyme activity was measured kinetically using a single saturating concentration (2 mm) of fluorescently labeled substrate. Results are reported as % activity relative to the wild type enzyme.
Compositional Analyses of Sulfatase-treated Heparin
20 μg of heparin was preincubated with 10 μm 6-O-sulfatase for 8 h at 30 °C in a 20-μl reaction volume that included 25 mm sodium acetate, pH 7.0, and 2 mm calcium acetate, pH 7.0. Following this preincubation, the enzyme was inactivated by heat denaturation at 95 °C for 10 min, and heparin was exhaustively digested overnight at 37 °C by the addition of 2 μl of a concentrated enzyme mixture containing heparinase I and III. Subsequent CE-based compositional analyses of heparinase-derived disaccharides were completed as described (17).
Electrospray Ionization (ESI)-Mass Spectrometry of Sulfated Glucosamine Monosaccharides
Electrospray ionization-mass spectrometry was performed in the negative ion mode using an Agilent 1100 Series VL LC/MSD trap. For simplicity, the samples were prepared by adding MeOH directly to the enzymatic reaction mixtures without purification, and directly injected into the source of the mass spectrometer using a syringe pump at a rate of ∼8 μl/min. The SPS function of the software (LC/MSD Trap Software 4.1 Build 143, MSD Trap Control Version 5.0 Build 65) was used to tune the instrument, with the target mass set to the mass of the substrate, the sample stability set to 50%, and the drive level set to 100%. Data were acquired over the scan range of m/z 100–2200 by accumulating 30,000 ions per scan. Capillary voltage was set to 3000 V. Nitrogen was used as the drying gas, whereas helium was used as the nebulizing gas, with flow rates of 5 and 15 liters/min, respectively. In each case a minimum of 10 spectra were averaged. Substrate specificity was determined using unlabeled glucosamide monosaccharide substrates with various substitutions at the 2-N, 3-OH, and 6-OH positions. Reactions were carried out with 2.5 mm substrate, 5 mm CaCl2, 1 μm enzyme, and 50 mm acetate buffer at pH 5.5 and 37 °C. Reactions were quenched by diluting the samples 1:4 in MeOH. The carrier solvent was H2O:MeOH (1:4, v/v). In the experiments, 4-MU-GlcNS was added prior to injection as the internal standard to monitor ionization and mass accuracy in source and trap.
Homology Modeling of 6-O-Sulfatase and Docking of Substrates
ClustalW (33) was used to perform multiple sequence alignment of 6-O-sulfatase with bacterial and lysosomal enzymes from the highly conserved sulfatase family (Fig. 1). The crystal structures of human arylsulfatase A (PDB code 1AUK), human arylsulfatase B (PDB code 1FSU), and P. aeruginosa arylsulfatase B (PDB code 1HDH) were obtained and structure-based multiple superposition was performed using CE-MC (30) and SuperPose (31) tools. A homology-based structural model of the 6-O-sulfatase was generated using the Homology module of InsightII version 2005 (Accelrys, San Diego, CA) using human arylsulfatase A as a template. The deletions in the modeled structure were closed by constrained minimization upon holding most of the structure rigid, except for regions close to the deletion site during 300 iterations of steepest descent and 500 iterations of conjugate gradient minimization without including charges. The loops and side chains of all residues were then allowed to move freely by performing 500 iterations of steepest descent minimization. The refined structure was then subjected to 500 iterations of steepest descent minimization without including charges and 500 iterations of conjugate gradient minimization including charges to obtain the final predicted model of the enzyme.
FIGURE 1.
Multiple sequence alignment of the sulfatases using ClustalW. The flavobacterial 6-sulfatase enzyme is a member of a large sulfatase family. The putative active site is boxed, with the critically modified Cys-80 noted by an asterisk. Invariant residues are shaded in dark gray, those with partial identity in light gray, and conservative substitutions in charcoal. Multiple sequence alignment was generated by ClustalW (1) using only select sequences identified from a BLASTP search of the protein data base. Enzymes with the following GenBank accession numbers are abbreviated as follows: F. heparinum 6-O-sulfatase (FH6S); F. heparinum 2-O-sulfatase (FH2S); Pseudomonas aeruginosa arylsulfatase (PARS, GenBank code CAA88421); human N-acetyl-galactosamine-6-sulfate sulfatase or chondroitin 6-sulfatase (HGal6S, GenBank code AAC51350); human cerebroside-3-sulfate sulfatase or arylsulfatase A (HARSA, GenBank code AAC51350); human N-acetylgalactosamine-4 sulfate sulfatase or arylsulfatase B (HARSB, GenBank code AAC51350).
The structures of GlcNS, GlcNS,6S, and GlcNS,3S,6S monosaccharides were obtained by appropriate modifications to the coordinates of the heparin structure (PDB 1HPN). These monosaccharide substrates were docked to the enzyme. To further verify the substrate specificity of the enzyme, the coordinates of chondroitin sulfate were obtained from its crystal structure (PDB code 1C4S) to generate the structural models of the monosaccharides GalNAc,6S and GalNAc,6S,4S. The initial orientation of the different substrates in the groove of the enzyme active site was facilitated by the highly conserved position of the cleavable sulfate group relative to Oγ-1 of the geminal diol as observed from the crystal structures of the sulfatase enzyme family. The glycosidic and exocyclic torsion angles were then adjusted manually upon fixing the 6-O-sulfate group to eliminate unfavorable steric contacts with the amino acids of the enzyme active site. Following this, the enzyme-substrate complexes were subjected to optimization with 400 steps of steepest descent followed by 500 steps of Newton-Raphson minimization including charges. To constrain the ring torsion angles to maintain the ring conformation of the monosaccharides during the process of energy minimization, a force constant of 7000 kcal/mol was utilized. Most of the enzyme was held rigidly, whereas regions constituting the active site were allowed to move freely.
The Viewer, Builder, and Discover modules of InsightII version 2005 (Accelrys) were used for visualization, structure building, and energy minimization, respectively. The potentials for all structures were assigned using the AMBER force field modified to include carbohydrates with sulfate and sulfamate groups. The AMBER force field (Amber95) provided with the Discover module was used to assign the potentials for both the enzyme and substrate. The parameters for sulfates and sulfamate groups in glycosaminoglycans described previously (32) were incorporated into this force field to assign the potentials for the sulfated monosaccharide substrates. A distance-dependent dielectric constant of 4*r and scaling of 0.5 for the p1–4 cross-terms were used in the Discover module for the AMBER force field-based simulations according to specifications in the InsightII manual.
RESULTS
Molecular Cloning and Recombinant Expression of F. heparinum Sulfatase Gene
The sulfatase gene described in this study was first identified through the screening of a genomic library with hybridization probes directed toward the flavobacterial 2-O-sulfatase (18). An overlapping phagemid clone identified during this process was expanded by chromosomal walking and restriction mapping. Sequence analyses of this genomic region revealed a sizeable open reading frame of 1647 base pairs (described hereafter as orfb). The gene sequence putatively encodes a protein of 545 amino acids in length starting at the initiating Met. The sequence does not possess an obvious Shine-Delgarno ribosomal binding site within 10 nucleotides of the initiating ATG codon. A closer examination of the sequence at the protein level revealed several important features. The flavobacterial orfb possesses an NH2-terminal hydrophobic signal peptide and the corresponding cleavage site sequence predicted by the von Heijne method for Gram-negative bacteria (26). The orfb gene product is characterized by a theoretical pI of 8.6 and also contains the canonical sulfatase domain as described by the Protein Family (PFAM) identifier PF 000884 (see “Discussion”).
The putative sulfatase gene was robustly expressed in E. coli as a soluble enzyme. Satisfactory expression of the soluble enzyme required the genetically engineered removal of the amino-terminal signal sequence of the protein. Exclusion of this domain had little deleterious effect on the specific activity of the enzyme (data not shown). At the same time, replacement of this NH2-terminal peptide with a histidine (His6) tag facilitated purification of the recombinant protein in essentially a single chromatographic step to greater than 80% purity (data not shown). Subsequent thrombin cleavage of the histidine tag was carried out as described under “Experimental Procedures.” These ΔNH2-terminal truncations (lacking both the native signal sequence and NH2-His6 tag) were used in all subsequent biochemical characterizations of the sulfatase. The apparent molecular mass of the recombinant protein based on SDS-PAGE was consistent with its theoretical molecular mass calculated from the amino acid composition (orfb gene product, 61,572 Da).
Biochemical Characterization of Recombinant HSGAG 6-O-Sulfatase: Preliminary Determination of Monosaccharide Substrate Specificity
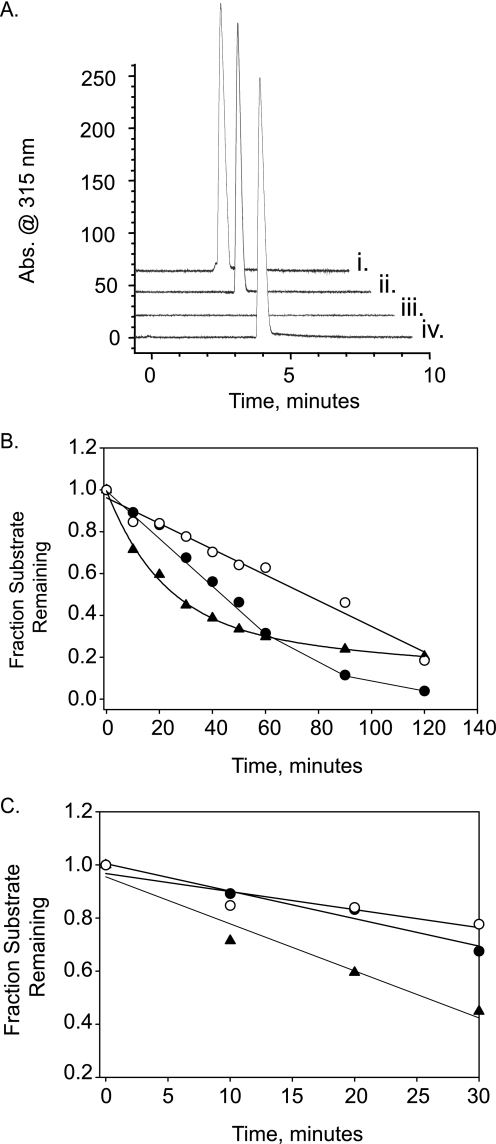
As a first step in biochemical characterization of the orfb sulfatase, we examined whether this enzyme, similar to the previously characterized 2-O-sulfatase (18), can function as a generic arylsulfatase. Both enzyme activities were tested against 4-catechol sulfate and 4-MU sulfate, two different aromatic sulfate esters commonly used as substrates to make this assessment. The 2-O-sulfatase exhibited an appreciable level of hydrolytic activity relative to a known arylsulfatase from A. aerogenes, which served as a positive control (data not shown). The orfb sulfatase partly hydrolyzed the 4-MU sulfate at a much slower but discernible rate. To further test our prediction that the orfb sulfatase acts on carbohydrates, in particular, heparin and heparan sulfate, we used a modified substrate wherein the sulfated hexosamine was linked 1→4 (α or β) to 4-MU. The presence of this chromophore allowed us to directly monitor desulfation of the monosaccharide by capillary electrophoresis. Four monosulfated substrates were tested, all of which were commercially available. These included the two “heparin” monosaccharides 4-MU-GlcNS,6S and 4-MU-GlcNS in addition to the 6-O-sulfated galactose sugars 4-MU-Gal6S and 4-MU-GalNAc,6S (corresponding to the monosaccharide constituents of keratan sulfate and chondroitin/dermatan sulfate, respectively). In this analysis, the orfb sulfatase was found to be specific for the 6-O-sulfated glucosamine (Fig. 2A) and did not act upon either of the two 6-O-sulfated galactose sugars. We further investigated the substrate specificity of this enzyme by examining the influence of various substitutions at the 2- amino, 3-OH, and 6-OH positions of the glucosamine. In these experiments, desulfation of non-derivatized monosaccharide substrates was detected and quantified by ESI-mass spectrometry. In this analysis, the enzyme acted specifically on the 6-O-sulfate position and required a substituted amine (acetate or sulfate) at the 2-amino position. A comparative kinetic analysis of the two corresponding substrates (GlcNAc,6S versus GlcNS,6S) indicated only a modest preference of the enzyme for the monosulfated substrate (Fig. 2C) based on initial kinetic profiles. The enzyme was also completely inhibited by the presence of a 3-O-sulfate group (data not shown). Based on these findings, we defined the orfb as a heparin/heparin sulfate 6-O-sulfatase. To investigate these experimental observations in greater detail, we proceeded to analyze the purified recombinant enzyme using a combination of more detailed biochemical and structural studies.
FIGURE 2.
Substrate specificity of recombinant 6-O-sulfatase. A, the specificity of the 6-O-sulfatase as heparin/heparan desulfating enzyme. Desulfation of 4-MU-Gal6S (i), 4-MU-GalNAc,6S (ii), or 4-MU-GlcNAc,6S (iii and iv) by the recombinant 6-O-sulfatase was followed by capillary electrophoresis. Exclusive desulfation of 4-MU-GlcNAc,6S by 6-O-sulfatase is evidenced by a singular disappearance of absorbance at 315 nm in electrophoretogram iii (that normally appears at ∼4 min). Minus enzyme control is shown in iv. Electrophoretograms are offset for illustrative purposes. B, time course analyses of recombinant 6-O-sulfatase activity using three different, 6-O-sulfated monosaccharide substrates (each at 2.5 mm). GlcNAc,6S (●), GlcNS,6S (○), and 4-MU-GlcNAc,6S (▴) are indicated. C, close-up of the initial kinetics out to 30 min. Desulfation kinetics were measured by ESI-mass spectrometry as described under “Experimental Procedures.”
The CE-based assay was also used to determine the pH optimum for the enzyme. The 6-O-sulfatase enzyme exhibited slightly acidic pH optima (between 5.5 and 6.5) and showed higher activity in acetate buffer when compared with sulfonate buffers, such as MES and MOPS when examined over this same pH range.
Optimization of in Vitro Reaction Conditions
Having identified a suitable chromogenic substrate for the recombinant sulfatase, we also used this substrate to further develop a fluorescence-based plate assay as the means to define the optimal in vitro reaction conditions for 6-O-sulfatase. We investigated parameters such as ionic strength and the effect of divalent metal ions as well as steady-state enzyme kinetics. We chose a coupled enzyme assay in which the recombinant sulfatase served as the primary (product limiting) enzyme and β-glucosidase served as the secondary enzyme. Use of this second enzyme permitted the indirect detection of relative sulfatase activity by means of the stoichiometric release of free 4-MU, which served as the fluorescent signal. This coupled assay for the 6-O-sulfatase was validated in control experiments demonstrating only modest hydrolysis by the glucosidase of the β1→4-MU glycosidic linkage of the sulfated glucosamine. The 6-O-sulfatase was sensitive to increasing ionic strength as measured by the addition of NaCl and 50% inhibition was observed at ∼200 mm NaCl with less than 20% activity remaining at 1 m NaCl relative to the zero NaCl control. Moreover, 6-O-sulfatase was inhibited by the addition of sulfate or phosphate, and between these anions, phosphate was clearly a more effective inhibitor with 50% inhibition of 6-O-sulfatase activity observed at ∼2 mm PO42− compared with ∼20 mm SO42−.
Structural Investigation of Substrate Binding and Enzyme Action of the 6-O-Sulfatase
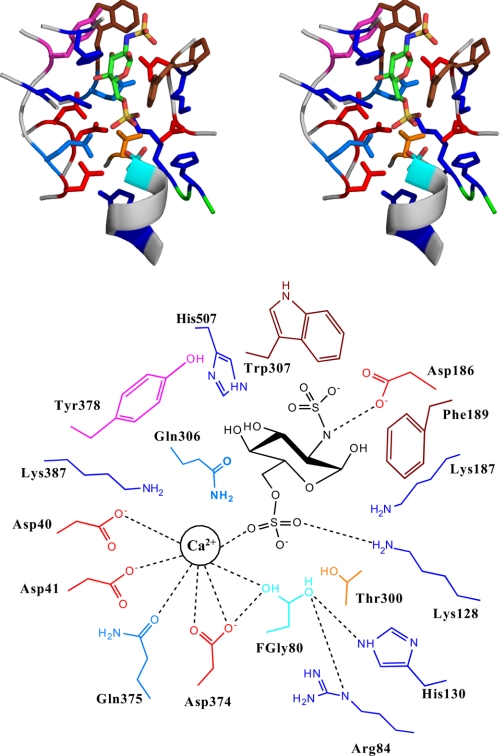
Having defined the basic biochemistry of the 6-O-sulfatase, we sought to provide a structural context to our findings. A homology based structural model of 6-O-sulfatase was constructed. A representative disulfated monosaccharide GlcNS,6S was docked into the putative active site of the enzyme to investigate the molecular interactions between the enzyme and the substrate (Fig. 3). The critical active site residues at the base of the sugar binding pocket involved in the positioning of the sulfate group and the sulfate-ester bond cleavage are structurally conserved among the different sulfatases (highlighted in bold in Table 1). The functional roles of these conserved residues in the sulfate-ester hydrolysis mechanism were assigned (Table 2) based on the analysis of the enzyme-substrate complex as well as on the assigned roles of their structurally conserved residues in the other sulfatases (33–35).
FIGURE 3.
Homology-based structural model of the 6-O-sulfatase-substrate complex. Top, stereo view of the active site of the enzyme (backbone shown as gray schematic) and the side chain of the critical active site amino acids. The amino acids are colored using the following scheme: Glu and Asp, red; His, Lys, and Arg, dark blue; Gln and Asn, light blue; Trp and Phe, brown; Ser and Thr, orange; active site l-C-α-formylglycine (FGly), cyan. The GlcNS,6S substrate is shown in stick representation and colored by atom (C, green; O, red; S, yellow; N, dark blue). Bottom, schematic of the active site (with the key amino acids labeled), the location of the divalent cation Ca+, and the substrate. The dotted line indicates the intermolecular enzyme-substrate interactions.
TABLE 1.
Structure-based comparison of sulfatase active site residues
Structural alignment of the modeled 6-O-sulfatase with the other sulfatases was obtained using the SuperPose tool (25). Residues that are highly conserved are displayed in boldface and non-conserved residues are listed in normal font. Regions of deletion are marked with the minus sign. Amino acids of 6-O-sulfatase that could potentially be involved in substrate binding are denoted with the asterisk.
| 6-O-Sulfatase (F. heparinum) | 2-O-Sulfatase (F. heparinum) | Arylsulfatase A (human) | Arylsulfatase B (human) | Arylsulfatase (P. aeruginosa) |
|---|---|---|---|---|
| Cys-80 | Cys-82 | Cys-69 | Cys-91 | Cys-51 |
| Arg-84 | Arg-86 | Arg-73 | Arg-95 | Arg-55 |
| Lys-128 | Lys-134 | Lys-123 | Lys-145 | Lys-113 |
| His-130 | His-136 | His-125 | His-147 | His-115 |
| Lys-387 | Lys-308 | Lys-302 | Lys-318 | Lys-375 |
| Gln-306 | Gln-237 | His-229 | His-242 | His-211 |
| Asp-40 | Asp-42 | Asp-29 | Asp-53 | Asp-13 |
| Asp-41 | Gln-43 | Asp-30 | Asp-54 | Asp-14 |
| Asp-374 | Asp-295 | Asp-281 | Asp-300 | Asp-317 |
| Gln-375 | His-296 | Asn-282 | Asn-301 | Asn-318 |
| Asp-186* | − | − | − | − |
| Lys-187* | − | − | − | − |
| Phe-189* | − | − | − | − |
| Thr-300* | − | − | − | − |
| Tyr-378* | − | Arg-288 | Ala-307 | Leu-325 |
| His-507* | − | His-405 | Ser-403 | − |
| Trp-307 | Lys-238 | Tyr-230 | Glu-243 | Trp-212 |
TABLE 2.
Functional assignment of 6-O-sulfatase active site residues
Amino acids listed in this table were identified by inspection of the 6-O-sulfatase model proposed in Fig. 3. The critical active site cysteine is shown in boldface.
| Amino acids | |
|---|---|
| Sulfate ester hydrolysis | |
| FGly-80 | Cys-80 in the active enzyme is modified to FGly and the FGly is further hydrated to form an aldehyde hydrate comprised of Oγ−1 and Oγ−2 atoms |
| Arg-84, His-130 | Stabilize the hydrated FGly by interaction with Oγ−2. His-130 is well positioned also for proton abstraction from Oγ−2 after the catalytic process for elimination of sulfate and regeneration of geminal diol |
| Lys-128, Lys187, Lys-387, Gln-306 | Positioned to interact with oxygen atoms of the 6-O-sulfate group to enhance electron density withdrawal from sulfate, hence contributing to the electrophilicity increase of the sulfur center. Also, Lys-387 is well positioned to protonate the oxygen atom of the leaving substrate |
| Asp-374, Asp-40, Asp-41, Gln-375, Thr-300 | Asp-374, Asp-40, Asp-41, and Gln-375 are well positioned to coordinate with a divalent metal ion such as Ca2+. Asp-374 also could donate proton and enhance nucleophilicity of Oγ−1. Thr-300 being weakly acidic could also participate instead of Gln-375 in metal ion coordination |
| Substrate positioning and specificity | |
| Phe-189, Tyr-378 | Positioned to stack with the pyranose ring of the Glc sugar |
| His-507, Asp-186, Tyr-378 | His-507 is positioned to interact with equatorial 4-hydroxyl of the Glc sugar. Asp-86 is positioned to interact with the N-atom of the Glc-NS or GlcNAc sugars |
| Trp-307, Pro-188 | Positioned to optimally interact with the acetyl group of the GlcNAc sugar |
Apart from the conserved residues, the other residues in the proximity of the active site are Asp-186, Lys-187, Pro-188, Phe-189, Thr-300, Trp-307, Tyr-378, and His-507. These residues are not structurally conserved in the other sulfatases and therefore they potentially play a unique role in the substrate specificity of the 6-O-sulfatase. Indeed, examination of the crystal structures for other, non-HSGAG sulfatases, indicates that there is no equivalent for these residues (Table 1). Furthermore, many of these residues are present as a part of the additional loop regions in the 6-O-sulfatase as compared with the other sulfatases. The presence of these additional loop regions makes the substrate-binding groove of 6-O-sulfatase more constricted as compared with the other sulfatases. This narrow substrate binding groove taken together with these additional residues are likely to restrict the processing of GlcNAc/NS,6S sugar present either as a monosaccharide or at the non-reducing end of a short oligosaccharide (exolytic processing of substrate).
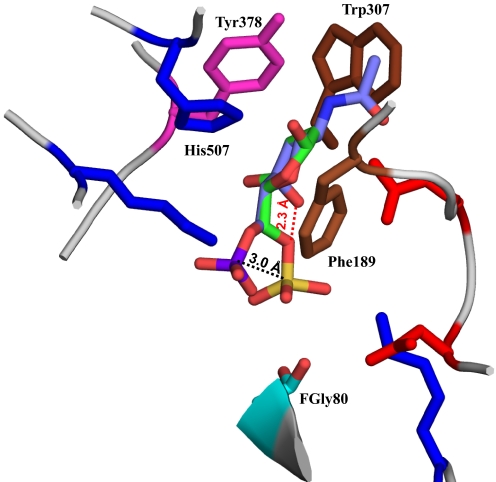
It was observed that Phe-189 and Tyr-378 are positioned to stack the pyranose sugar ring and thus lock the substrate in the active site. Asp-186 is positioned to make favorable contacts with the amine group in both GlcNS,6S and GlcNAc,6S substrates. Due to its role in substrate binding, mutation of Asp-186 to alanine resulted in a modest decrease in 6-O-sulfatase activity relative to the wild type (Table 3), consistent with the fact that multiple contacts are important for orientation of substrate and stabilization of its binding. His-507 on the other hand interacts with the equatorial 4-OH of the Glc pyranose sugar. Among the GlcNS,6S and GlcNAc,6S substrates, the N-acetyl group containing substrate is likely to make a higher number of favorable contacts because of the planarity of the acetyl group and the stacking of the CH3 group with the hydrophobic Trp-307 and Pro-188 residues. This observation provides a structural basis for the observed kinetic preference of the monosulfated GlcNAc,6S substrate. Furthermore, the position of the 3-OH is such that a glucosamine sugar containing a 3-O-sulfate group (such as GlcNS/NAc,3S,6S) would have unfavorable steric hindrance with the Trp-307 residue.
TABLE 3.
Assessment of relative activity of 6-O-sulfatase active site mutants
| Mutant | Classification | % Activitya | |
|---|---|---|---|
| After 0.5 h | After 5 h | ||
| Wild type | Control | 100 | 100 |
| Cys-80 → Ala | Active site (FGly) | 0 | 0 |
| Asp-374 → Ala | Ca2+ coordination | 0 | 0 |
| Asp-186 → Ala | Interaction with 2 n GlcNS or GlcNAc | 53 | 92 |
aActivity was measured using the coupled enzyme assay under maximum (saturating substrate) conditions as described under “Experimental Procedures.”
To understand the specificity of this enzyme toward α-d-glucosamine sugar in HSGAGs versus β-d-galactosamine sugar in chondroitin and dermatan sulfate, a GalNAc,6S sugar was docked into the active site (Fig. 4). The ring stacking constraints imposed by hydrophobic residues Phe-189 and Tyr-378 caused the pyranose ring of GalNAc,6S sugar to coincide with that of GlcNAc,6S sugar. However, axial orientation of the 4-OH in GalNAc,6S (as compared with its equatorial orientation in GlcNAc,6S) causes the 6-O-sulfate group to move away from the active site. If the 6-O-sulfate group of GalNAc,6S has to coincide with that of GlcNAc,6S sugar, then the C6-C5 bond will be cis relative to the C4-O4 bond, which is not energetically favorable given the unfavorably close proximity of O6 and O4 atoms in this conformation. These observations provide structural insights into the specificity of the 6-O-sulfatase enzyme toward HSGAG oligosaccharide fragments comprising the GlcNAc/NS,6S sugar in the reducing end.
FIGURE 4.
Structural basis for substrate specificity of 6-O-sulfatase. Shown in the figure is a close-up view of the active site with GlcNAc,6S (colored by atom as described in the legend to Fig. 3) superimposed on GalNAc,6S (colored by atom C, violet; O, red; N, blue; S, magenta). Only some of the key active site amino acids are shown for clarity. Due to the axial orientation of the C4-O4 bond, the position of the 6-O-sulfate in GalNAc,6S is such that the sulfate group is 3 Å away from the optimal position of the corresponding 6-O-sulfate in the GlcNS,6S substrate. The red dotted line shows the unfavorable proximity of the O4 atom of GalNAc,6S and the O6 atom if the 6-O-sulfate of the GalNAc6S coincided with that of GlcNS,6S.
Role of Divalent Metal Ions in Activity of 6-O-Sulfatase
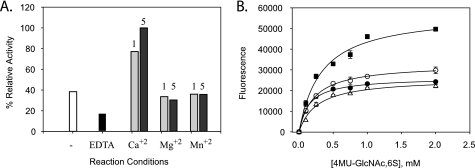
Our next step was to investigate the interactions of the active site residues with a divalent metal ion such as Ca2+ that has been shown to play a critical role in sulfatase activity of many arylsulfatases (33, 35). In each of the arylsulfatases, the metal ion coordinates with the oxygen atoms of the sulfate group of the respective substrate. Additionally, a cluster of four highly conserved acidic amino acids have been observed to coordinate with this divalent metal ion. The corresponding metal ion coordinating amino acids in the 6-O-sulfatase are identified based on the structural model as Asp-40, Asp-41, Asp-374, and Gln-375 (Table 1). These four amino acids are well positioned to coordinate with the Ca2+ ion (Fig. 3). The other amino acid present near the vicinity of this tetrad is Thr-301, which is also weakly acidic and could participate in metal ion coordination. Based on these observations, we predict that Ca2+ ions would substantially influence the activity of 6-O-sulfatase and that there would be a reduction in the activity in the absence of Ca2+ ions.
We set out to test this hypothesis by examining the effect of Ca2+ and other divalent metal ions on enzyme activity. 6-O-Sulfatase activity was indeed found to be enhanced 2–3-fold (Table 4) by the presence of calcium in a concentration-dependent manner. Moreover, as hypothesized, the enzyme was somewhat active even in the presence of 1 mm EDTA (Fig. 5A). These results validate the hypothesis made from the structural model. Interestingly, however, the divalent metal activation for the enzyme was specific to calcium and inclusion of Mg+2 or Mn+2 was determined to have only negligible effects. To examine this metal selectivity, we measured the potential for enzyme inhibition in the presence of a calcium-specific chelator (EGTA) but found that it had no appreciable effect on the specific activity of 6-O-sulfatase. In an attempt to determine the mechanism by which calcium exerts its effect on the enzyme we followed up these metal ion experiments by measuring the effect of calcium on enzyme steady-state kinetics (Fig. 5B). Consistent with our previous results, the initial rate of enzyme activity was affected by calcium in a concentration-dependent fashion and was largely manifested as a kcat effect (Table 4). The likely role of Asp-374 in coordinating calcium as predicted by the model was confirmed by site-directed mutagenesis (Table 3). Thus, through a combination of structural analysis and biochemical experiments; we were able to determine a role for Ca2+ ion coordination in the enzyme activity of 6-O-sulfatase.
TABLE 4.
Steady state kinetic parameters for 6-O-sulfatase using 4-MU monosaccharide substrates
| Addition | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | ×102 | |
| None | 2.5 | 146 | 1.7 |
| 0.5 mm Ca+2 | 3.3 | 217 | 1.5 |
| 5 mm Ca+2 | 6.8 | 327 | 2.1 |
| 1 mm EDTA | 2.5 | 264 | 0.95 |
FIGURE 5.
Effect of divalent metals on 6-O-sulfatase activity. A, calcium specific activation of 6-O-sulfatase activity and inhibition by EDTA (A). The divalent metal effect was not observed when calcium was replaced by either Mg2+ or Mn2+ (at either 1 or 5 mm concentrations); open bars, no divalent metals added; black bars, 1 mm EDTA added; light gray bars, 1 mm divalent metal; stippled gray bars, 5 mm divalent metal. B, effect of calcium on steady-state kinetics for the 6-O-sulfatase as described under “Experimental Procedures” at various concentrations of Ca2+ or in the presence of 1 mm EDTA. Substrate saturation plots were fitted to pseudo-first order Michaelis-Menten kinetics by non-linear regression analyses. Symbols indicate 0.5 mm Ca+2 (●), 1 mm Ca+2 (○), 5 mm Ca+2 (■), and 1 mm EDTA (Δ).
Experimental Validation of Exolytic Mode of Action by 6-O-Sulfatase
The structural analysis performed on our model indicates that the 6-O-sulfatase should be predominantly exolytic, given the highly specific orientation of the active-site residues. To verify this hypothesis, we performed experiments to address the possibility of this enzyme acting exolytically/endolytically on heparin or heparan sulfate-derived oligosaccharides. Of note is the fact that in the natural degradation pathway, 6-O-sulfatase would likely act downstream of the heparinases and possibly the Δ4,5-glycuronidase (17). Thus, these HSGAG oligosaccharide substrates could possess either an uronic acid (even number of saccharide units) or a hexosamine (odd number oligosaccharide) at the non-reducing end. In the former case, the uronic acid would likely be unsaturated due to the preceding action of heparin lyases, which cleave the GAG chain through a β-eliminative catalytic mechanism. In the latter case, loss of the unsaturated hexuronic acid would result from the hydrolytic action of the glycuronidase. To address this important issue, the 6-O-sulfatase enzyme was initially tested against a panel of unsaturated heparin disaccharides such as ΔU±2SHNAc,6S and ΔU±2SHNS,6S. For these experiments, standard reaction conditions were chosen as defined in the monosaccharide studies. None of the unsaturated disaccharides were desulfated by 6-O-sulfatase (data not shown). The inability of this enzyme to do so was confirmed in a related experiment in which all possible heparin disaccharides were first generated by pre-treating heparin with heparinase I and III prior to adding the sulfatases to the same reaction tube. The converse experiment was also conducted in which unfractionated heparin was preincubated with the 6-O-sulfatase for an extended period of time (8 h) followed by the addition of heparinase I and III. In this particular sequence, sulfatase pretreatment had no effect on the compositional profile of the heparinase-derived cleavage products (data not shown). These experiments categorize the 6-O-sulfatase as an obligatorily exolytic enzyme, confirming the hypothesis derived from observation of the active-site topology.
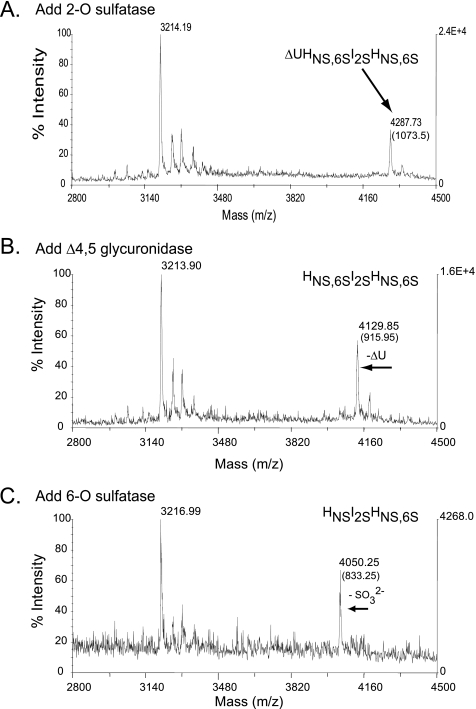
To further verify the mode of 6-O-sulfatase processing, we proceeded to examine the possibility of 6-O-sulfatase acting on the non-reducing end of saturated, odd-numbered oligosaccharides. We addressed this using a combination of two structurally related sulfated trisaccharides, HNS,6SIHNS,6S and HNS,6SI2SHNS,6S. Each of these trisaccharides was generated from the corresponding heparin-derived tetrasaccharide ΔU2SHNS,6SI±2SHNS,6S (Fig. 6, data not shown for ΔU2SHNS,6SIHNS,6S) by the tandem use of the 2-O-sulfatase (Fig. 6A) and the Δ4,5-glycuronidase (Fig. 6B) prior to the addition of 6-O-sulfatase. Desulfation was followed by matrix-assisted laser desorption ionization-MS and the 6-O-sulfatase was found to singularly desulfate both trisaccharides (Fig. 6C). The results presented for exolytic desulfation of oligosaccharides at the non-reducing end are consistent with the substrate specificity data pertaining to desulfation of monosaccharide substrates. Taken together, our structural and biochemical results indicate that 6-O-sulfatase is a highly specific, exolytic enzyme that acts on the non-reducing end of GlcNS,6S or GlcNAc,6S of saturated, odd-numbered oligosaccharide substrates and that its activity is enhanced by the presence of divalent Ca2+ ions.
FIGURE 6.
Sequential degradation of an HSGAG tetrasaccharide using recombinantly expressed flavobacterial enzymes. The ability of the 6-O-sulfatase to hydrolyze the non-reducing end of an oligosaccharide is demonstrated in the context of exo-sequencing the heparin-derived tetrasaccharides ΔU2SHNS,6SI2SHNS,6S (shown here) and structurally related ΔUHNS,6SI2SHNS,6S lacking a 2-O-sulfate at the internal iduronic acid position (data not shown). Sequential treatment of the HSGAG tetrasaccharide was physically assessed after each enzyme step (A–C) by matrix-assisted laser desorption ionization-MS. Masses listed in each panel represent either peptide alone (∼3216 Da) or oligosaccharide-peptide complex. The net mass of the oligosaccharide is listed in parentheses. A, addition of the 2-O-sulfatase; B, subsequent addition of the Δ4,5-glycuronidase; C, subsequent addition of the 6-O-sulfatase (note loss of a sulfate represented by a shift in net molecular mass from ∼915 to ∼835 Da).
DISCUSSION
In this study, we describe the cloning, biochemical characterization, molecular modeling, and structure-function analysis of a sulfatase gene from the F. heparinum genome. A BLASTP sequence homology search of the flavobacterial gene against the protein data base unambiguously identified the gene product as a member of a large sulfatase family. The 6-O-sulfatase protein sequence possesses the signature PFAM sulfatase motif C/SXPXRXXXXS/TG as well as the highly conserved sequence LTG (at the +9 through +11 positions relative to this motif). The two flavobacterial sulfatases that we have now cloned (i.e. the 2-O-sulfatase and orfb gene product) share only a limited overall homology to one another, The flavobacterial sulfatase does show a strong sequence homology (greater than 50%) to the mucin-desulfating sulfatase encoded by the enteric bacterium Prevotella strain RS2 (MdsA gene). In addition to mucin, this particular enzyme is specific for free N-acetylglucosamine-6-O-sulfate (36). Our previous studies with 2-O-sulfatase and other enzymes from the same system have confirmed the presence of a cysteine-specific active site (Cys-80) and it is at this conserved cysteine (and not serine) that the critical co- or post-translational oxidation to an l-C-α-formylglycine occurs (37, 38). This obligatory requirement for the covalently modified cysteine has also been demonstrated by site-directed mutagenesis; mutation of Cys-80 to Ala completely abolished 6-O-sulfatase activity (Table 4).
Beyond the predicted function as inferred from structural homology to other known sulfatases, we set out to empirically confirm its putative function, first, by examining the ability of the enzyme to act as a so-called “arylsulfatase,” second, to act within the context of HSGAG degradation and finally, by performing an exhaustive atomic analysis of the enzyme structural model active-site topology. We also experimentally validated the role of key active site residues by mutating Cys-80 → Ala or Asp-374 → Ala (as described previously in Ref. 24). These mutants showed no activity (Table 4) in comparison to the wild type enzyme and hence validate the critical role of these amino acids in the catalytic activity of the enzyme.
Although the 6-O-sulfatase was found to be a poor “generic” arylsulfatase on the basis of a rather nonspecific but commonly used biochemical screen, our results obtained by more structurally directed monosaccharide substrates have unequivocally confirmed that we had indeed cloned the heparan N-acetylglucosamine-6-O-sulfatase. These results demonstrate the exclusivity of the recombinant enzyme in terms of the singular position of the sulfate that is hydrolyzed. Moreover, this work goes beyond this basic description and identifies putatively the important structural determinants of enzyme specificity. In particular, the results presented identify the critical spatial orientation of the C4 hydroxyl as an additional structural determinant of substrate specificity, thus making the two flavobacterial sulfatases uniquely heparin/heparan sulfate-degrading enzymes. The HSGAG specificity of this enzyme points to the likely existence of a unique flavobacterial gene (or set of genes) encoding for the 6-O-desulfation of galactose/galactosamine. By analogy, distinct enzymes for desulfating these galacto-sugars do exist in the chondroitin sulfate/dermatan sulfate and keratan sulfate-specific lysosomal degradation pathways of higher eukaryotes. At the same time, there have been no reports that we are aware of describing the purification of either galactose or GalNac-6-desulfating activity directly from F. heparinum.
In addition to describing the critical substrate specificity for this sulfohydrolase, we have also defined important parameters related to their optimal use in vitro. These include pH and the role of divalent cations, namely calcium. In regard to pH, the slightly acidic pH optima demonstrated for 6-O-sulfatase is consistent with our previous observations for the pH optima of the 2-O-sulfatase (18) and Δ4,5-glycuronidase (17). Collectively, this profile also generally distinguishes the flavobacterial HSGAG degrading enzymes from their lysosomal counterparts, which, by virtue of their subcellular localization are most active at pH 4.5. Unlike the activity of 2-O-sulfatase that was not enhanced by the presence of a calcium ion coordination center, however, the intrinsic activity of 6-O-sulfatase was increased ∼2–3-fold in the presence of calcium ions and in a concentration-dependent fashion. This calcium dependence is clearly inferred from our structural model.
The Ca2+ ion was not an absolute requirement for enzymatic activity. Our structural model of the 6-O-sulfatase further helped us to identify specific residues that are well positioned to interact directly with the 6-O-sulfate group and potentially act as a catalytic base to confer hydrolytic activity, even in the absence of a divalent metal ion (e.g. Asp-374). This putative structure-activity assignment to Asp-374 is supported by our mutagenesis studies.
Pivotal in the potential use of the 6-O-sulfatase enzyme for controlled desulfation of heparin/heparan sulfate oligosaccharides is the question of its endolytic versus exolytic potential. By definition, the former mode of action would predict their ability to hydrolyze internally located sulfates within either a disaccharide or oligosaccharide chain. Our combined structural modeling and biochemical characterization results argue against any endolytic potential for this enzyme. On the other hand, an exolytic mode of action would necessarily require this enzyme to sequentially follow Δ4,5-glycuronidase hydrolysis of terminal uronic acids if, in fact, it is to act on the non-reducing end of these saccharides. The data presented here confirm this prediction, i.e. by demonstrating the ability of the enzyme to hydrolyze the non-reducing end of heparin-derived oligosaccharides, albeit with certain structural constraints, on the non-reducing end of the saccharide, namely a requirement of direct access to a sulfated hexosamine that is not sterically hindered by the presence of an intervening uronate.
The molecular modeling of 6-O-sulfatase, together with the biochemical characterization described herein, provide an insightful understanding of important structure-function relationships. Taken together with our understanding of the substrate specificity of the other flavobacterial sulfatases (including the N-sulfamidase described in the accompanying paper (39)) this work also provides a practical framework toward the use of these enzymes and discrete analytical tools for elucidating the HSGAG fine structure and generating HSGAG-derived oligosaccharides of defined length and structure.
This work was supported, in whole or in part, by National Institutes of Health Grant GM57073.
- HSGAG
- heparin/heparan sulfate glycosaminoglycan
- ΔU
- Δ4,5-unsaturated uronic acid
- GlcNS,6S
- N-, 6-O-disulfated α-D-glucosamine
- GlcNS,3S,6S
- N-, 6-O-, 3-O-trisulfated α-d-glucosamine
- GlcNAc,6S
- N-acetylated, 6-O-sulfated α-d-glucosamine
- GalNAc,6S
- N-acetylated, 6-O-sulfated β-d-galactosamine
- 4-MU
- 4-methylumbelliferyl
- CE
- capillary electrophoresis
- LC/MSD
- liquid chromatography/mass selective detector
- MES
- 4-morpholineethanesulfonic acid
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- ESI
- electrospray ionization
- MS
- mass spectrometry
- ORF
- open reading frame
- PDB
- Protein data bank.
REFERENCES
- 1.Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 2.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 3.Shriver Z., Liu D., Sasisekharan R. (2002) Trends Cardiovasc. Med. 12, 71–77 [DOI] [PubMed] [Google Scholar]
- 4.Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 6.Prince J. M., Klinowska T. C., Marshman E., Lowe E. T., Mayer U., Miner J., Aberdam D., Vestweber D., Gusterson B., Streuli C. H. (2002) Dev. Dyn. 223, 497–516 [DOI] [PubMed] [Google Scholar]
- 7.Lai J. P., Chien J. R., Moser D. R., Staub J. K., Aderca I., Montoya D. P., Matthews T. A., Nagorney D. M., Cunningham J. M., Smith D. I., Greene E. L., Shridhar V., Roberts L. R. (2004) Gastroenterology 126, 231–248 [DOI] [PubMed] [Google Scholar]
- 8.Häcker U., Nybakken K., Perrimon N. (2005) Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Shriver Z., Pope R. M., Thorp S. C., Duncan M. B., Copeland R. J., Raska C. S., Yoshida K., Eisenberg R. J., Cohen G., Linhardt R. J., Sasisekharan R. (2002) J. Biol. Chem. 277, 33456–33467 [DOI] [PubMed] [Google Scholar]
- 10.Vivès R. R., Lortat-Jacob H., Fender P. (2006) Curr. Gene Ther. 6, 35–44 [DOI] [PubMed] [Google Scholar]
- 11.Dietrich C. P., Silva M. E., Michelacci Y. M. (1973) J. Biol. Chem. 248, 6408–6415 [PubMed] [Google Scholar]
- 12.Nakamura T., Shibata Y., Fujimura S. (1988) J. Clin. Microbiol. 26, 1070–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohse D. L., Linhardt R. J. (1992) J. Biol. Chem. 267, 24347–24355 [PubMed] [Google Scholar]
- 14.Kertesz M. A. (2000) FEMS Microbiol. Rev. 24, 135–175 [DOI] [PubMed] [Google Scholar]
- 15.Sasisekharan R., Bulmer M., Moremen K. W., Cooney C. L., Langer R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3660–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godavarti R., Davis M., Venkataraman G., Cooney C., Langer R., Sasisekharan R. (1996) Biochem. Biophys. Res. Commun. 225, 751–758 [DOI] [PubMed] [Google Scholar]
- 17.Myette J. R., Shriver Z., Kiziltepe T., McLean M. W., Venkataraman G., Sasisekharan R. (2002) Biochemistry 41, 7424–7434 [DOI] [PubMed] [Google Scholar]
- 18.Myette J. R., Shriver Z., Claycamp C., McLean M. W., Venkataraman G., Sasisekharan R. (2003) J. Biol. Chem. 278, 12157–12166 [DOI] [PubMed] [Google Scholar]
- 19.Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 20.Uchimura K., Morimoto-Tomita M., Bistrup A., Li J., Lyon M., Gallagher J., Werb Z., Rosen S. D. (2006) BMC Biochem. 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusnati M., Oreste P., Zoppetti G., Presta M. (2005) Curr. Pharm. Des. 11, 2489–2499 [DOI] [PubMed] [Google Scholar]
- 22.Ernst S., Venkataraman G., Winkler S., Godavarti R., Langer R., Cooney C. L., Sasisekharan R. (1996) Biochem. J. 315, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaya D., Tocilj A., Li Y., Myette J., Venkataraman G., Sasisekharan R., Cygler M. (2006) J. Biol. Chem. 281, 15525–15535 [DOI] [PubMed] [Google Scholar]
- 24.Raman R., Myette J. R., Shriver Z., Pojasek K., Venkataraman G., Sasisekharan R. (2003) J. Biol. Chem. 278, 12167–12174 [DOI] [PubMed] [Google Scholar]
- 25.Bateman A., Birney E., Durbin R., Eddy S. R., Howe K. L., Sonnhammer E. L. (2000) Nucleic Acids Res. 28, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 27.Beil S., Kehrli H., James P., Staudenmann W., Cook A. M., Leisinger T., Kertesz M. A. (1995) Eur. J. Biochem. 229, 385–394 [DOI] [PubMed] [Google Scholar]
- 28.Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S. D. (2002) J. Biol. Chem. 277, 49175–49185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkataraman G., Shriver Z., Raman R., Sasisekharan R. (1999) Science 286, 537–542 [DOI] [PubMed] [Google Scholar]
- 30.Guda C., Lu S., Scheeff E. D., Bourne P. E., Shindyalov I. N. (2004) Nucleic Acids Res. 32, W100–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiti R., Van Domselaar G. H., Zhang H., Wishart D. S. (2004) Nucleic Acids Res. 32, W590–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huige C. J. M., Altona C. (1995) J. Comput. Chem. 16, 56–79 [Google Scholar]
- 33.von Bülow R., Schmidt B., Dierks T., von Figura K., Usón I. (2001) J. Mol. Biol. 305, 269–277 [DOI] [PubMed] [Google Scholar]
- 34.Boltes I., Czapinska H., Kahnert A., von Bülow R., Dierks T., Schmidt B., von Figura K., Kertesz M. A., Usón I. (2001) Structure 9, 483–491 [DOI] [PubMed] [Google Scholar]
- 35.Lukatela G., Krauss N., Theis K., Selmer T., Gieselmann V., von Figura K., Saenger W. (1998) Biochemistry 37, 3654–3664 [DOI] [PubMed] [Google Scholar]
- 36.Roberton A. M., Rosendale D. I., Wright D. P. (2000) Methods Mol. Biol. 125, 417–426 [DOI] [PubMed] [Google Scholar]
- 37.Dierks T., Miech C., Hummerjohann J., Schmidt B., Kertesz M. A., von Figura K. (1998) J. Biol. Chem. 273, 25560–25564 [DOI] [PubMed] [Google Scholar]
- 38.Dierks T., Dickmanns A., Preusser-Kunze A., Schmidt B., Mariappan M., von Figura K., Ficner R., Rudolph M. G. (2005) Cell 121, 541–552 [DOI] [PubMed] [Google Scholar]
- 39.Myette J. R., Soundararajan V., Behr J., Shriver Z., Raman R., Sasisekharan R. (2009) J. Biol. Chem. 284, 35189–35200 [DOI] [PMC free article] [PubMed] [Google Scholar]