Abstract
Sulfated polysaccharides such as heparin and heparan sulfate glycosaminoglycans (HSGAGs) are chemically and structurally heterogeneous biopolymers that that function as key regulators of numerous biological functions. The elucidation of HSGAG fine structure is fundamental to understanding their functional diversity, and this is facilitated by the use of select degrading enzymes of defined substrate specificity. Our previous studies have reported the cloning, characterization, recombinant expression, and structure-function analysis in Escherichia coli of the Flavobacterium heparinum 2-O-sulfatase and 6-O-sulfatase enzymes that cleave O-sulfate groups from specific locations of the HSGAG polymer. Building on these preceding studies, we report here the molecular cloning and recombinant expression in Escherichia coli of an N-sulfamidase, specific for HSGAGs. In addition, we examine the basic enzymology of this enzyme through molecular modeling studies and structure-function analysis of substrate specificity and basic biochemistry. We use the results from these studies to propose a novel mechanism for nitrogen-sulfur bond cleavage by the N-sulfamidase. Taken together, our structural and biochemical studies indicate that N-sulfamidase is a predominantly exolytic enzyme that specifically acts on N-sulfated and 6-O-desulfated glucosamines present as monosaccharides or at the nonreducing end of odd-numbered oligosaccharide substrates. In conjunction with the previously reported specificities for the F. heparinum 2-O-sulfatase, 6-O-sulfatase, and unsaturated glucuronyl hydrolase, we are able to now reconstruct in vitro the defined exolytic sequence for the heparin and heparan sulfate degradation pathway of F. heparinum and apply these enzymes in tandem toward the exo-sequencing of heparin-derived oligosaccharides.
Introduction
The N-sulfate group is characteristic and unique to heparin sulfate glycosaminoglycans (HSGAGs),4 which are commonly occurring polysaccharides predominant in proteoglycans (1). HSGAGs are linear polymers with variable repeating disaccharide units and diverse chemical heterogeneity due to the variable positions of O- and N-linked sulfates (2, 3). Other factors contributing to the structural diversity of HSGAGs include the presence of N-linked acetates and possible epimerization at the C-5 carboxylates. The structure-function relationship of this diversity plays out in the dynamic regulation by HSGAGs of various signaling pathways (4), including cell death (5, 6), intercellular communication, cell growth and differentiation (7), and adhesion and tissue morphogenesis (8). Microbial pathogenesis has also been shown to depend upon the HSGAGs that are present as structurally defined binding epitopes on cell surfaces and as part of the extracellular matrix (9, 10).
GAG degradation follows an obligatory sequence of depolymerization steps involving multiple enzymes acting in tandem to cleave the HSGAG chain. Most of these enzymes act exolytically (11, 12) but may differ in the extent of their processivity, e.g. when comparing functionally similar enzymes between eukaryotic lysosomal and bacterial systems (13). Previously cited work in our laboratory has led to the molecular cloning and biochemical characterization of several HSGAG-degrading enzymes derived from the Gram-negative soil bacterium Flavobacterium heparinum (Pedobacter heparinus). Although earlier work described the enzymology of the heparin lyases (14–17), more recent work has focused on downstream enzymes such as an unsaturated glucuronyl hydrolase (Δ4,5-glycuronidase) and the 2-O- and 6-O-sulfatases (18–20). By their very nature, these latter enzymes provide an attractive subject of study and application given the following: 1) their respective substrate specificities, 2) their relevance as tools to probe multiple biological processes, and 3) a solid biochemical foundation of their structure-function relationships that include several protein structures (21, 22) and a detailed understanding of their common catalytic mechanism (23). Taken together, these factors make the enzymes a useful “tool kit” for the structural characterization of heparin and heparan sulfate.
Toward this end, we describe in the preceding paper (19) the cloning, recombinant expression, structural and biochemical characterization of enzymatic action, and substrate specificity of the F. heparinum 6-O-sulfatase enzyme. In this paper, we extend this work through the cloning of the N-sulfamidase gene from F. heparinum and the bacterial expression of the gene product. A biochemical and structural characterization of the enzyme is also described, including a determination of the substrate specificity and metal ion requirements for optimal enzyme activity. This sulfamidase enzyme is unique because it hydrolyzes the sulfamate linkage (N–S) and not a sulfoester as is described for the other HSGAG sulfohydrolases. Using a homology-based structural model of the N-sulfamidase enzyme, we have outlined a possible mechanism for the N–S bond cleavage and the critical amino acids involved in both the catalytic activity and substrate specificity of the enzyme. Finally, taken together with the reported substrate specificities for the previously characterized F. heparinum 2-O-sulfatase, the 6-O-sulfatase (described in the preceding paper, see Ref. 19) and the unsaturated glucuronyl hydrolase, we are now able to reconstruct in vitro the complete and defined exolytic sequence for the heparin and heparan sulfate degradation pathway of F. heparinum and apply these enzymes in tandem toward the exo-sequencing of HSGAG-derived oligosaccharides.
EXPERIMENTAL PROCEDURES
Reagents
Glucosamine and galactosamine monosaccharides and aryl sulfate substrates 4-catechol sulfate and 4-methyllumbelliferyl sulfate were purchased from Sigma. Exo-glucosidases were purchased from MP Biomedicals (Irvine, CA). N-Sulfated fluorogenic glycopyranoside derivatives were obtained through Toronto Research Chemicals (Toronto, Ontario, Canada). Fluorescent glucopyranoside substrates 4-methyllumbelliferyl-α/β-d-glucopyranoside (4-MU-α-d-Glc and 4-MU-β-d-Glc) were purchased from EMD Biosciences, Inc. (San Diego). PCR enzymes, TOP10 chemically competent cells, and oligonucleotide primers were obtained from Invitrogen. Materials for genomic library construction and screening and site-directed mutagenesis were obtained from Stratagene (La Jolla, CA). Additional reagents for molecular cloning were purchased from New England Biolabs (Beverly, MA) or the listed manufacturers.
Molecular Cloning of Flavobacterial N-Sulfamidase
PCR from a λZAPII flavobacterial genomic library originally screened using DNA hybridization probes specific to the 2-O-sulfatase (18) and subsequently used for the 6-O-sulfatase (19) were used to clone the sulfamidase gene. Library construction, hybridization screening, and phage excision were performed as outlined. Two overlapping clones were expanded by chromosomal walking and Southern restriction mapping using the Lambda DASH II genomic cloning kit (Stratagene). This was used for the ligation of size-fractionated genomic DNA generated by partial digestion of Sau3AI. The DNA was purified from a high titer lysate using standard techniques, and three successive rounds of plaque purification were performed on 2-O-sulfatase-positive clones from an amplified library. Recombinant phage DNA was subcloned into pBluescript SK± for sequencing the DNA. The putative N-sulfamidase gene is described throughout this paper as orfc. The coding sequence of orfc was identified by the canonical Protein Families Database (24) sulfatase family identifier (CXPXRXXXX(S/T)G) and subsequently PCR-amplified using the primer set given by 5′-TCT AGA CAT ATG AAA TTT AAC AAA TTG AAA TAT TTC-3′ (forward) and 5′-GGA TCC TCG AGT TAC TTC AAA TAA TTG TAA CTG GAA T-3′ (reverse). Subcloning of the amplified gene into T7-based bacterial expression vector pET28a (Novagen) was performed as an NdeI-XhoI cassettes described for the 6-O-sulfatase (19).
Bacterial Expression and Protein Purification
Similar to the setup in the 2-O-sulfatase (18) and 6-O-sulfatase (19) expression systems that we had designed earlier, the Escherichia coli strain BL21(DE3) and one-step affinity purification by nickel chelation chromatography likewise were used. von Hejne computational method was used to make the prediction of the NH2-terminal signal sequence and putative cleavage site for the protein (25). The engineering and expression of the truncated protein lacking the signal sequence were as described for the complete gene with the exceptions of substituting the internal 5′ primer with 5′-TCT AGA CAT ATG TCC TGC ACT TCG CCG GAA-3′ (NdeI site underlined) and beginning the orfc gene sequence at Ser-21. Site-specific protease cleavage using the thrombin cleavage capture kit (Novagen) was performed to remove the His6 tag with proteolysis conditions as described for other recombinantly expressed flavobacterial heparin-degrading enzymes. Ultrafiltration was used to concentrate the enzymes, and cleaved proteins were dialyzed against 4 liters of 50 mm Tris, pH 7.5, and 0.1 m NaCl, 4 °C overnight using a 3-ml Slide-a-Lyzer cassette with a 10,000 molecular weight cutoff (Pierce). The Bradford assay (Bio-Rad) was used to determine final protein concentrations and was confirmed by UV absorption spectroscopy with theoretical molar extinction coefficients (ϵ280) of 86,340 (53,193 Da) for the NH2-terminally truncated orfc (N-sulfamidase). This value was calculated for the thrombin-cleaved protein lacking the His6 purification tag. To retain full enzyme activity for months, the enzyme was stored at 4 °C and 10 mg/ml concentration.
Site-directed Mutagenesis of Putative N-Sulfamidase Active Site Residues
Based on the homology modeling of the N-sulfamidase (described below), three residues were initially chosen for mutagenesis: Cys-80, Asp-40, and Lys-158. Site-directed mutagenesis was completed by thermal cycling using the QuickChangeTM method (Stratagene, La Jolla California) as described in the accompanying paper (19) and using the following oligonucleotide primer pairs: 5′-AT ATC CTG ATG ATC ATG TCC GcT AAC CAA TCC TGG AAC CAC G-3′, and 5′-CG TGG TTC CAG GAT TGG TTA gCG GAC ATG ATC ATC AGG ATA T-3′ (D40A); 5′-CT TTT TGC AGT TCA CCT TCC gcT ACG CCC GCA AGG GCT G-3′ and 5′ CA GCC CTT GCG GGC GTA gcG GAA GGT GAA CTG CAA AAA G-3′ (C80A); 5′-G TTT AAA AGT TTT GGC GCA TTT TTA gcA GAT AAA AAA GAA GGT CCC TG-3′ and 5′-CAG GGA CCT TCT TTT TTA TCT gcT AAA AAT GCG CCA AAA CTT TTA AAC-3′ (K158A). Base pair changes are noted in lowercase boldface type. The same pET28 (His6) vector used for bacterial expression of the wild type N-sulfamidase-sulfatase was likewise used as the template for linear amplification. Parental DNA was restricted using DpnI. Targeted mutations were confirmed by direct DNA sequencing of both strands.
Arylsulfatase, N-Sulfamidase, and Coupled Enzyme Assays; Biochemical Protocols and Kinetic Studies
The 4-catechol sulfate and 4-methylumbelliferyl sulfate were two chromogenic substrates used to independently measure arylsulfatase activity. The catechol substrate assay was conducted as described (26). Fluorimetric arylsulfatase assay using 4-methylumbelliferyl sulfate was also generally as described previously (27), and detection of fluorescent methylumbelliferone was measured using a SpectraMax microtiter plate reader (GE Healthcare) set at excitation and emission wavelengths of 360 and 440 nm, respectively. Fluorescence intensity was corrected against background (minus enzyme control). In both assays, 0.5 unit of arylsulfatase from Aerobacter aerogenes was the positive control.
For the pilot N-sulfamidase assay, initial assessment of substrate specificity and pH optima was made using a capillary electrophoresis-based assay for detection of desulfated products. The fluorescently derivatized monosulfated gluco- and galactopyranosides used as substrates to test enzyme activity included 4-MU-GlcNAc,6S, 4-MU-GlcNS, 4-MU-GalNAc,6S, and 4-MU-Gal6S. Standard reactions included 1 mm substrate, 1–10 μm enzyme, 50 mm sodium acetate, pH 5.5–6.5, and 5 mm CaCl2 in a 20-μl reaction volume. Exhaustive reactions involving overnight incubations at 30 °C were used for pilot experiments. N-Sulfamidase was inactivated by heat denaturation at 95 °C for 10 min, followed by a 10-fold dilution into water, and the products were resolved by capillary electrophoresis using a 25-cm long, 75-μm inner diameter fused silica capillary (Agilent Technologies). Substrate desulfation was measured as a percentage of substrate depletion relative to a minus enzyme control as monitored by the loss of UV absorbance at 315 nm detected at ∼4 min. A standard capillary electrophoresis buffer included 50 mm Tris and 10 μm dextran sulfate (average molecular mass of 10,000 Da) adjusted to pH 2.0 with phosphoric acid. The effect of pH was measured by capillary electrophoresis using the following three sets of buffers with overlapping pH ranging from 4.5 to 8.0: 50 mm sodium citrate at 4.5, 5.0, and 5.5; 50 mm MES at 5.5, 6.0, 6.5, and 7.0; and 50 mm MOPS at 6.5, 7.0, 7.5, and 8.0. For N-sulfamidase, the reactions included 4-MU-GlcNS, 50 mm buffer, and 5 mm CaCl2 in a 20-μl reaction volume, and this assay was initiated by addition of 2 μl of 10× enzyme stock to 18 μl of preheated reaction mixture. All the reactions were performed at 30 °C for 30 min each. The ability of N-sulfamidase to desulfate unsaturated heparin and chondroitin disaccharides was assessed essentially as described for CE-based compositional analyses of enzymatically generated glycosaminoglycan di- and tetrasaccharides (28).
A fluorimetrically based plate assay was used for indirect measurement of enzyme activity, in which the N-sulfamidase-driven desulfation of the glucopyranoside 1→4 methylbelliferone substrate was coupled to the glucosidase-controlled hydrolysis of the stereospecific 1→4 glycosidic linkage between the pyranose ring and the adjoining fluorophore. Release of the 4-MU was monitored spectroscopically as described above for the arylsulfatase assay. N-Sulfamidase hydrolysis of 4-MU-α-d-GlcNS at the 2-amino position was coupled to α-glucosidase (catalogue number 153487, MP Biomedicals), and the efficacy of this coupled assay was contingent on the intrinsically poor ability of the enzyme for hydrolyzing the glycosidic bond when the adjoining glucosamine is modified by a sulfate.
The coupled N-sulfamidase assay was generally described for the 6-O-sulfatase (19) but with the following modifications: 4-MU-GlcNS as substrate, 50 mm sodium acetate at pH 6.0 (instead of 5.5), and 1 μm enzyme. For the second enzyme step, 5 units of α-glucosidase were added. Enzyme incubation was carried out for 22 h at 37 °C. The obvious difference in enzyme efficacies between α-glucosidase versus β-glucosidase is reflected in the substantially longer incubation times required for the α-glucosidase to quantitatively hydrolyze the glycosidic α1→4 linkage between the fluorophore and the desulfated glucosamine. All other reaction conditions were as described for the coupled 6-O-sulfatase/β-glucosidase assay (19). Michaelis-Menten kinetics were extrapolated from Vo versus substrate concentration plots fit by nonlinear regression to pseudo first-order kinetics, and all obtained data represent mean of three experimental trials.
The coupled enzyme assay was also used for the assessment of enzyme activity for select site-directed mutants. Enzyme activity was measured kinetically using a single saturating concentration (2 mm) of fluorescently labeled substrate. Results are reported as % activity relative to the wild type enzyme.
Compositional Analyses of Sulfamidase-treated Heparin
10 μm N-sulfamidase was preincubated with 20 μg of heparin for 8 h at 30 °C in a 20-μl reaction, which included 25 mm sodium acetate, pH 7.0, and 2 mm calcium acetate, pH 7.0. The enzyme was then inactivated by heat denaturation at 95 °C for 10 min, and the heparin was completely digested at 37 °C by the addition of 2 μl of concentrated enzyme mixture containing heparinase I and III. The CE-based compositional analyses of heparinase-derived disaccharides were performed as described (29).
Sequential Degradation of Heparin Oligosaccharide by Flavobacterial Exo-enzymes
The purified penta- and hexasulfated tetrasaccharides ΔU2SHNS,6SI±2SHNS,6S were a gift from Dr. I. Capila (Momenta Pharmaceuticals, Inc.). The following enzyme sequence was performed: 2-O-sulfatase, Δ4,5-glycuronidase, 6-O-sulfatase, and N-sulfamidase. The enzyme was heat-inactivated after each step, and 20-μl aliquots were removed prior to the addition of the next enzyme. The initial reaction conditions included 20 mm Tris, pH 7.2, and 60 nmol of tetrasaccharide in a 120-μl reaction volume, and enzyme reactions were carried out at 30 °C. The following specific conditions were also used: 1) 2-O-sulfatase, 1 μm enzyme, 6 h; 2) Δ4,5-glycuronidase, 1 μm enzyme, 6 h; 3) 6-O-sulfatase, 5 μm enzyme, 5 mm CaCl2, 12–15 h; 4) N-sulfamidase, same conditions as for 6-O-sulfatase. MALDI-mass spectrometry is an established method that was used for determination of the molecular masses of enzyme products (30).
The APTS derivatization protocol was adapted from Chen and Evangelista (31). Briefly, 2 μl of 100 mm APTS in 25% acetic acid (v/v) was mixed with 10 μl of 1 m sodium cyanoborohydride in tetrahydrofuran and 1 μmol of saccharide. The reaction mixture was incubated at 75 °C for 2 h and was diluted 1:100 prior to CE analysis. Capillary electrophoresis/laser-induced fluorescence was performed on a Beckman Coulter ProteomeLab PA 800 with a 488-nm argon laser-induced fluorescence module. Samples were loaded onto a N-CHO capillary (50-μm inner diameter × 65-cm total length) using 0.5 p.s.i. of pressure at the anode for 20 s. Electrophoretic separations were performed using a 20-kV potential in a 100 mm sodium borate, pH 10.2 buffer for 15 min at 25 °C. Fluorescence emission spectra were collected using a 520-nm narrow band filter.
Electrospray Ionization-Mass Spectrometry of Sulfated Glucosamine Monosaccharides
Electrospray ionization-mass spectrometry was performed generally as described for the 6-O-sulfatase (19) but with a few modifications. Reactions were carried out with 100 μm substrate, 2 mm CaCl2, 5 μm enzyme, and 50 mm sodium acetate buffer, pH 6.0, at 37 °C overnight.
Homology Modeling of N-Sulfamidase and Docking of Substrates
Multiple sequence alignment of N-sulfamidase with human arylsulfatase A (Protein Data Bank code 1AUK), human arylsulfatase B (Protein Data Bank code 1FSU), and P. aeruginosa arylsulfatase B (Protein Data Bank code 1HDH) was performed using ClustalW (Fig. 1). The crystal structures of these enzymes were obtained from the Protein Data Bank and structure-based multiple superposition was obtained using CE-MC (32) and SuperPose (33). The Homology module of Insight II molecular simulations package (Accelrys, San Diego) was used to obtain a structural model of N-sulfamidase with PDB 1AUK as template. The loop regions of the structure were modeled using the homology module of InsightII. The deletions were closed using the Discover module of Insight II, wherein most of the structure was held rigid except for the regions in the proximity of the deletion site, followed by 200 iterations of steepest descent and 300 iterations of conjugate gradient minimization. The loop regions and side chains of all residues were then allowed to move freely by performing 300 iterations of steepest descent minimization. This was followed by further refinement of the structure by subjecting it to 500 iterations of steepest descent minimization without including charges and 500 iterations of conjugate gradient minimization, including charges to obtain the final predicted model of N-sulfamidase.
FIGURE 1.
Structure-based multiple sequence alignment of the sulfatases using ClustalW. The flavobacterial N-sulfamidase enzyme is a member of a large sulfatase family. The putative active site is boxed, with the critically modified cysteine 80 noted by an asterisk. Invariant residues are shaded in dark gray; those with partial identity in light gray, and conservative substitutions in charcoal. Multiple sequence alignment was generated by ClustalW using only select sequences identified from a BLASTP search of the protein data base. Enzymes with the following GenBankTM accession numbers are abbreviated as follows: F. heparinum N-sulfamidase (Nsulf); F. heparinum 6-O-sulfatase (FH6S); F. heparinum 2-O-sulfatase (FH2S); P. aeruginosa arylsulfatase (PARS, GenBankTM code CAA88421); human N-acetylgalactosamine-6-sulfate sulfatase or chondroitin-6-sulfatase (HGal6S, GenBankTM code AAC51350); human cerebroside-3-sulfate sulfatase or arylsulfatase A (HARSA, GenBankTM code AAC51350); human N-acetylgalactosamine-4 sulfate sulfatase or arylsulfatase B (HARSB, GenBankTM code AAC51350).
The coordinates of a heparin-derived hexasaccharide were obtained from co-crystal structure of FGF-1 heparin complex (Protein Data Bank code 2AXM). The hexasaccharide coordinates were imported into the Builder module of InsightII to generate the monosaccharide (GlcNS, GlcNS,3S, and GlcNS,6S) and disaccharide (ΔU±2SHNS±6S) substrates for N-sulfamidase. As with our previous studies on other sulfatases, the initial docking of the substrates in the enzyme groove was facilitated by the highly conserved position of the cleavable NHSO3− group relative to the O-γ1 of the geminal diol. The enzyme-substrate complexes were then subjected to optimization with 300 steps of steepest descent followed by 400 steps of Newton-Raphson minimization, including charges, using the Discover module of Insight II. In this step, a force constant of 7000 kcal/mol was utilized to constrain the ring torsion angles to maintain the ring conformation of the substrates. The Amber force field was used to assign potentials to enzyme and sugar substrates for the energy minimization as described in the accompanying paper (19).
RESULTS
Molecular Cloning and Recombinant Expression of F. heparinum N-Sulfamidase Gene
Screening of a genomic library with hybridization probes directed at the flavobacterial 2-O-sulfatase (26) revealed an overlapping phagemid clone that was then expanded by chromosomal walking and restriction mapping. An open reading frame of 1524 bp (orfc) was identified by sequence analysis of this genomic region, and this was found to encode a protein of 500 amino acids in length. An examination of the putative orfc gene product revealed that it possesses an NH2-terminal hydrophobic signal peptide and corresponding cleavage site sequence predicted by the von Heijne method for Gram-negative bacteria (25). The gene product also possesses a canonical sulfatase domain as described by the Protein Families Database identifier PF 000884 (see under “Discussion”). The orfc gene product was robustly expressed in E. coli as a soluble enzyme by removal of the amino-terminal signal sequence, as detailed earlier for the 6-O-sulfatase (19). The apparent molecular mass of 53,193 Da for the protein based on SDS-PAGE was consistent with the theoretical molecular weight calculated from putative amino acid composition based on translation of orfc.
Biochemical Characterization of N-Sulfamidase and Optimization of in Vitro Reaction Conditions
We first considered the possibility of the orfc gene product functioning as a generic arylsulfatase by testing it against 4-catechol sulfate and 4-MU sulfate, which are two different aromatic sulfate esters commonly used as substrates to assess generic sulfatase activity. It was found that the orfc gene product exhibited only negligible activity relative to a known arylsulfatase from A. aerogenes that served as a positive control. To further test whether more robust activity could be determined against heparin and heparan sulfate, we used capillary electrophoresis wherein the two commercially available “heparin” monosaccharides 4-MU-GlcNAc,6S and 4-MU-GlcNS and the 6-O-sulfated galactose sugars 4-MU-Gal6S and 4-MU-GalNAc,6S (corresponding to the monosaccharide constituents of keratan sulfate and chondroitin/dermatan sulfate, respectively) were tested. This analysis showed that orfc gene product could only hydrolyze the glucosamine sulfated at the 2-amino position. As such, we hereafter refer to the orfc gene product as the N-sulfamidase (data not shown).
We further investigated the substrate specificities of N-sulfamidase by examining the influence of various substitutions at the 2-amino, 3-OH, and 6-OH positions of the glucosamine, using electrospray ionization-mass spectrometry. In this analysis, we found the N-sulfamidase activity at the 2-amino position was absolutely abolished when a 6-O-sulfate was also present (Fig. 2). Reciprocally related to this observation, in the accompanying paper (19) we determined that the 6-O-sulfatase enzyme required a substituted amine (acetate or sulfate) at the 2-amino position. These findings imply that the 6-O-sulfatase and N-sulfamidase process substrates in a strictly sequential fashion with the former preceding the latter. Additionally, N-sulfamidase was completely inhibited by the presence of a 3-O-sulfate.
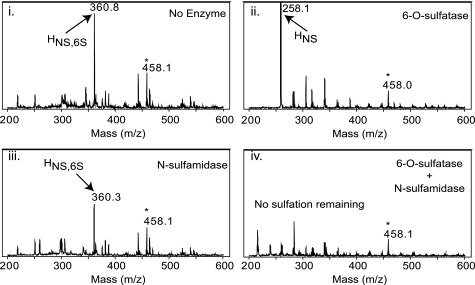
FIGURE 2.
Obligatory substrate-product relationship of N-sulfamidase. Desulfation of the disulfated monosaccharide HNS,6S (GlcNS,6S) by the enzymes was followed by electrospray mass spectrometry. Panel i, substrate only shown here as the sodium adduct of a single ion species (M − 1) with molecular mass of 360.8 Da; panel ii, treatment of GlcNS,6S substrate with 6-O-sulfatase desulfates the 6-O position; panel iii, inability of N-sulfamidase to hydrolyze the original disulfated monosaccharide (compare with panel i); panel iv, co-treatment of the disulfated substrate with both 6-O-sulfatase and N-sulfamidase enzymes showing the disappearance of all sulfated monosaccharides and demonstrating a prerequisite 6-O-desulfation by the 6-O-sulfatase prior to sulfate hydrolysis at the 2-amino position by the N-sulfamidase. Internal standard (458.1 Da) used to monitor ionization efficiency and mass calibration is noted by an asterisk.
The CE-based assay was also used to determine the pH optimum for the enzyme. N-Sulfamidase exhibited a slightly acidic pH optima (between 5.5 and 6.5) but was active over a broad range, especially above pH 7.0. The enzyme showed higher activity in acetate buffer when compared with sulfonate buffers such as MES and MOPS when examined over this same pH range.
Having identified a suitable chromogenic substrate for measuring N-sulfamidase activity, we likewise used this substrate to develop a fluorescence-based, coupled enzyme assay as the means to define the optimal in vitro reaction conditions and enzyme kinetic parameters as generally described for the 6-O-sulfatase (19), with the exception that the secondary enzyme that was used was an α-glucosidase. These experiments demonstrated only modest hydrolysis of the 1→4-MU glycosidic linkage of the N-sulfated glucosamine by the α-glucosidase in the absence of an N-sulfamidase preincubation. In this assay, the N-sulfamidase exhibited a sensitivity to increasing ionic strength with 50% inhibition observed at ∼200 mm NaCl and less than 20% activity remaining at 1 m NaCl relative to the zero NaCl control. Addition of sulfate or phosphate anions did not inhibit the action of N-sulfamidase.
We also investigated the action of the N-sulfamidase toward HSGAG disaccharides that possess a uronic acid at the nonreducing end. N-Sulfamidase was initially tested against a panel of unsaturated heparin disaccharides represented by ΔU±2SHNS±6S. For these experiments, standard reaction conditions were chosen as defined in the monosaccharide studies. None of the unsaturated disaccharides were desulfated by the enzyme (data not shown). The inability of N-sulfamidase to do so was confirmed in a related experiment in which all possible heparin disaccharides were first generated by pretreating heparin with heparinase I and III prior to adding N-sulfamidase to the same reaction tube. The converse experiment was also conducted in which unfractionated heparin was preincubated with N-sulfamidase for an extended period of time (8 h) followed by the addition of heparinase I and III. In this sequence, sulfatase pretreatment had no effect on the compositional profile of the heparinase-derived cleavage products (data not shown). These results confirm that the enzyme requires an N-sulfated (and non-O-sulfated) glucosamine at the nonreducing end of the chain. Taken together, these data also strongly suggest that the N-sulfamidase like the 6-O-sulfatase acts in an exclusively exolytic manner.
Structural Basis for Substrate Specificity and Enzymatic Activity of N-Sulfamidase
The molecular surface of the N-sulfamidase structural model revealed a groove region that was much wider than the 2-O-sulfatase that we have studied earlier (20), as well as the 6-O-sulfatase described in the accompanying paper (19). The theoretical model of the N-sulfamidase complexed with GlcNS substrate was used to identify the critical residues potentially involved in positioning of the substrate and/or catalysis (Fig. 3). Based on this theoretical model, the proposed roles of the putative active site residues are summarized in Table 1. The residues Arg-84 and Asp-40 that stabilize the FGly in the resting state in the active site are highly conserved among the different sulfatases. Furthermore, there is a pocket of oxygen atoms formed by Asp-40, Asn-41, Asp-246, Asn-247, the geminal diol of FGly, and N-sulfate group that can potentially coordinate a divalent metal ion such as Ca2+. The Ca2+ coordination formed by these residues resembles a similar arrangement in the other sulfatases (such as 6-O-sulfatase in the accompanying manuscript (19)) whose activity is critically influenced by Ca2+ ions. Surprisingly, two critical His residues that are highly conserved among the other sulfatases are absent in the putative active site of the N-sulfamidase. In the case of the 6-O-sulfatase homology model, His-130 is proposed to stabilize the FGly, whereas His-507 is proposed to be involved in the catalytic desulfation of the O-sulfate group. The absence of these critical His residues could be central to conferring the specificity of the N-sulfamidase enzyme to solely cleave the N–S bond (as against the O–S bond) in GlcNS containing HSGAG substrates. Other O-sulfated hexosamine sugars such as GlcNAc,6S, GalNAc,6S, GalNAc,4S, and GalNAc,4S,6S would therefore not be processed by N-sulfamidase, although these substrates might be accommodated in the active site. The modeled structural complex of N-sulfamidase with a 6-O-sulfate containing GlcNS,6S substrate showed that when the N-sulfate group is oriented toward the FGly residue, the 6-O-sulfate would make unfavorable steric contacts with Trp-249 in the active site (data not shown). This observation points to the obligatory sequential activity of the 6-O-sulfatase acting first to desulfate the 6-O-sulfate group, which is then followed by the action of N-sulfamidase. Similarly the complex of N-sulfamidase with GlcNS,3S showed that the 3-O-sulfate group would also have unfavorable steric contacts with Ile-188 and Asp-189 residues in the active site.
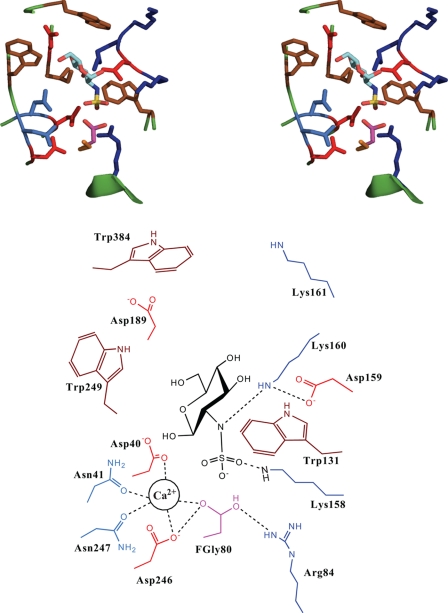
FIGURE 3.
Theoretical structural model of N-sulfamidase-GlcNS substrate. Shown on top is the stereo view of the active site of N-sulfamidase with the docked GlcNS substrate. The side chains of the key residues are shown and colored as follows: Arg and Lys, blue; Asp and Glu, red; Trp, Leu, and Ile, brown; Asn and Gln, light blue; and FGly, purple. The Glc-NS substrate is colored by atom as follows: C, cyan; O, red; N, blue; and S, yellow. Shown at the bottom is the schematic of the active site with the key amino acids labeled for clarity. Also shown in the schematic is the location of the divalent Ca2+ ion and its interactions with the active site and the substrate.
TABLE 1.
Functional assignment of N-sulfamidase active site residues
Amino acids listed in this table were identified by molecular interaction analysis of the N-sulfamidase model depicted in Fig. 3 as well as the proposed enzymatic cleavage mechanism shown in Fig. 4.
| Active site residues | Proposed functional role |
|---|---|
| Cys-80 | The active site cysteine is modified into the hydrated form of the FGly-O-γ1 (20) and participates in nucleophilic attack on the sulfate group |
| Arg-84 | This primarily stabilizes the hydrated FGly by interaction with O-γ2. This is also well positioned for proton abstraction from O-γ2 after the catalytic process for elimination of sulfate and regeneration of diol |
| Asp-159 | The negatively charged carboxylate groups of these residues are likely to abstract the proton from the NH3+ group of Lys-160 |
| Lys-160 | Upon loss of the proton to Asp-159, the nitrogen of this lysine residue is likely to be sulfated by the NHSO3− group, thereby breaking the existing nitrogen-sulfur bond. This process is mediated by the calcium ion. After this step, the sulfated lysine will be desulfated by the FGly residue. This lysine is also likely to increase electrophilicity of sulfur center by coordinating with the oxygen atoms of the sulfate group |
| Asp-246, Asn-247, Asp-40, Asn-41 | Asp-246, Asp-247, Asp-40, and Asn-41 are well positioned to coordinate with the divalent Ca2+ metal ion. The calcium ion is likely to play the crucial role of coordinating with the nitrogen of NHSO3− group, hence helping Lys-160 to break the N–S bond successfully. Hence, this tetrad is important for optimal N-sulfamidase function. Furthermore, Asp-40 is also understood to donate a proton and enhance nucleophilicity of O-γ1 |
Using the homology model, we also investigated the structural rationale for the predominant exolytic action of N-sulfamidase. The unsaturated ΔU±2SHNS±6S disaccharides were docked into the active site in an attempt to optimally position the N-sulfate group for catalysis. In so doing, the ΔU sugar and its sulfate group now made unfavorable steric contacts with several residues in the active site, including Trp-249, Leu-255, Ile-188, Trp-131, Trp-384, and Pro-363. This structural constraint imposed by the model is consistent with our observation that the N-sulfamidase will only process a GlcNS sugar at the nonreducing end of an oligosaccharide substrate (data not shown).
Specificity of N-Sulfamidase toward Longer Oligosaccharide Substrates
To experimentally validate the proposed exolytic activity of N-sulfamidase, we studied its activity on two structurally related sulfated trisaccharides (HNS,6SIHNS,6S and HNS,6SI2SHNS,6S). Each of these was generated from the corresponding tetrasaccharide ΔU2SHNS,6SI±2SHNS,6S by the tandem use of the 2-O-sulfatase and the Δ4,5-glycuronidase prior to the addition of 6-O-sulfatase and N-sulfamidase. Desulfation of the resultant trisaccharide was followed by MALDI-mass spectrometry (Fig. 4; data not shown for ΔU2SHNS,6SIHNS,6S). In this experiment, we observed that the 6-O-sulfatase was able to singularly desulfate both trisaccharides (Fig. 4, C and E), as detailed in the accompanying paper (19). The N-sulfamidase, however, was not able to do so, presumably because of the presence of the interfering 6-O-sulfate. In contrast, a pretreatment of the pentasulfated trisaccharide with the 6-O-sulfatase followed by N-sulfamidase treatment resulted in both the loss of a detectable pentasulfated trisaccharide mass and a dramatic reduction of the tetrasulfated trisaccharide mass (corresponding to the predicted substrate for the N-sulfamidase). Unfortunately, the formation of the 2N-desulfated trisaccharide product was not readily observable in this experiment (possibly because of inefficient ionization or poor complexation with the basic RG15 peptide that normally facilitates the flight of the intact sulfated saccharide).
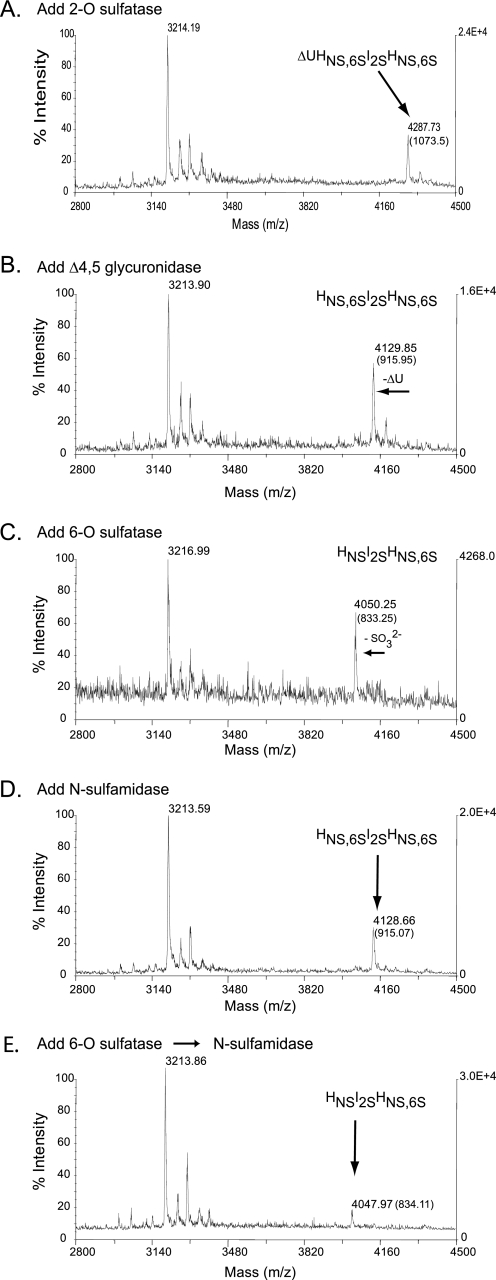
FIGURE 4.
Sequential degradation of an HSGAG tetrasaccharide using recombinantly expressed flavobacterial enzymes. Sequential treatment of the HSGAG tetrasaccharide (ΔU2SHNS,6SI2SHNS,6S) was physically assessed after each enzyme step (A–E) by MALDI-mass spectrometry. Masses listed in each panel represent either peptide alone (∼3216 Da) or oligosaccharide-peptide complex. The net mass of the oligosaccharide is listed in parentheses. A, addition of the 2-O-sulfatase; B, subsequent addition of the Δ4,5-glycuronidase; C, subsequent addition of the 6-O-sulfatase (note loss of a sulfate represented by a shift in net molecular mass from ∼915 to ∼835 Da); D, addition of N-sulfamidase directly after the Δ4,5-glycuronidase step (note the lack of any desulfation); E, addition of the N-sulfamidase subsequent to the 6-O-sulfatase (6-O then NS). The results in E are equivocal inasmuch as a double desulfated species with a net molecular mass of ∼755 Da was not clearly detected in this experiment.
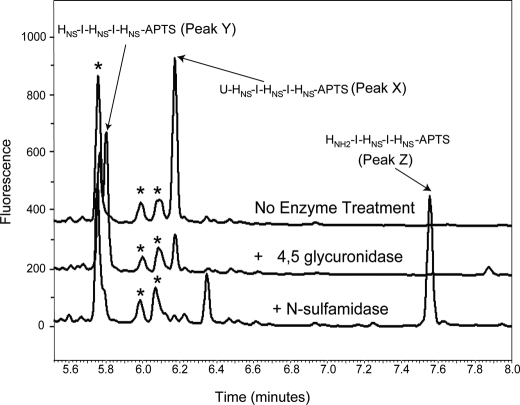
For this reason, we used an orthogonal CE-based method in an attempt to confirm the ability of the N-sulfamidase to hydrolyze odd-numbered oligosaccharides (lacking a 6-O-sulfate at their respective nonreducing ends) as strongly inferred in the previous experiment. Toward this end, the trisulfated pentasaccharide HNSIHNSIHNS was generated by Δ4,5-glycuronidase treatment of the purified hexasaccharide ΔUHNSIHNSIHNS and followed by incubation with the sulfamidase. All of the saccharides (untreated, Δ4,5 alone, and Δ4,5 followed by N-sulfamidase) were subsequently fluorescently labeled at their reducing end through reductive amination. End labeling of the sugars permitted their detection by laser-induced fluorescence following resolution of the products by capillary electrophoresis (Fig. 5). At each step in the experiment, saccharide peak assignment was inferred by observing discrete electrophoretic shifts in peak migration times as a function of exo-enzyme treatment and in accordance with their expected elution times based on differential charge densities. In this experiment, a unique peak (peak Z) eluting at 7.6 min appears following the sequential treatment of the starting material with the Δ4,5-glycuronidase and the N-sulfamidase. The late elution time of this peak is consistent with it possessing a lower sulfate density than either the trisulfated hexasaccharide starting material (peak X) or the pentasulfated pentasaccharide (peak Y, intermediate) resultant from digestion with the unsaturated glycuronidase. Such a dramatically shifted elution time is also suggestive of the presence of a positively charged amine that would reduce the formal negative charge of the sulfated oligosaccharide run under the electrophoresis conditions described. From this analysis, it appears that the N-sulfamidase does desulfate oligosaccharides devoid of the 6-O-sulfate group on the terminal hexosamine, and it does so in a predictably exolytic fashion.
FIGURE 5.
Sequential degradation of an HSGAG hexasaccharide. The structurally defined, heparin-derived hexasaccharide ΔUHNSIHNSIHNS was treated with Δ4,5-glycuronidase alone or with Δ4,5-glycuronidase followed by the N-sulfamidase. Resultant oligosaccharide products were fluorescently labeled at the reducing end with APTS by reductive amination as described under “Experimental Procedures.” Oligosaccharides were resolved by capillary electrophoresis and detected by laser-induced fluorescence. The N-desulfated pentasaccharide HNH2IHNSIHNS is observed as a unique peak appearing at ∼7.6 min (peak Z). Oligosaccharide impurities are noted by an asterisk.
Role of Calcium Metal Ions in Catalysis by N-Sulfamidase
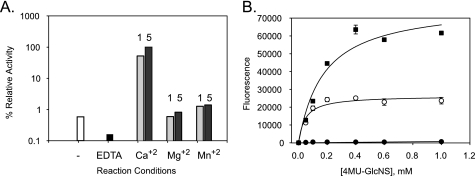
N-Sulfamidase enzyme was found to be activated by the presence of calcium in a concentration-dependent manner (Table 2). Interestingly, the enzyme was found to be almost inactive in the absence of calcium, unlike previously analyzed sulfatases (Fig. 6A). For example, the 6-O-sulfatase described in the accompanying paper (19) was activated 2–3-fold by the presence of calcium but was somewhat active even in the presence of 1 mm EDTA. Furthermore, the divalent metal activation was found to be specific to calcium; inclusion of Mg2+ or Mn2+ had only negligible effects. To further examine this metal selectivity, we measured the potential for enzyme inhibition in the presence of the calcium-specific chelator EGTA. As expected, EGTA inhibited calcium-dependent N-sulfamidase activity (at 5 mm Ca2+) in a concentration-dependent manner, with 50% inhibition occurring at ∼3 mm EGTA (data not shown). In an attempt to determine the mechanism by which calcium exerts its effect on N-sulfamidase, we followed up these metal ion experiments by next measuring the effect of calcium on enzyme steady-state kinetics (Fig. 6B). Consistent with our previous results, the initial rate of the enzyme was significantly affected by calcium in a concentration-dependent fashion with both kinetic parameters being affected proportionally (Table 2).
TABLE 2.
Steady-state kinetic parameters using 4-MU monosaccharide substrates
| Addition |
N-Sulfamidase |
||
|---|---|---|---|
| kcat | Km | kcat/Km (×102) | |
| min−1 | μm | ||
| None | NDa | ND | ND |
| 0.5 mm Ca2+ | 5.1 | 45 | 11.3 |
| 5 mm Ca2+ | 21.9 | 178 | 12.3 |
| 1 mm EDTA | ND | ND | ND |
a ND means not determined due to lack of activity.
FIGURE 6.
Effect of calcium metal ions on N-sulfamidase activity. A, calcium-specific requirement for N-sulfamidase activity and inhibition by EDTA wherein the divalent metal effect was not observed when calcium was replaced by either Mg2+ or Mn2+ (at either 1 or 5 mm concentrations). The following is the key: open bars (no divalent metals added); black bars (1 mm EDTA added); light gray bars (1 mm divalent metal); stippled gray bars (5 mm divalent metal). B, effect of calcium on steady-state kinetics of N-sulfamidase observed while varying concentrations of Ca2+ or in the presence of 1 mm EDTA. Substrate saturation plots were fitted to pseudo first-order Michaelis-Menten kinetics by nonlinear regression analyses. 0.5 mm Ca2+ (●), 1 mm Ca2+ (○), 5 mm Ca2+ (■). For clarification, the EDTA result (showing a lack of activity) is omitted.
Proposed Mechanism for Nitrogen-Sulfur Bond Cleavage by N-Sulfamidase Enzyme
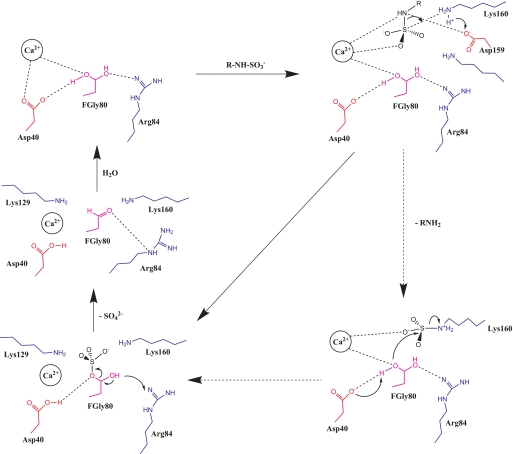
The combination of our biochemical studies and the structural model of the N-sulfamidase active site and its interactions with the substrate led us to propose a mechanism for the N–S bond cleavage. This mechanism is novel given the uniqueness of N-sulfamidase active site and that it is the only enzyme from P. heparinus that cleaves N–S bond in HSGAGs. The proposed mechanism of N-sulfamidase action has been depicted as a step-by-step process pictorially (Fig. 7). In the resting state, the active site cysteine that is understood to be modified into the hydrated form of the FGly-O-γ1 (20) is likely to be stabilized by interactions with the carboxylate anion group of Asp-40 and the amino group of Arg-84 as shown. Furthermore, the divalent calcium ion has been proposed to coordinate with the diol form of FGly-O-γ1 and also with the negatively charged oxygen atom of the NHSO3− group upon addition of a suitable substrate R-NH-SO3−. Subsequently, the carboxylate anionic group of Asp-159 is well poised to abstract the proton from the NH3+ group of Lys-160 residues. The nitrogen-sulfur bond cleavage is mediated by Ca2+ ion and Lys-160 resulting in the transfer of the sulfate group to the formyl glycine. Alternatively, it is possible for the amine group of the deprotonated Lys-160 to form a bond with the sulfate group (potential transition state) that would in turn facilitate the transfer of this group to the formyl glycine (Fig. 7, dotted arrows). This crucial step that is unique to N-sulfamidase is likely to be motivated by the interaction of the Ca2+ ion with the nitrogen atom of the NHSO3− group and simultaneous protonation of this nitrogen atom by the NH3+ group of Lys-158 as shown. This form of nitrogen-sulfur bond cleavage in the presence of metallic cations has been proposed for sulfonamide reactions (34). In our proposed mechanism, the divalent calcium cation and the basic Lys-160 residue provide the three interactions of this process, whereas in the sulfonamide mechanism proposed, a trivalent metal cation was used. The carboxylate group of Asp-40 is positioned to abstract a proton from the geminal diol leading to acquisition of the SO3− group from the sulfated form of Lys-160 as depicted. Following these distinguishing steps, the rest of the mechanism is likely to be similar to the other sulfatases (35). The basic Arg-84 residue has been indicated to interact with FGly-80 leading to the efflux of SO42− and formation of the aldehyde. This in turn is hydrolyzed back into the diol state, and the enzyme is ready to catalyze another desulfation process as and when a new substrate is encountered. The functions assigned to the key amino acids that participate in the proposed mechanism are summarized (Table 1).
FIGURE 7.
Proposed mechanism for cleavage of nitrogen-sulfur bond by N-sulfamidase. The color coding of the amino acids is as mentioned in Fig. 3 legend. The mechanism is depicted in parts for clarity, and all interactions are shown as broken lines. Transfer of protons and electrons are shown by arrows to denote directionality of transfer. The alternate pathway where Lys-160 potentially mediates the sulfate transfer to the formylglycine is shown with dotted arrow.
An insight into the proposed mechanism for N-sulfamidase action based upon our structural model provides possible rationale for the observed behavior. First, calcium ion is absolutely required for coordinating the nitrogen-sulfur bond cleavage as outlined above. Without the presence of the divalent cation and the coordination of nitrogen of the N-SO3− group, the enzyme cannot act. Therefore, it is clear that the activity of N-sulfamidase is very much dependent on calcium ion concentration and that the enzyme becomes inactive in the absence of the cation. Moreover, it is well know that magnesium ions (Mg2+) almost always exhibit octahedral coordination geometry with six being the preferred coordination number. Also, manganese ions (Mn2+) prefer coordinating with four ligands thereby exhibiting predominantly tetrahedral coordination geometry. These are fewer ligands than calcium (Ca2+) that is most commonly found coordinated by seven or eight ligands. The optimal coordination number in metal ions is a function of the ion size typically, because close packing of maximum possible ligands around the ions is generally attempted. This analysis of the enzyme-substrate interactions explains the absolute necessity for calcium ions for N-sulfamidase action and also helps to rationalize the inability of other divalent cations such as manganese and magnesium to promote enzyme activity.
DISCUSSION
In this paper, we have described the cloning, characterization, molecular modeling, and structure-function analysis of the flavobacterial N-sulfamidase enzyme. The N-sulfamidase (EC 3.10.1.1), commonly known as N-sulfoglucosamine sulfohydrolase, described here is one of only two nitrogen-sulfur cleaving enzymes currently identified by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB), the other enzyme being cyclamate sulfohydrolase (EC 3.10.1.2). Although significant progress has been made in understanding enzymatic O-sulfate cleavage mechanisms (35) involving oxygen-sulfur bond cleavage, surprisingly little is known about the mechanisms of enzymatic action by the corresponding N-sulfate cleaving hydrolases that specifically break the normally stable nitrogen-sulfur bonds that are unique to HSGAGs. As such, the investigation presented here provides valuable insight into the unique properties of this enzyme.
One of the important observations that motivated our structure-function studies of N-sulfamidase was the notable absence of key histidines that have been implicated as integral to the function of O-sulfatases (35). The absence of these key histidines could potentially govern the specificity of N-sulfamidase to cleave nitrogen-sulfur as against oxygen-sulfur bond. This observation prompted us to further investigate the mechanism of nitrogen-sulfur bond cleavage by this unique active site of N-sulfamidase. Using the theoretical structural model of the enzyme-substrate complex, we propose a new mechanism for the N–S bond cleavage by N-sulfamidase that was different from oxygen-sulfur cleavage of O-sulfatases. To our knowledge, this study is the first to propose such a mechanism of enzymatic nitrogen-sulfur bond cleavage. We also experimentally validated the role of key active site residues by C80A or D40A as described previously (20). These mutants showed no activity (Table 3) in comparison with the wild type enzyme and hence validate the critical role of these amino acids in the catalytic activity of the enzyme.
TABLE 3.
Assessment of relative activity of N-sulfamidase active site mutants
| Mutant | Classification | % Activitya |
|
|---|---|---|---|
| After 0.5 h | After 3 h | ||
| Wild type | Control | 100 | 100 |
| C80A | Active site (FGly) | 0 | 0 |
| D40A | Ca2+ coordination and catalytic role | 0 | 0 |
a Activity measured using the coupled enzyme assay under maximum (saturating substrate) conditions as described under “Experimental Procedures.”
Using this and previous studies, we have been able to reconstruct the complete F. heparinum HSGAG degradation pathway in vitro through a biochemical description of the respective substrate specificities for each of the cloned enzymes. As such, we are also able to place the activity of N-sulfamidase in a sequential context related to the F. heparinum HSGAG degradation pathway as it presumably exists in vivo, i.e. a degradation pathway that begins with the heparin lyases (heparinases), which leads to small oligosaccharides with ΔU uronic acid at the nonreducing end. These oligosaccharides are then acted upon by the exolytic Δ4,5-glycuronidase and the sulfatases. Previously, we established that 2-O-sulfatase action must precede the Δ4,5-glycuronidase cleavage because the presence of a 2-O-sulfate group inhibits the glycuronidase enzyme (18, 29). In the accompanying paper (19) and this study, we further establish that the 6-O-sulfatase enzyme should act prior to N-sulfamidase because the presence of the 6-O-sulfate group inhibits the sulfamidase enzyme. Interestingly, we still have to assign within this sequence the functional position of the 3-O-sulfatase activity reported in the literature (36, 37). The observations from this study and the accompanying paper (19) point to 3-O-sulfatase preceding the 6-O-sulfatase and N-sulfamidase. Therefore, the complete HSGAG degradation pathway (after action by the depolymerizing heparinases) is likely to proceed in the following sequence of enzymatic action: 2-O-sulfatase, Δ4,5-glycuronidase, 3-O-sulfatase (putative), 6-O-sulfatase, and N-sulfamidase.
Other questions related to the concerted activity of the N-sulfamidase enzyme in vivo also remain. Chief among them is the question of what precise form the substrates for these end-of-the-line sulfatases actually take. Is it reasonable to assume, for example, the “natural” substrates for the N-sulfamidase are actually monosaccharides? This assumption is at least consistent with the sequentially exolytic nature of the flavobacterial HSGAG-degrading pathway that we have described. It is also in line with the HSGAG structure-activity relationships and the active site architecture implied from these relationships. Ultimately, the ability of these enzymes to act on longer oligosaccharides in a manner predicted by their substrate specificities is of great practical value toward the use of these enzymes as discrete analytical tools for elucidating HSGAG fine structure.
N-Sulfated glucosamines are unique to heparin and heparan sulfate, and it is not surprising that the mechanism proposed in this paper is somewhat unique to this sulfate ester. The distinct chemistry and atomic interactions obtained from an analysis of our molecular model explained the empirically observed properties such as the substrate specificity, the novel mechanism of nitrogen-sulfur cleavage, the activity of the enzyme only in the presence of calcium ions but not other divalent metal cations, the order of enzyme processivity, and the inhibition of the enzyme by the presence of secondary sulfates within the glucosamine. As form follows function, these distinctions naturally play out at the level of enzyme structure and, in the case of the lysosomal N-sulfamidase, these are perceptible even at the primary sequence level where there is only about 10–25% identity to O-sulfatases. We also point out that even when one compares the heparan sulfamidase between divergent organisms such as flavobacterium and mammals, discrete structural differences are likely given the reversed order within the degradation sequence in which the sulfatase enzymes act. In the lysosomal pathway, the N-sulfamidase is a relatively early enzyme that precedes the 6-O-sulfatase, although our results indicate a reverse order for the flavobacterial enzymes. As such, the lysosomal heparin N-sulfamidase naturally possesses broader substrate specificity relative to the functional homologue from F. heparinum. It follows that the relative active site topologies should also differ, especially as it pertains to additional residues for the lysosomal enzyme that must accommodate secondary sulfate interactions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM57073.
- HSGAG
- heparin/heparan sulfate glycosaminoglycan
- ΔU
- Δ4,5 unsaturated uronic acid
- GlcNS,6S
- N- and 6-O-disulfated α-d-glucosamine
- GlcNAc,6S
- N-acetylated, 6-O-sulfated α-d-glucosamine
- GalNAc,6S
- N-acetylated, 6-O-sulfated β-d-galactosamine
- 4-MU
- 4-methylumbelliferyl
- APTS
- aminopyrene-1,4,6-trisulfonate
- FGly
- l-C-α-formylglycine
- MES
- 4-morpholineethanesulfonic acid
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- MALDI
- matrix-assisted laser desorption ionization
- ORF
- open reading frame
- CE
- capillary electrophoresis.
REFERENCES
- 1.Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 2.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 3.Sasisekharan R., Raman R., Prabhakar V. (2006) Annu. Rev. Biomed. Eng. 8, 181–231 [DOI] [PubMed] [Google Scholar]
- 4.Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prince J. M., Klinowska T. C., Marshman E., Lowe E. T., Mayer U., Miner J., Aberdam D., Vestweber D., Gusterson B., Streuli C. H. (2002) Dev. Dyn. 223, 497–516 [DOI] [PubMed] [Google Scholar]
- 6.Lai J. P., Chien J. R., Moser D. R., Staub J. K., Aderca I., Montoya D. P., Matthews T. A., Nagorney D. M., Cunningham J. M., Smith D. I., Greene E. L., Shridhar V., Roberts L. R. (2004) Gastroenterology 126, 231–248 [DOI] [PubMed] [Google Scholar]
- 7.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 8.Häcker U., Nybakken K., Perrimon N. (2005) Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Shriver Z., Pope R. M., Thorp S. C., Duncan M. B., Copeland R. J., Raska C. S., Yoshida K., Eisenberg R. J., Cohen G., Linhardt R. J., Sasisekharan R. (2002) J. Biol. Chem. 277, 33456–33467 [DOI] [PubMed] [Google Scholar]
- 10.Vivès R. R., Lortat-Jacob H., Fender P. (2006) Curr. Gene Ther. 6, 35–44 [DOI] [PubMed] [Google Scholar]
- 11.Dietrich C. P., Silva M. E., Michelacci Y. M. (1973) J. Biol. Chem. 248, 6408–6415 [PubMed] [Google Scholar]
- 12.Nakamura T., Shibata Y., Fujimura S. (1988) J. Clin. Microbiol. 26, 1070–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kertesz M. A. (2000) FEMS Microbiol. Rev. 24, 135–175 [DOI] [PubMed] [Google Scholar]
- 14.Sasisekharan R., Venkataraman G., Godavarti R., Ernst S., Cooney C. L., Langer R. (1996) J. Biol. Chem. 271, 3124–3131 [DOI] [PubMed] [Google Scholar]
- 15.Shriver Z., Liu D., Hu Y., Sasisekharan R. (1999) J. Biol. Chem. 274, 4082–4088 [DOI] [PubMed] [Google Scholar]
- 16.Sasisekharan R., Leckband D., Godavarti R., Venkataraman G., Cooney C. L., Langer R. (1995) Biochemistry 34, 14441–14448 [DOI] [PubMed] [Google Scholar]
- 17.Ernst S., Venkataraman G., Winkler S., Godavarti R., Langer R., Cooney C. L., Sasisekharan R. (1996) Biochem. J. 315, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myette J. R., Shriver Z., Claycamp C., McLean M. W., Venkataraman G., Sasisekharan R. (2003) J. Biol. Chem. 278, 12157–12166 [DOI] [PubMed] [Google Scholar]
- 19.Myette J. R., Soundararajan V., Shriver Z., Raman R., Sasisekharan R. (2009) J. Biol. Chem. 284, 35177–35188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raman R., Myette J. R., Shriver Z., Pojasek K., Venkataraman G., Sasisekharan R. (2003) J. Biol. Chem. 278, 12167–12174 [DOI] [PubMed] [Google Scholar]
- 21.Bond C. S., Clements P. R., Ashby S. J., Collyer C. A., Harrop S. J., Hopwood J. J., Guss J. M. (1997) Structure 5, 277–289 [DOI] [PubMed] [Google Scholar]
- 22.Boltes I., Czapinska H., Kahnert A., von Bülow R., Dierks T., Schmidt B., von Figura K., Kertesz M. A., Usón I. (2001) Structure 9, 483–491 [DOI] [PubMed] [Google Scholar]
- 23.Dierks T., Miech C., Hummerjohann J., Schmidt B., Kertesz M. A., von Figura K. (1998) J. Biol. Chem. 273, 25560–25564 [DOI] [PubMed] [Google Scholar]
- 24.Bateman A., Birney E., Durbin R., Eddy S. R., Howe K. L., Sonnhammer E. L. (2000) Nucleic Acids Res. 28, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 26.Beil S., Kehrli H., James P., Staudenmann W., Cook A. M., Leisinger T., Kertesz M. A. (1995) Eur. J. Biochem. 229, 385–394 [DOI] [PubMed] [Google Scholar]
- 27.Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S. D. (2002) J. Biol. Chem. 277, 49175–49185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkataraman G., Shriver Z., Raman R., Sasisekharan R. (1999) Science 286, 537–542 [DOI] [PubMed] [Google Scholar]
- 29.Myette J. R., Shriver Z., Kiziltepe T., McLean M. W., Venkataraman G., Sasisekharan R. (2002) Biochemistry 41, 7424–7434 [DOI] [PubMed] [Google Scholar]
- 30.Rhomberg A. J., Ernst S., Sasisekharan R., Biemann K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4176–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F. T., Evangelista R. A. (1995) Anal. Biochem. 230, 273–280 [DOI] [PubMed] [Google Scholar]
- 32.Guda C., Lu S., Scheeff E. D., Bourne P. E., Shindyalov I. N. (2004) Nucleic Acids Res. 32, W100–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiti R., Van Domselaar G. H., Zhang H., Wishart D. S. (2004) Nucleic Acids Res. 32, W590–W594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hankin D. M., Danopoulos A. A., Wilkinson G., Sweet T. K., Hursthouse M. B. (1996) J. Chem. Soc. Dalton Trans. 21, 4063–4069 [Google Scholar]
- 35.Hanson S. R., Best M. D., Wong C. H. (2004) Angew. Chem. Int. Ed. Engl. 43, 5736–5763 [DOI] [PubMed] [Google Scholar]
- 36.Lindahl U., Bäckström G., Thunberg L., Leder I. G. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 6551–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce J. S., McLean M. W., Long W. F., Williamson F. B. (1985) Eur. J. Biochem. 148, 359–365 [DOI] [PubMed] [Google Scholar]