Abstract
The antithrombotic benefits of warfarin are countered by a narrow therapeutic index that contributes to excessive bleeding or cerebrovascular clotting and stroke in some patients. This article reviews the current literature describing warfarin sensitivity genotyping and compares the results of that review to the findings of our study in 189 patients at Mayo Clinic conducted between June 2001 and April 2003. For the review of the literature, we identified relevant peer-reviewed articles by searching the Web of Knowledge using key word warfarin-related adverse event. For the 189 Mayo Clinic patients initiating warfarin therapy to achieve a target international normalized ratio (INR) in the range of 2.0 to 3.5, we analyzed the CYP2C9 (cytochrome P450 2C9) and VKORC1 (vitamin K epoxide reductase complex, subunit 1) genetic loci to study the relationship among the initial warfarin dose, steady-state dose, time to achieve steady-state dose, variations in INR, and allelic variance. Results were compared with those previously reported in the literature for 637 patients. The relationships between allelic variants and warfarin sensitivity found in our study of Mayo Clinic patients are fundamentally the same as in those reported by others. The Mayo Clinic population is predominantly white and shows considerable allelic variability in CYP2C9 and VKORC1. Certain of these alleles are associated with increased sensitivity to warfarin. Polymorphisms in CYP2C9 and VKORC1 have a considerable effect on warfarin dose in white people. A correlation between steady-state warfarin dose and allelic variants of CYP2C9 and VKORC1 has been demonstrated by many previous reports and is reconfirmed in this report. The allelic variants found to most affect warfarin sensitivity are CYP2C9*1*1-VKORC1BB (less warfarin sensitivity than typical); CYP2C9*1*1-VKORC1AA (considerable variance in INR throughout initiation); CYP2C9*1*2-VKORC1AB (more sensitivity to warfarin than typical); CYP2C9*1*3-VKORC1AB (much more sensitivity to warfarin than typical); CYP2C9*1*2-VKORC1AB (much more sensitivity to warfarin than typical); CYP2C9*1*3-VKORC1AA (much more sensitivity to warfarin than typical); and CYP2C9*2*2-VKORC1AB (much more sensitivity to warfarin than typical). Although we were unable to show an association between allelic variants and initial warfarin dose or dose escalation, an association was seen between allelic variant and steady-state warfarin dose. White people show considerable variance in CYP2C9 allele types, whereas people of Asian or African descent infrequently carry CYP2C9 allelic variants. The VKORC1AA allele associated with high warfarin sensitivity predominates in those of Asian descent, whereas white people and those of African descent show diversity, carrying either the VKORC1BB, an allele associated with low warfarin sensitivity, or VKORC1AB or VKORC1AA, alleles associated with moderate and high warfarin sensitivity, respectively.
ADE = adverse drug event; AEI = American Enterprise Institute; ASPE = allele-specific primer extension; CYP2C9 = cytochrome P450 2C9; INR = international normalized ratio; PCR = polymerase chain reaction; SNP = single-nucleotide polymorphism; VitKH2 = vitamin K hydroquinone; VKOR = vitamin K epoxide reductase; VKORC1 = VKOR complex, subunit 1
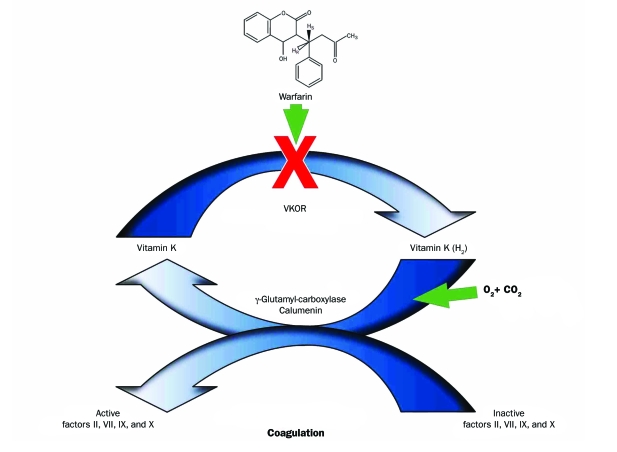
Warfarin is a well-accepted therapy used for the prevention of stroke in patients with atrial fibrillation, for prophylaxis of venous thromboembolism and pulmonary embolism in patients with prosthetic heart valves and myocardial infarction, and for prevention of pulmonary embolism or deep venous thrombosis in patients undergoing orthopedic surgery or with a history of venous or arterial thromboembolism. Many reviews and clinical practice guidelines (summarized by Ansell et al,1 Baglin et al,2 Flockhart et al,3 Husted et al,4 and Singer et al5) outline the substantial benefits of warfarin therapy for the prevention of strokes in these patients. The American Enterprise Institute (AEI)-Brookings Joint Center for Regulatory Studies Report, which reviewed the use of warfarin in the United States, estimates that approximately 2 million US citizens are prescribed warfarin annually.6 In 2003, a total of 21.2 million prescriptions were written for oral warfarin in the United States.7 Warfarin interferes with coagulation by inhibiting regeneration of the reduced form of vitamin K, a cofactor in the γ-glutamyl-carboxylase—mediated activation of coagulation factors II, VII, IX, and X (Figure 1). The oxidized, inactive form of vitamin K is converted to the reduced form of vitamin K by vitamin K epoxide reductase (VKOR); warfarin inhibits VKOR, resulting in less of the reduced form of vitamin K available to support the factor carboxylation required to sustain the coagulation cascade.8
FIGURE 1.
The coagulation cascade requires vitamin K in the reduced form (vitamin K [H2]) as a cofactor for γ-glutamyl-carboxylase to convert inactive factors II, VII, IX, and X to the active forms that are required for coagulation. Vitamin K (H2) is oxidized during this process to vitamin K epoxide. To conserve vitamin K (H2), the enzyme vitamin K epoxide reductase (VKOR) converts vitamin K epoxide back into vitamin K (H2). Warfarin inhibits VKOR, decreasing vitamin K (H2) availability, diminishing activatable factors II, VII, IX, and X and thus inhibiting coagulation. CO2 = carbon dioxide; O2 = oxygen.
Warfarin has a narrow therapeutic index that contributes either to therapeutic failure (potentially leading to stroke or other complications) or therapeutic excess (potentially leading to bleeding and hemorrhage). Many surveys have described the incidence of warfarin-related adverse drug events (ADEs). A recent report representative of these surveys estimates that more than 36,000 patients with warfarin-related ADEs were seen by emergency departments in the United States in 2006.9 The AEI-Brookings Report6 estimates that warfarin-related ADEs result in 43,000 visits to US hospital emergency departments annually.
INCIDENCE OF WARFARIN-RELATED ADEs
Early reports of warfarin-related ADEs suggested an overall bleeding rate of 7.6 to 16.5 per 100 patient-years. Major or life-threatening bleeding events occur at a rate of 1.3 to 2.7 per 100 patient-years.10-12 Landefeld and Beyth13 identified the annual risk of warfarin-related bleeding as ranging between 0.6% and 9.6%, approximately 5 times the frequency observed in patients not treated with warfarin. The risk of bleeding during the first month of warfarin therapy was reported to be 10 times higher than the risk after the first year of therapy. Bleeding in the gastrointestinal tract, soft tissue, and urinary tract was most common; intracranial bleeding was noted to be rare. The most important risk factors associated with warfarin-related bleeding were advanced age, female sex, history of a gastrointestinal bleeding event, and comorbid conditions, such as hypertension, cerebrovascular disease, atrial fibrillation, heart disease, renal insufficiency, liver disease, and alcoholism.
In a retrospective study of 261 patients with a median age of 69 years who began taking warfarin and continued receiving therapy for longer than 4 weeks in 1987-1989, Gitter et al14 reported the incidence of warfarin-related bleeding at 8% per year of warfarin therapy and the incidence of thromboembolism at 3% per year of warfarin therapy. From 1992-1994, Samsa et al15 performed a similar evaluation of 2481 patients with a median age of 68.8 years who had begun receiving warfarin therapy. Using the international normalized ratio (INR) as the indicator of treatment success, only 44% of patients achieved satisfactory INR; most reported INR values below the effective level, indicating a tendency to underdose anticoagulation therapy. In an evaluation of 11,526 patients, Go et al16 reported that the incidence of warfarin-related major thromboembolism was 1.3%, concluding that this low warfarin ADE rate was related to success in maintaining INR within the acceptable limit (2<INR>3).
In a study of age, Hirschl et al17 observed that patients older than 70 years experienced an anticoagulant-related bleeding rate of 12%, whereas those younger than 70 years had a bleeding rate of 7%. However, when all anticoagulation-related events were normalized, Hirschl et al17 concluded that age did not put patients at greater risk of anticoagulation-related adverse events. In a study of 221 patients comparing anticoagulation management by a primary care physician with management by an anticoagulation clinic, Wilson et al18 observed no difference in major bleeding events (1%-2%), no difference in thromboembolic events (1%-2%), and no difference in death rate (4%-5%) between the 2 management environments. However, Wilson et al18 noted that abnormal INRs (<1.5 or >3.5) were observed twice as frequently in the primary care environment. Wallvik et al19 reached a similar conclusion in an evaluation of 2731 patients, observing a major bleeding incidence of 3.9% in the primary care setting and 4.1% in the coagulation clinic setting. The incidence of fatal hemorrhage was 0.6% in both settings.
In a review of 26,345 records in the US National Registry of Atrial Fibrillation, Shireman et al20 found a 5.4% incidence of anticoagulation-related major bleeding episodes associated with women older than 70 years with a recent minor bleeding event, anemia, diabetes, and/or alcohol abuse. The incidence among men older than 65 years without a recent minor bleeding event, anemia, diabetes, or alcohol abuse was 0.9%. In a community study, Gurwitz et al21 reported an incidence of anticoagulation-related events of 25.5 per 100 residents receiving warfarin therapy per month. Flaherty et al22 extrapolated the findings from a multiyear study to the entire US population. He predicted an incidence of anticoagulation-associated intracerebral hemorrhage of between 0.8 per 100,000 patient-years in 1988 to 4.4 per 100,000 patient-years in 1999. In a review of the Food and Drug Administration Adverse Event Reporting System for the years 1998-2004, Wysowski et al23 reported that the incidence of warfarin-related bleeding events ranged from 10% to 16%.
Leigh and White24 estimated the cost of warfarin-related ADEs using a semi-Markov model. In a population of 10,000 patients aged 70 years who had been prescribed warfarin for atrial fibrillation, the cumulative cost associated with all warfarin-related stroke and hemorrhage events was estimated to be $18.3 million per 10,000 persons during a 5-year period.
Gladstone et al25 noted that atrial fibrillation—related strokes are severe, with a 1-year mortality rate of 50%, and that warfarin anticoagulation reduces the relative risk of ischemic stroke by 65% and the risk of death by 25%. This group reported that, among patients in the Registry of the Canadian Stroke Network, only 40% had been prescribed warfarin; 30%, combined antiplatelet therapy; and 29%, no antithrombotic drug. Of those taking warfarin, three-fourths had a subtherapeutic INR (INR <2.0) at the time of stroke admission. Overall, only 10% of patients with acute stroke were therapeutically anticoagulated (INR >2.0) at admission. In patients with stroke who had a history of atrial fibrillation and had had a previous transient ischemic attack or ischemic stroke (n=323), only 18% were taking warfarin with therapeutic INR at the time of admission for stroke, 39% were taking warfarin with subtherapeutic INR, and 15% were not receiving antithrombotic therapy. In an editorial, Gattellari et al26 speculated that physician fear of warfarin overdosing leads to either underdosing or failure to prescribe warfarin at all, resulting in 60% of all patients presenting with stroke being inadequately anticoagulated.
This large body of literature, accumulated for the purpose of review by search of Web of Knowledge for keyword warfarin-related adverse events, suggests that the incidence of major bleeding episodes in warfarin-treated patients is 1% to 2% higher than in untreated individuals; the incidence of minor bleeding events, 8% to 15% higher; and the incidence of thromboembolism, 1% higher. These generalizations are consistent with recent findings by Crowther et al.27
INTERNATIONAL NORMALIZED RATIO
Kirkwood28 introduced the INR to standardize prothrombin time measurements across laboratories. Shortly thereafter, Furie et al29 demonstrated the use of INR to monitor anticoagulation therapy. Kornberg et al30 and Le et al31 identified an ideal or target INR associated with warfarin therapy in large numbers of patients. Palareti et al12 and the Stroke Prevention in Atrial Fibrillation Investigators32 associated decreased incidence of bleeding with an INR less than 3 and a decreased incidence of stroke with an INR greater than 2. These works formed the basis for clinical practice standards (2<INR>3)33 that have been universally adopted for managing warfarin therapy.
MECHANISM OF ACTION AND METABOLIC FATE
In the normal coagulation cascade, factors II, VII, IX, and X undergo γ-glutamyl-carboxylation to become functional. The enzyme catalyzing this reaction, γ-glutamyl-carboxylase, requires reduced vitamin K hydroquinone (VitKH2) as a source of protons to complete carboxylation. For this reason, factors II, VII, IX, and X are referred to as the vitamin K—dependent coagulation factors. Vitamin K epoxide is a byproduct of γ-carboxylation. The enzyme VKOR recycles vitamin K epoxide to VitKH2 to sustain formation of coagulation factors that can be activated in the maintenance of the coagulation process. Warfarin is a potent inhibitor of VKOR. Anticoagulation occurs when warfarin inhibits VKOR, decreasing the amount of VitKH2 available to sustain functional vitamin K—dependent coagulation factors.1,8
The VKORC1 (VKOR complex, subunit 1) gene located on chromosome 16 at band p11.2 encodes the VKOR enzyme, a small transmembrane protein of the endoplasmic reticulum. Transcription of VKORC1 occurs primarily in the liver; however, smaller amounts of VKORC1 are present in the heart and pancreas. A polymorphism within the promoter region of VKORC1, specifically a guanine to adenine conversion at position −1639 (−1639 G>A, rs9923231), is associated with decreased warfarin dosing in white people and people of Asian descent.34 This may result from decreased production of VKORC1 mRNA by the −1639A allele and reduced expression of the enzyme VKOR.35 As a result of the reduced expression of VKOR, less warfarin is needed in patients carrying the −1639A promoter variant to maintain the target INR. Population studies have shown that the VKORCI −1639A allele predominates in people of Asian descent but is less common in white people and those of African descent. The VKORCI −1639G allele associated with greater VKORC1 expression and activity predominates in those of African descent. Several studies have cast doubt that the −1639G allele is responsible for warfarin dosing variability in people of African origin.36-38 Other alleles in VKORC1 may affect warfarin dosing, such as the mild to moderate increase in warfarin dose associated with the Asp36Tyr amino acid substitution.39
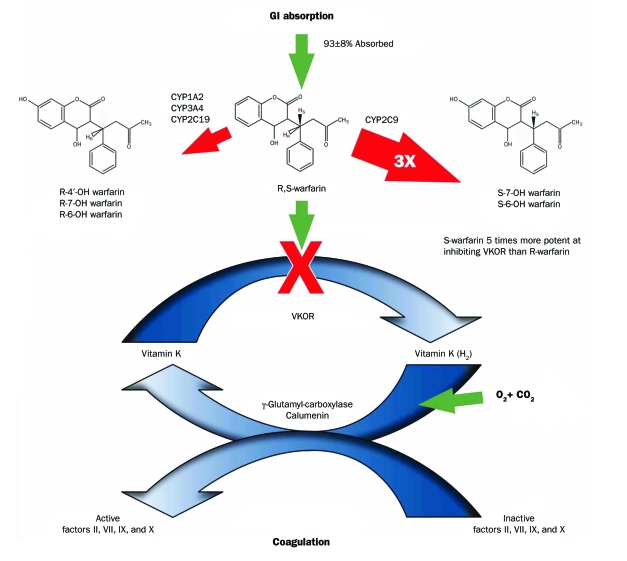
The combined interaction of the cytochrome P450 2C9 isoenzyme (CYP2C9) and VKORC1 on warfarin's anticoagulant activity is shown in Figure 2. Warfarin is a racemic molecule owing to the asymmetric nature of the first aliphatic carbon in the molecule. Clinically available warfarin is a mixture of 50% R-warfarin and 50% S-warfarin. Cytochrome P450 2C9 catalyzes the metabolism of S-warfarin into the inactive metabolites 6-hydroxy-S-warfarin and 7-hydroxy-S-warfarin. Cytochrome P450 1A2, 2C19, and 3A4 are responsible for the metabolism of R-warfarin into inactive 6-hydroxy-R-warfarin and 7-hydroxy-R-warfarin. Metabolic elimination of S-warfarin is 3 times faster than that of R-warfarin. Compared with R-warfarin, S-warfarin is approximately 5 times more active as a VKOR inhibitor. Because R-warfarin has considerably less anticoagulant activity, its metabolic fate is considered to have less consequence in the overall warfarin activity scheme.
FIGURE 2.
The coagulation cascade is inhibited by warfarin (see Figure 1). Warfarin, an enantiomeric mixture of equal concentrations of R- and S-forms, is 93%±8% absorbed from the gastrointestinal (GI) tract. Hepatic enzymes metabolize warfarin. Cytochrome P450 isomer 2C9 (CYP2C9) selectively converts S-warfarin into inactive hydroxylated metabolites. Cytochrome P450 isomers 1A2, 3A4, and 2C19 (CYP1A2, CYP3A4, and CYP2C19) selectively metabolize R-warfarin into inactive hydroxylated metabolites. The pace of metabolism for S-warfarin is approximately 3 times (3×) faster than for R-warfarin. The potency of S-warfarin at inhibiting vitamin K epoxide reductase (VKOR) is approximately 5 times that of R-warfarin. CO2 = carbon dioxide; O2 = oxygen; OH = hydroxide.
Metabolism of warfarin is catalyzed by CYP2C9, encoded for by the CYP2C9 gene. Hepatic CYP2C9 is responsible for the metabolism of the more active isomer of warfarin (S-warfarin) into inactive products. Several polymorphisms of the CYP2C9 gene have been identified that decrease the activity of the enzyme. This decreased enzymatic activity causes increased serum warfarin levels consistent with patient overmedication, increasing the INR above the therapeutic target level and accounting for increased bleeding incidents in some patients. In these patients, lower doses are required.17 The CYP2C9 gene is located on chromosome 10 at band q23.33. Clinically important nucleotide variants in CYP2C9 occur at positions 430, 818, and 1075. A change of cytosine to thymidine at position 430 (430C>T, rs1799853), known as the *2 allele, encodes a protein with cysteine instead of arginine at residue 144 with reduced enzymatic activity. A change of adenine to cytosine at position 1075 (1075A>C, rs1057910), known as *3, encodes a protein with leucine instead of isoleucine at residue 359 with minimal enzymatic activity. A deletion of adenine at position 818 (818 delA, rs9332131), known as *6, results in a protein with minimal enzymatic activity. A variant of CYP4F2 (7253232 C>T rs2108622) on chromosome 19 at band q13.12 also affects warfarin metabolism.
EFFECT OF ALLELIC VARIANTS ON WARFARIN ACTIVITY
The AEI-Brookings Report6 estimates that implementation of routine genetic testing for warfarin allelic variants could reduce the number of warfarin-related major bleeding events by 85,000 and reduce strokes by 17,000 annually in the United States, resulting in a reduction of $1.1 billion annually in health care spending. On the basis of these rather dramatic claims, a review of studies evaluating the role of genetic testing for warfarin sensitivity seems in order.
Aithal et al40 were the first to report an association between CYP2C9 genotype and warfarin dose; the odds ratio for persons with a low warfarin dose requirement having 1 or more CYP2C9 variant alleles compared with the normal population was 6.2. Patients in the low-dose group were more likely to have difficulties at the time of induction of warfarin therapy and have increased risk of major bleeding complications when compared with randomly selected clinic controls. Taube et al41 confirmed the finding of Aithal et al,40 reporting the mean maintenance dose of warfarin in 560 patients. Those with wild-type CYP2C9 for both alleles (*2 and *3) were receiving a maintenance warfarin dose of 5 mg, whereas the dose of warfarin in patients with variant alleles was between 61% and 86% of that in wild-type patients. Margaglione et al42 reported similar observations in 180 patients followed up in a coagulation clinic. The warfarin dose was higher (5.6 mg/d) in the CYP2C9*1 haplotype than in the CYP2C9*2 haplotype (4.7 mg/d) or the CYP2C9*3 haplotype (4.0 mg/d). One patient carrying both the CYP2C9*2 and *3 haplotypes maintained therapeutic INR with a warfarin dose of 1.8 mg/d. Bleeding complications were more frequent among patients with the CYP2C9*2 or *3 haplotype than in the patient with CYP2C9*1 haplotype.
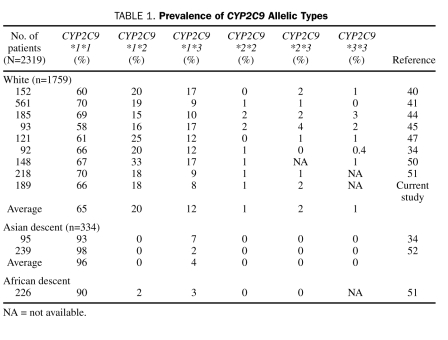
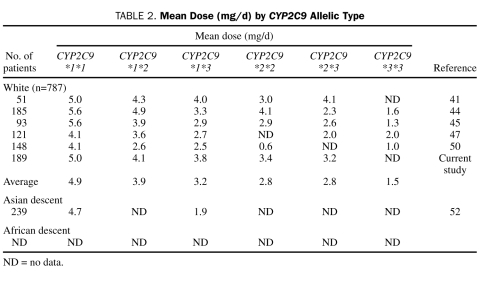
Loebstein et al,43 Higashi et al,44 Scordo et al,45 Linder et al,46 Kamali et al,47 Wadelius et al,48 and Peyvandi et al49 all reported similar findings. Studies articulating CYP2C9 allelic frequencies and associated warfarin doses among various ethnic groups are summarized in Table 1. Doses associated with these genotypes are summarized in Table 2.
TABLE 1.
Prevalence of CYP2C9 Allelic Types
TABLE 2.
Mean Dose (mg/d) by CYP2C9 Allelic Type
Sanderson et al53 performed a meta-analysis of 9 previously reported studies. Studies meeting inclusion criteria included 2775 patients. Of the study patients, 20% carried a variant allele: 12% had CYP2C9*2 and 8% had CYP2C9*3. The mean difference in daily warfarin dose in patients with CYP2C9*2 was −0.85 mg, and CYP2C9*3 reduction was −1.92 mg. The relative bleeding risk was 1.9 for CYP2C9*2 and 1.8 for CYP2C9*3. Overall, it was concluded that patients with CYP2C9*2 and CYP2C9*3 alleles have lower mean daily warfarin doses and a greater risk of bleeding.
Caldwell et al54 reported a CYP2C9 variant in CYP4F2 associated with warfarin dose in white patients who were receiving a stable dose of warfarin. Takeuchi et al55 estimated that the CYP4F2 accounted for 1.5% of the effect on warfarin dose. Combined, CYY2C9, VKORC1, and CYP4F2 explain 56% to 64% of the gene variant effect on warfarin metabolism and mechanism of action.56
Li et al57 were the first to report an association between VKORC1 allelic variants and vitamin K response in a human cell line. D'Andrea et al58 reported on 147 patients followed up from the start of anticoagulation with warfarin, investigating whether VKORC1 variants affected doses of drug prescribed to acquire the target anticoagulation intensity. Two common polymorphisms were found, 1173C>T in intron 1 (also known as c.174-136C>T, rs9934438) and 3730G>A (also known as c.*134G>A, rs7294) in the 3′ untranslated region. Regardless of the presence of confounding variables, the mean adjusted daily dose of warfarin required to maintain stable INR was higher (6.2 mg) among patients with the VKORC1 1173 CC genotype than that of patients carrying the CT (4.8 mg) or the TT genotype (3.5 mg). The VKORC1 and CYP2C9 genetic variants investigated accounted for about a third of the interindividual warfarin dose variability. Rieder et al59 identified 10 common noncoding VKORC1 single-nucleotide polymorphisms (SNPs) and inferred 5 major haplotypes in 368 patients (119 white, 96 of African descent, 120 of Asian descent), declaring a low-dose haplotype group (A) and a high-dose haplotype group (B). The mean maintenance dose of warfarin differed significantly among the 3 haplotype group combinations, at 2.7 mg/d for A/A, 4.9 mg/d for A/B, and 6.2 mg/d for B/B. They found that VKORC1 haplotype groups A and B explained approximately 25% of the variance in dose. People of Asian descent had a higher proportion of group A haplotypes and those of African descent had a higher proportion of group B haplotypes. These findings are consistent with data from a previous study that also showed an association between an SNP in intron 1 of VKORC1 (1173 C>T, or c.174-136C>T, or the SNP at position 6484 in the Rieder study) and the warfarin dose.58 The B haplotype appeared to generate twice as much RNA as the A haplotype. Key haplotypes in Group A (low warfarin dose) were H1 CCGATCTCTG and H2 CCGAGCTCTG; in Group B (high warfarin dose), they were H7 TCGGTCCGCA, H8 TAGGTCCGCA, and H9 TACGTTCGCG. Veenstra et al60 studied 69 Hong Kong Chinese patients for the relationship between VKORC1 allelic variants and warfarin dose. Patients carrying at least 1 copy of a VKORC1 group B haplotype required a significantly higher stable warfarin dose (n=16; 5.2 mg/d) than patients who were homozygous for group A haplotypes (n=53; 2.9 mg/d).
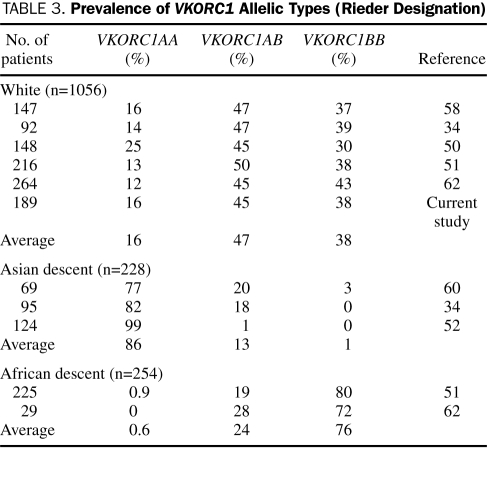
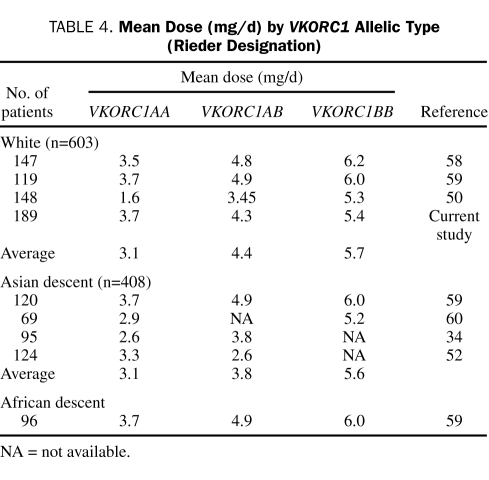
Because several different alleles in VKORC1 are associated with VKOR expression, it is important to develop a common nomenclature. The basis of such a nomenclature is suggested by the associations described by Rieder et al59: A haplotype represents more warfarin sensitivity (ie, less VKOR expressed) and B haplotype represents less warfarin sensitivity. McClain et al61 reported that the clinically relevant variants are −1639G>A in the promoter region and 1173C>T (c.174-136C>T), 1542G>C (c.283+124G>C, rs8050894), 2255T>C (c.283+837T>C, rs2359612), 3730G>A (c.*134G>A) in the intronic and 3′ untranslated regions. Applying the VKORC1 haplotype nomenclature developed by Rieder et al,59 A haplotype is described by −1639A, 1173T, 1542C, 2255C, or 3730A and B haplotype is described by −1639G, 1173C, 1542G, 2255T, or 3730G. Studies describing VKORC1 allelic frequencies and associated warfarin doses are summarized in Table 3. Doses associated with these genotypes are summarized in Table 4.
TABLE 3.
Prevalence of VKORC1 Allelic Types (Rieder Designation)
TABLE 4.
Mean Dose (mg/d) by VKORC1 Allelic Type (Rieder Designation)
Veenstra et al60 reported on the combined effect of CYP2C9 and VKORC1 allelic variants. Among patients with the VKORC1AA haplotype in a Hong Kong Chinese population, 4 (5.8%) were heterozygous for CYP2C9*3 and had a lower dose requirement (1.9 mg/d) than patients who exhibited the CYP2C9*1/*1 genotype (3.0 mg/d). Veenstra et al found that VKORC1 and CYP2C9 explained 31.0% and 7.9% of the variability in warfarin dose, respectively. Sconce et al63 studied 297 patients with stable anticoagulation and a target INR of 2.0 to 3.0. Genetic analyses for CYP2C9 (*2 and *3 alleles) and VKORC1 correlated with INR and plasma R- and S-warfarin concentrations. The mean warfarin daily dose requirement was highest in patients with the allelic variant CYP2C9 *1/*1 compared with those with the variant *2 and *3 alleles and highest in patients with the VKORC1 BB genotype compared with those with the AB genotype and the AA genotype. Age, height, and CYP2C9 genotype significantly contributed to S-warfarin and total warfarin clearance, whereas only age and body size significantly contributed to R-warfarin clearance. The multivariate regression model, which included the variables of age, CYP2C9 and VKORC1 genotype, and height, produced the best model for estimating warfarin dose.
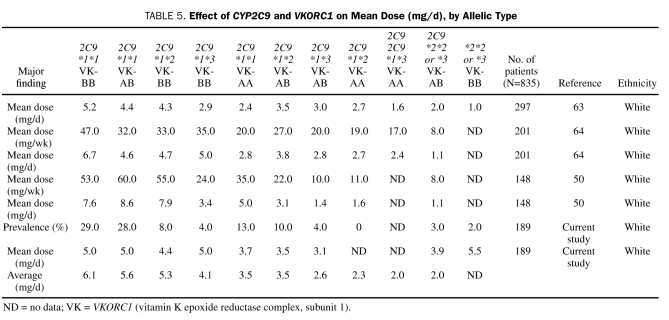
Wadelius et al,64 Gage et al,65 Bodin et al,66 Voora et al,67 Hillman et al,68 Kimura et al,69 Loebstein et al,39 and Borgiani et al50 reported similar findings when genotypes were distributed by race. Studies articulating combined CYP2C9 and VKORC1 allelic frequencies and associated warfarin doses are summarized in Table 5.
TABLE 5.
Effect of CYP2C9 and VKORC1 on Mean Dose (mg/d), by Allelic Type
Findings summarized in Tables 1, 2, 3, 4 and 5 represent peer-reviewed publications found by searching Web of Knowledge using key words warfarin dose related to genotype, CYP, and VKORC and selecting articles that reported warfarin dose in units of mg/d, mg/wk, or mg/kg. Numerous articles on this topic report risk associations between genotype and warfarin; although extremely valuable in understanding the relationship between warfarin dose and genotype, these articles were not included in the data summarized in Tables 1, 2, 3, 4 and 5.
Studies evaluating prospective warfarin sensitivity genotyping to predict optimal warfarin dose have been reported.70 Anderson et al71 evaluated pharmacogenetic-guided dosing of warfarin in 206 patients who were beginning warfarin therapy; these study patients were randomized to pharmacogenetic-guided or standard dosing. Buccal swab DNA was genotyped for CYP2C9*2 and CYP2C9*3 and VKORC1A (promoter polymorphism −1639G>A)/VKORCB. Standard dosing followed an empirical protocol, whereas pharmacogenetic-guided dosing followed a regression equation that included the 3 genetic variants and age, sex, and weight. A research pharmacist, unblinded to treatment strategy, managed the dose adjustments. Patients were followed up for as long as 3 months. Pharmacogenetic-guided doses more accurately approximated stable doses, resulting in smaller and fewer dosing changes. The percentage of out-of-range INRs did not differ significantly between treatment arms. Carriers of multiple variant alleles were at increased risk of an INR of 4 or less. Gage et al72 used an algorithm based on the independent predictors of therapeutic dose: VKORC1 genotype, body surface area, CYP2C9*3, CYP2C9*2, age, INR, amiodarone use, smoking status, race, and current thrombosis. This pharmacogenetic equation explained 53% to 54% of the variability in the warfarin dose in a validation cohort of 292 patients. Caraco et al73 studied 96 patients treated prospectively with warfarin and knowledge of warfarin sensitivity genotype. Patients treated in the genotyping arm of this study reached target INR 2.8 days earlier and stable warfarin dose 19 days earlier compared with 96 control patients matched for age, sex, body weight, body mass index, smoking status, indications for treatment, clinical characteristics, and co-medications treated without knowledge of genotype. The faster rate of initial anticoagulation was driven by a 28% higher daily dose in the study group. Study group patients spent more time within therapeutic INR (80.4% vs 63.4%, respectively) and experienced fewer episodes of minor bleeding (3.2% vs 12.5%, respectively).
A recent study74 involving 5052 patients created a warfarin-dosing protocol that more accurately identified patients with unusually high or low warfarin doses (<21 mg/wk or >49 mg/wk) than did a standard clinical evaluation. Following a pharmacogenetically tailored dosing protocol, these patients achieved stable INR faster than similar patients guided by standard clinical dosing protocols. Warfarin dosing recommendations were fundamentally similar to those outlined in Table 5.
DESCRIPTION OF MAYO CLINIC STUDY
Warfarin therapy has been managed by standardized antithrombotic guidelines at Mayo Clinic since before 1995. We thought it would be instructive to evaluate a large group of patients who were prescribed warfarin and managed using standardized antithrombotic guidelines and examine whether genetic information could identify patients whose dose deviated significantly from the norm. Knowledge gained from such an evaluation might provide guidance for future patient management, including warfarin sensitivity genotyping.
Mayo Clinic patients treated in the Chronic Anticoagulation Management Service of the Mayo Clinic Thrombophilia Center or the Coagulation Clinic of Community Internal Medicine during the years 1995-2003 were recruited to a cross-sectional study. The study period was selected because it represented a time when INR evaluation and warfarin management followed a common approach. Enrolled patients were new to warfarin anticoagulation therapy and had to complete at least 10 weeks of warfarin therapy. Candidates for the study were excluded if: (1) they had received warfarin therapy within the past 10 years; (2) they took concurrent medications known to increase or decrease the anticoagulant effect of warfarin; (3) clinical evidence (diminished or excessive vitamin K intake or absorption) suggested altered body vitamin K stores; or (4) clinical or laboratory evidence indicated hyperthyroidism or hypothyroidism, hepatic disease or dysfunction, or renal disease or dysfunction. A total of 900 patients were identified as candidates for the study. Review of medical records eliminated 470 patients because they were either taking interfering medications or had previously been prescribed warfarin. The remaining 430 patients were invited to participate. Of those, 251 (58%) agreed to participate and 207 (48%) provided blood samples for DNA testing. Of these patients, 15 were eliminated from the study because they underwent procedures during the first 10 weeks that required the discontinuation of warfarin therapy. Of the patients recruited during 2002-2003, 44 declined to provide blood because the point-of-care INR had been implemented at that time and they did not wish to undergo another venipuncture. Four were not included because genotyping was incomplete. Ultimately, 189 patients were included in the study, all of whom had an INR evaluated by the routine (not point-of-care) method throughout the 10-week study period. This study was approved by the Mayo Clinic Institutional Review Board, and participants signed written informed consent.
Initiation of warfarin was defined as the day warfarin therapy was started with the goal to achieve an INR in the target range of 2.0 to 3.5. It was understood that that dose was likely to be adjusted during the subsequent 10 weeks to achieve the target INR. After obtaining informed consent, we collected the following demographic information and baseline data: date of birth, sex, race, weight, height, indication for oral anticoagulation therapy, date of initiation of oral anticoagulation therapy, daily warfarin dose, and all INR measurements during the subsequent 10 weeks. After fasting and at the time of venipuncture for collection of a blood sample for clinically indicated INR testing, an extra 3.0-mL blood sample (ethylenediaminetetraacetic acid solution) was collected for leukocyte genomic DNA extraction, storage, and genotyping at the CYP2C9 and VKORC1 loci. Warfarin dose, dose changes, INR, and concomitant medications were recorded throughout the 10-week study period.
Genotyping for CYP2C9 and VKORC1 promoter polymorphisms used multiplex polymerase chain reaction (PCR) and multiplex allele-specific primer extension (ASPE) with a universal tag-sorting system (Tm Bioscience, Austin, TX) performed on the Luminex100 xMAP platform (Luminex, Austin, TX). Briefly, each sample was amplified in a single multiplex (7-plex) PCR reaction, resulting in amplicon sizes for the allelic regions of interest of 315 bp (CYP2C9*2), 586 bp (CYP2C9*3, *4, and *5), 319 bp (CYP2C9*6), and 43 bp (VKORC1—1639 promoter polymorphism). The PCR products were treated with shrimp alkaline phosphatase to inactivate unincorporated nucleotides and with exonuclease I to degrade leftover primers. An aliquot of the treated PCR product was used in the multiplex ASPE reaction, incorporating biotin-labeled deoxycytidine triphosphate and 10 universally tagged primers. The ASPE products were then sorted by hybridization to the Luminex universal bead array and conjugated with streptavidin, R-phycoerythrin reporter. Samples were analyzed using the Luminex100 xMAP IS instrument, and a fluorescent signal was generated for each of the alleles. The median fluorescence intensity values were used to determine whether wild-type and/or polymorphic alleles were present. Genotypes were assigned on the basis of the alleles detected. Dye-terminator DNA sequencing was used to verify CYP2C9 allelic variants and the −1639 promoter polymorphism in VKORC1. Concordance between the genotyping and sequencing was obtained in all cases with the exception of one study participant who had a CYP2C9 429C>T transition that resulted in no genotyping call made for the adjacent CYP2C9*2 allele (CYP2C9 430C>T) in the Luminex genotyping assay. The 429C>T change is a synonymous coding SNP with no change in function or amino acid sequence in the CYP2C9 protein. In 4 specimens, DNA quality was inadequate, resulting in incomplete genotyping results; data from these 4 patients were not included in the study.
Study patient histories were reviewed for warfarin dose and INR from the day of warfarin initiation to 10 weeks after initiation of treatment. Warfarin dose was prescribed and adjusted on the basis of the anticoagulation practice in place at the time (dependent on such variables as age, sex, body weight, and INR).30,31 Because many patients were treated with variable warfarin doses from day to day, all warfarin doses were summed for an entire week, normalized by patient weight, and expressed in units of milligrams of warfarin per kilogram of body weight per week (mg/kg/wk). When reported, daily dose was the weekly warfarin dose sum in milligrams divided by 7 days. At a later date and at a time when patients were in the clinic for routine evaluation of INR by phlebotomy (note: point-of-care INR testing had not been implemented at the time these patients were being treated), an extra blood sample was collected under an Institutional Review Board—approved protocol for genotype evaluation.
Data corresponding to warfarin dose and INR values during weeks 2 to 10 were used for the analysis; because INR or warfarin dose values from week 1 were either not recorded in the medical record or not performed in approximately one-third of patients, information from week 1 was not included in the review. A separate multiple linear regression model (INR value as dependent-response variable) with dose and week as the predictor variables was examined for each patient. On the basis of the model coefficient estimates for each patient, the dose corresponding to a “prediction of an INR of 2.5” was computed (eg, as in the linear calibration approach). This yielded the dose for a given patient that would be expected to provide an INR value of 2.5. These dose estimates (for all patients) were then assessed in an analysis of covariance model (estimated dose to provide an INR of 2.5 as the response) to evaluate the association of required dose with allelic type. Covariates included in this model were age, sex, and weight.
RESULTS OF MAYO CLINIC STUDY
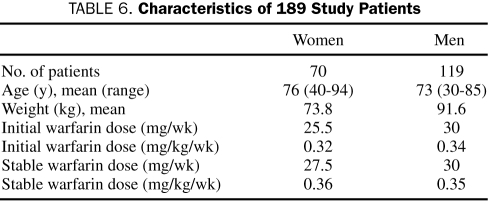
Genotyping was performed on 189 white patients enrolled in this Institutional Review Board—approved study. Patient demographics and median warfarin doses are described in Table 6.
TABLE 6.
Characteristics of 189 Study Patients
Of the study patients, 9 (5%) experienced elevated INRs (>3.5) of more than 1-week duration during weeks 2 through 5. All but 1 of these patients had allelic types associated with warfarin sensitivity (CYP2C9*2 and/or *3, plus VKORC1AA or VKORC1AB genotype), 4 experienced subtherapeutic INRs (<2.0) of more than 1-week duration during weeks 2 through 5, and all had allelic types associated with considerable warfarin sensitivity (CYP2C9*2 and *3, with VKORC1AB genotype).
Of these 189 patients, 1 experienced frequent minor bleeding episodes while maintaining an INR in the range of 2 to 3. This patient was a white woman aged 75 years who weighed 52 kg, was homozygous for CYP2C9*2, and had the VKORC1AB genotype. Using INR to guide warfarin dose adjustment, this patient's stable warfarin dose was achieved at 2.5 mg/d or 0.36 mg/kg/wk. She continued to experience minor bleeding episodes throughout her therapeutic regimen.
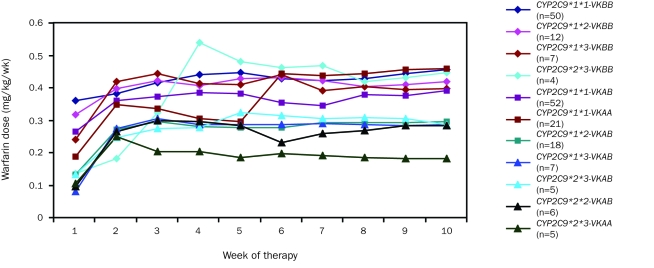
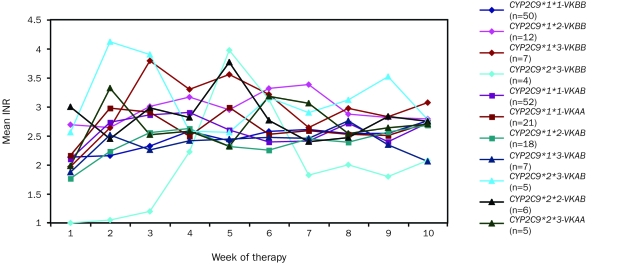
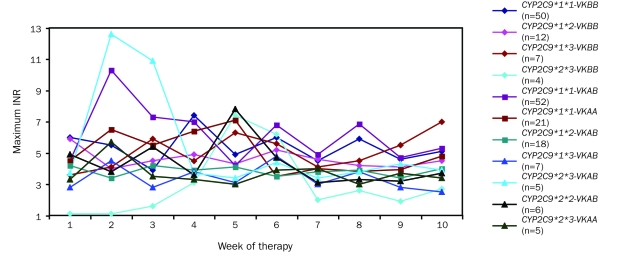
Figures 3, 4 and 5 display the patterns of mean warfarin dose and mean and maximum INR by week of therapy, differentiated by allelic types. Warfarin dose was achieved through weekly clinical observation guided by INR.
FIGURE 3.
Mean warfarin dose (mg/kg/wk) by week of therapy in all patients, differentiated by allelic type. Graph insert identifies allelic types and the number of patients with each allelic type. K = VKORC1 (vitamin K epoxide reductase complex, subunit 1).
FIGURE 4.
Mean observed international normalized ratio (INR) by week of therapy in all patients, differentiated by allelic type. Graph insert identifies allelic types and the number of patients with each allelic type. VK = VKORC1 (vitamin K epoxide reductase complex, subunit 1).
FIGURE 5.
Maximum observed international normalized ratio (INR) by week of therapy in all patients, differentiated by allelic type. Graph insert identifies allelic types and the number of patients with each allelic type. VK = VKORC1 (vitamin K epoxide reductase complex, subunit 1).
With the exception of the patients with CYP2C9*1*1 VKORC1AB and CYP2C9*2*3 VKORC1AB allelic types, stable warfarin dose was achieved by the third week. Patients with the CYP2C9*1*1 VKORC1AB allelic type showed considerable dose variability throughout the 10-week study period (Figure 3).
Mean INR tended to stabilize in the optimal range (2<INR>3) by week 10 (Figure 4). Most patients carrying the CYP2C9*1*1 VKORC1BB, CYP2C9*1*1 VKORC1AB, and CYP2C9*1*1 VKORC1AA allelic types achieved stable, optimal INR by week 3. Patients in the CYP2C9*1*2 VKORC1BB, CYP2C9*1*3 VKORC1BB, CYP2C9*2*3 VKORC1AB, and CYP2C9*2*3 VKORC1AB allelic groups exhibited variable INR throughout the 10-week period. Patients with the CYP2C9*2*3 VKORC1BB allelic type tended to be either under or over optimal INR.
Maximum INR exceeded the safe threshold of 5 in a number of cases. Patients carrying the CYP2C9*1*3 VKORC1BB, CYP2C9*1*1 VKORC1AA, and CYP2C9*2*3 VKORC1AB allelic types represent groups who experienced high INR of more than 1-week duration. Patients with the CYP2C9*2*3 VKORC1BB allelic type started therapy with several weeks of low INR but experienced significant INR elevations in weeks 5 and 6.
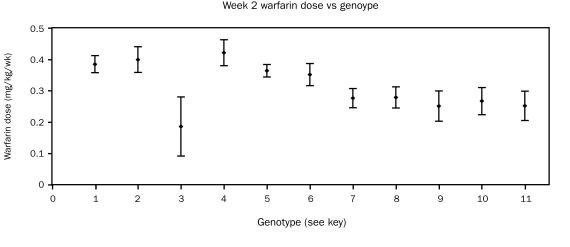
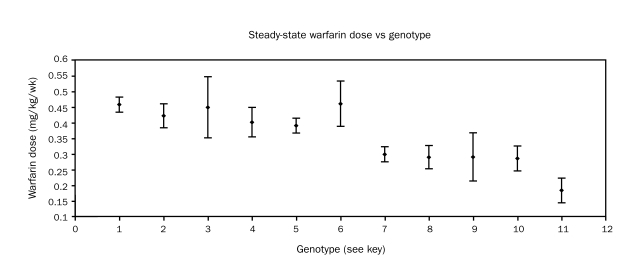
Figures 6 and 7 display the mean ± SEM warfarin dose in week 2 and week 10, differentiated by allelic types. A tendency for lower weekly doses in the allelic types 7 through 11 was noted as early as week 2 of therapy. Patients with the CYP2C9*1*1 VKORC1AB allelic type showed dose variance more than twice that of other allelic types (Figure 6).
FIGURE 6.
Mean ± SEM warfarin dose (mg/kg/wk) in the second week of therapy in all patients, differentiated by allelic type. Key: 1 = 2C9*1*1VKORC1BB (n=50); 2 = 2C9*1*2VKORC1BB (n=12); 3 = 2C9*1*3VKORC1BB (n=7); 4 = 2C9*2*3VKORC1BB (n=4); 5 = 2C9*1*1VKORC1AB (n=52); 6 = 2C9*1*1VKORC1AA (n=21); 7 = 2C9*1*2VKORC1AB (n=18); 8 = 2C9*1*3VKORC1AB (n=7); 9 = 2C9*2*3VKORC1AB (n=5); 10 = 2C9*2*2VKORC1AB (n=6); 11 = 2C9*2*3VKORC1AA (n=5)
FIGURE 7.
Mean ± SEM warfarin dose (mg/kg/wk) in the tenth week of therapy in all patients, differentiated by allelic type. Key: 1 = 2C9*1*1VKORC1BB (n=50); 2 = 2C9*1*2VKORC1BB (n=18); 3 = 2C9*1*3VKORC1BB (n=7); 4 = 2C9*2*3VKORC1BB (n=4); 5 = 2C9*1*1VKORC1AB (n=52); 6 = 2C9*1*1VKORC1AA (n=21); 7 = 2C9*1*2VKORC1AB (n=18); 8 = 2C9*1*3VKORC1AB (n=7); 9 = 2C9*2*3VKORC1AB (n=5); 10 = 2C9*2*2VKORC1AB (n=6); 11 = 2C9*2*3VKORC1AA (n=5)
The association of weekly dose with allelic type appeared even stronger at week 10 than at week 2 of therapy. Patients with the CYP2C9*1*1 VKORC1AB allelic type continued to show dose variability more than twice that of other allelic types (Figure 7). An analysis of covariance of the (log-transformed) week 10 doses indicated an overall association of allelic type with dose (P=.01), adjusting for age and sex. However, specific pairwise comparisons with CYP2C9*1*1 VKORC1AB did not detect associations of dose with this allelic type vs each of the others (P>.10; Dunnett test). There was a significant association between the week 10 dose and the dose estimated to provide the target INR value of 2.5; however, the analysis of covariance did not detect an overall significant association of allelic type with this dose (P=.13).
Further exploratory analyses examined pairwise differences among allelic subtypes in the dose calculated for each patient to provide the target INR value of 2.5. These comparisons suggested allelic types 2C9*1*2 VKORC1AB and 2C9*1/*2 and *2/3 VKORC1AA differed from allelic subgroup 2C9*1*1 VKORC1BB (P<.05; Dunnett test).
DISCUSSION
Warfarin is a well-accepted therapy for many conditions. However, its clinical benefits are countered by a narrow therapeutic index that contributes to excessive bleeding or cerebrovascular clotting and stroke in some patients. Because of concerns about bleeding, physicians tend to underdose warfarin, reducing its therapeutic efficacy.25
Many reports document some relationship between warfarin dose and warfarin sensitivity genotype. Furthermore, there is considerable consensus in the findings among research groups from different regions (Tables 1, 2, 3, 4 and 5). The 2 genes showing greatest interaction with varying warfarin dose are CYP2C9 and VKORC1. Combined, polymorphisms in these 2 alleles plus age, sex, and weight account for 56% to 64% of warfarin dose variation.56,73-75
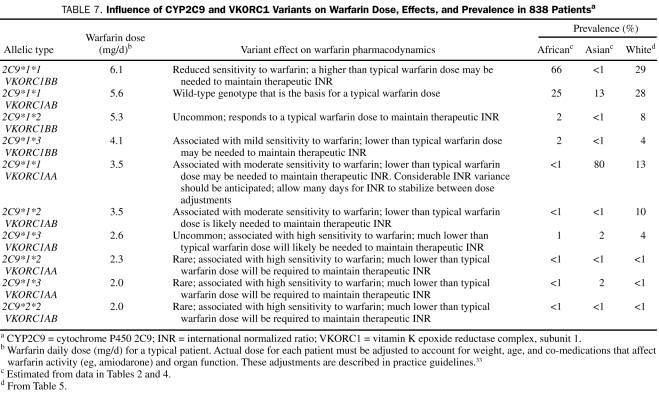
Table 7 summarizes the effects of CYP2C9 and VKORC1 variants on warfarin dose, the effects these variants have on sensitivity to warfarin, and variant prevalence as observed and reported in 3 previously published peer-reviewed reports plus results from the current report (835 patients total). The findings from these 4 unrelated studies describing the association between warfarin daily dose and genotype show considerable agreement. The summary in Table 7 is offered as guidance for warfarin dosing on the basis of warfarin sensitivity genotype evaluation until such time that practice guidelines are presented by professional societies. Results from a recent large study of white patients (N=1480)56 are consistent with our findings.
TABLE 7.
Influence of CYP2C9 and VKORC1 Variants on Warfarin Dose, Effects, and Prevalence in 838 Patientsa
People of African and Asian descent have very different CYP2C9 allelic frequencies than do white people (Tables 1, 2, 3 and 4). White people show considerable variability in CYP2C9, whereas people of African and Asian descent are predominantly of the CYP2C9*1*1 genotype, predisposing them to normal warfarin metabolism. White people and people of African descent have greater allelic diversity of VKORC1; those of Asian descent are predominantly of the VKORC1AA genotype,76 the allelic type associated with increased sensitivity to warfarin; and those of African descent are predominantly of the VKORC1BB genotype, the allelic type associated with decreased sensitivity to warfarin. Thus, polymorphisms in CYP2C9 have a larger effect on warfarin dose in white people, whereas VKORC1 polymorphisms affect warfarin dose in white people and people of African and Asian descent, with highest prevalence in those of Asian descent. We also observed that all white patients with homozygous CYP2C9*2*2 alleles had the VKORC1AB allelic type.
Considering the previously discussed associations between warfarin dose and genotype, it is important to recall that the 189 Mayo patients described in this article were managed on the basis of age, sex, body weight, and INR without access to genotype information. It seems reasonable to assume that most patients prescribed warfarin can be managed adequately on the basis of INR alone. Unfortunately, a physician cannot predict which patients will have the warfarin-sensitive genotype on the basis of physical examination findings or other non-laboratory—based parameters, perhaps with the exception of prolonged INR elevations for several weeks. Many reports show an association between prolonged INR elevation and warfarin sensitivity genotype. Unfortunately, a prolonged INR elevation predisposes patients to bleeding, and a prolonged subtherapeutic INR predisposes patients to stroke, both undesirable events; a prolonged aberrant INR is not a clinically advantageous tool to guide warfarin dose.
The one Mayo patient who experienced frequent minor bleeding episodes while maintaining an INR in the range of 2.0 to 3.0 was a white woman aged 78 years who weighed 52 kg, was homozygous for CYP2C9*2, had a VKORC1AB genotype, and was stabilized on a warfarin dose of 2.5 mg/d or 0.36 mg/kg/wk. The patient continued to experience minor bleeding episodes throughout her therapy. Adaptation of the data described in Table 7 suggests the optimal dose for this patient is less than 2 mg/d of warfarin.
Recently, Ansell et al1 reported practice recommendations for the management of vitamin K antagonists (warfarin and others) on behalf of the American College of Chest Physicians. These guidelines advise against the use of genotyping to guide warfarin therapy until such time that randomized prospective studies demonstrate a benefit. Such studies are currently under way at a number of academic medical centers in the United States and in Europe.77-79 Recently, several academic centers have offered warfarin dosing algorithms that include genomic information to aid in optimizing warfarin dose (Linder MW, personal communication, 2009). One of these aids is available via the Internet to all clinicians at www.warfarindosing.org.72
CONCLUSION
A correlation between steady-state warfarin dose and allelic variants of CYP2C9 and VKORC1 has been demonstrated by many previous reports and is reconfirmed in this report. Although we were unable to show a statistically significant relationship between allelic variants and initial warfarin dose or dose escalation, an association was demonstrated between allelic variant and steady-state warfarin dose.
White people show considerable variance in CYP2C9 allele types, whereas those of Asian and African descent infrequently carry CYP2C9 allelic variants. The VKORC1AA allele associated with high warfarin sensitivity predominates in those of Asian descent, whereas white people and people of African descent show diversity, carrying either the VKORC1BB allele associated with low warfarin sensitivity or the VKORC1AB or VKORC1AA alleles associated with moderate or high warfarin sensitivity, respectively. People of African descent predominantly carry the VKORC1BB allele associated with decreased warfarin sensitivity. We estimate that 86% of those of Asian descent and 16% of white people carry the 2C9*1*1 VKORC1AA variant, predisposing them to wide swings in INR throughout drug initiation. An additional 9% of white people and a lesser percentage of those of African descent carry the combination of CYP2C9 and VKORC1 allelic types associated with high sensitivity to warfarin. Unfortunately, the literature describing the incidence of the VKORC1 allele type in those of African descent is sparse. That limited literature suggests that 76% carry allelic variants associated with warfarin sensitivity requiring a higher than typical dose to maintain therapeutic INR.
Footnotes
This research was funded by a grant from the Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN.
REFERENCES
- 1.Ansell J, Hirsh J, Hylek E, et al. Pharmacology and management of the vitamin K antagonists. Chest 2008;133(suppl 6):160S-198S [DOI] [PubMed] [Google Scholar]
- 2.Baglin TP, Cousins D, Keeling DM, Perry DJ, Watson HG. Recommendations from the British Committee for Standards in Haematology and National Patient Safety Agency. BJH 2006;136:26-29 [DOI] [PubMed] [Google Scholar]
- 3.Flockhart DA, O'Kane D, Williams MS, et al. ACMG Working Group on Pharmacogenetic Testing of CYP2C9. VKORC1 Alleles for Warfarin Use Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med. 2008;10(2):139-150 [DOI] [PubMed] [Google Scholar]
- 4.Husted SE, Ziegler BK, Kher A. Long-term anticoagulant therapy in patients with coronary artery disease. Eur Heart J. 2006April;27(8):913-919 Epub 2006 Jan 9 [DOI] [PubMed] [Google Scholar]
- 5.Singer DE, Albers GW, Dalen JE, et al. Antithrombotic therapy in atrial fibrillation: evidence-based clinical practice guidelines, American College of Chest Physicians (8th Edition). Chest 2008;133(6 suppl):546S-592S [DOI] [PubMed] [Google Scholar]
- 6.McWilliams A, Lutter R, Nardinelli C.Health care savings from personalizing medicine using genetic testing: the case of warfarin. Working Paper 06-23 November 2006. http://www.reg-markets.org/publications/abstract.php?pid=1127. http://www.reg-markets.org/publications/abstract.php?pid=1127 Accessed September 3, 2009.
- 7.Marketos M. The top 200 generic drugs in 2003 (by unit). Drug Topics 2004March8;148:82 [Google Scholar]
- 8.Reynolds K, Valdes R, Jr, Hartung BR, Linder MW. Individualizing warfarin therapy. Per Med. 2007;4(1):11-31 [DOI] [PubMed] [Google Scholar]
- 9.Budnitz DS, Pollock DA, Weidenbach KN, Mendelsohn AB, Schroeder TJ, Annest JL. National surveillance of emergency department visits for outpatient adverse drug events. JAMA 2006;296(15):1858-1866 [DOI] [PubMed] [Google Scholar]
- 10.van der Meer FJ, Rosendaal FR, Vandenbroucke JP, Briët E. Bleeding complications in oral anticoagulant therapy. Arch Intern Med. 1993;153(13):1557-1562 [DOI] [PubMed] [Google Scholar]
- 11.Cannegieter SC, Rosendaal FR, Wintzen AR, van der Meer FJ, Vandenbroucke JP, Briët E. Optimal oral anticoagulant therapy in patients with mechanical heart valves. N Engl J Med. 1995;333(1):11-17 [DOI] [PubMed] [Google Scholar]
- 12.Palareti G, Leali N, Coccheri S, et al. Italian Study on Complications of Oral Anticoagulant Therapy Bleeding complications of oral anticoagulation treatment: an inception-cohort, prospective collaborative study (ISCOAT). Lancet 1996;348(9025):423-428 [DOI] [PubMed] [Google Scholar]
- 13.Landefeld CS, Beyth RJ. Anticoagulant-related bleeding: clinical epidemiology, prediction, and prevention. Am J Med. 1993;95(3):315-328 [DOI] [PubMed] [Google Scholar]
- 14.Gitter MJ, Jaeger TM, Petterson TM, Gersh BJ, Silverstein MD. Bleeding and thromboembolism during anticoagulation therapy: a population-based study in Rochester, Minnesota. Mayo Clin Proc. 1995;70(8):725-733 [DOI] [PubMed] [Google Scholar]
- 15.Samsa GP, Matchar DB, Goldstein LB, et al. Quality of anticoagulation management among patients with atrial fibrillation: results of a review of medical records from 2 communities. Arch Intern Med. 2000;160(7):967-973 [DOI] [PubMed] [Google Scholar]
- 16.Go AS, Hylek EM, Borowsky LH, Phillips KA, Selby JV, Singer DE. Warfarin use among ambulatory patients with nonvalvular atrial fibrillation: the Anticoagulation and Risk Factors in Atrial Fibrillation (ATRIA) study. Ann Intern Med. 1999;131(12):927-934 [DOI] [PubMed] [Google Scholar]
- 17.Hirschl M, Pluschnig U, Kundi M, Katzenschlager R. Oral anticoagulation in older patients with vascular or cardiovascular diseases: aged over 70 years: same risk? Same benefit? Int Angiol. 2003;22(4):370-375 [PubMed] [Google Scholar]
- 18.Wilson SJ, Wells PS, Kovacs MJ, et al. Comparing the quality of oral anticoagulation management by anticoagulation clinics and by family physicians: a randomized controlled trial [published correction appears in CAMJ. 2004;170(4):451] CMAJ 2003;169(4):293-298 [PMC free article] [PubMed] [Google Scholar]
- 19.Wallvik J, Själander A, Johansson L, Bjuhr R, Jansson JH. Bleeding complications during warfarin treatment in primary healthcare centres compared with anticoagulation clinics. Scand J Prim Health Care 2007;25(2):123-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shireman TI, Mahnken JD, Howard PA, Kresowik TF, Hou Q, Ellerbeck EF. Development of a contemporary bleeding risk model for elderly warfarin recipients. Chest 2006;130(5):1390-1396 [DOI] [PubMed] [Google Scholar]
- 21.Gurwitz JH, Field TS, Radford MJ, et al. The safety of warfarin therapy in the nursing home setting. Am J Med. 2007June;120(6):539-544 Epub 2007 Apr 26 [DOI] [PubMed] [Google Scholar]
- 22.Flaherty ML, Kissela B, Woo D, et al. The increasing incidence of anticoagulant-associated intracerebral hemorrhage. Neurology 2007;68(2):116-121 [DOI] [PubMed] [Google Scholar]
- 23.Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007;167(13):1414-1419 [DOI] [PubMed] [Google Scholar]
- 24.Leigh JP, White RH. An economic model of adverse events and the costs for oral anticoagulants used for atrial fibrillation. Curr Med Res Opin. 2007;23(9):2071-2081 [DOI] [PubMed] [Google Scholar]
- 25.Gladstone DJ, Bui E, Fang J, et al. Potentially preventable strokes in high-risk patients with atrial fibrillation who are not adequately anticoagulated. Stroke 2009January;40(1):235-240 Epub 2008 Aug 28 [DOI] [PubMed] [Google Scholar]
- 26.Gattellari M, Worthington J, Zwar N. Warfarin: an inconvenient truth [editorial]. Stroke 2009January;40(1):5-7 Epub 2008 Aug 28 [DOI] [PubMed] [Google Scholar]
- 27.Crowther MA, Ageno W, Garcia D, et al. Oral vitamin K versus placebo to correct excessive anticoagulation in patients receiving warfarin. Ann Intern Med. 2009;150(5):293-300 [DOI] [PubMed] [Google Scholar]
- 28.Kirkwood TB. Calibration of reference thromboplastins and standardisation of the prothrombin time ratio. Thromb Haemost. 1983;49(3):238-244 [PubMed] [Google Scholar]
- 29.Furie B, Liebman HA, Blanchard RA, Coleman MS, Kruger SF, Furie BC. Comparison of the native prothrombin antigen and the prothrombin time for monitoring oral anticoagulant therapy. Blood 1984;64(2):445-451 [PubMed] [Google Scholar]
- 30.Kornberg A, Francis CW, Pellegrini VD, Jr, Gabriel KR, Marder VJ. Comparison of native prothrombin antigen with the prothrombin time for monitoring oral anticoagulant prophylaxis. Circulation 1993;88(2):454-460 [DOI] [PubMed] [Google Scholar]
- 31.Le DT, Weibert RT, Sevilla BK, Donnelly KJ, Rapaport SI. The International Normalized Ratio (INR) for monitoring warfarin therapy: reliability and relation to other monitoring methods. Ann Intern Med. 1994;120(7):552-558 [DOI] [PubMed] [Google Scholar]
- 32.Stroke Prevention in Atrial Fibrillation Investigators Adjusted-dose warfarin versus low-intensity, fixed-dose warfarin plus aspirin for high-risk patients with atrial fibrillation: Stroke Prevention in Atrial Fibrillation III randomised clinical trial. Lancet 1996;348(9028):633-638 [PubMed] [Google Scholar]
- 33.Health Care Guideline Institute for Clinical Systems Improvement Web site Antithrombotic therapy supplement (guideline) September2007: http://www.icsi.org/guidelines_and_more/gl_os_prot/cardiovascular/antithrombotic_therapy_supplement__guideline__14045/antithrombotic_therapy_supplement_guideline.html Accessed September 4, 2009
- 34.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005July1;14(13):1745-1751 Epub 2005 May 11 [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 2008August15;112(4):1013-1021 Epub 2008 Jun 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kealey C, Chen Z, Christie J, et al. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics 2007;8(3):217-225 [DOI] [PubMed] [Google Scholar]
- 37.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007May;81(5):742-747 Epub 2007 Feb 28 [DOI] [PubMed] [Google Scholar]
- 38.Kimmel SE, Christie J, Kealey C, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2008February;8(1):53-60 Epub 2007 Feb 27 [DOI] [PubMed] [Google Scholar]
- 39.Loebstein R, Dvoskin I, Halkin H, et al. A coding VKORC1 Asp-36Tyr polymorphism predisposes to warfarin resistance. Blood 2007March15;109(6):2477-2480 Epub 2006 Nov 16 [DOI] [PubMed] [Google Scholar]
- 40.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999;353(9154):717-719 [DOI] [PubMed] [Google Scholar]
- 41.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 2000;96(5):1816-1819 [PubMed] [Google Scholar]
- 42.Margaglione M, Colaizzo D, D'Andrea G, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000;84(5):775-778 [PubMed] [Google Scholar]
- 43.Loebstein R, Yonath H, Peleg D, et al. Interindividual variability in sensitivity to warfarin: nature or nuture? Clin Pharmacol Ther. 2001;70(2):159-164 [DOI] [PubMed] [Google Scholar]
- 44.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287(13):1690-1698 [DOI] [PubMed] [Google Scholar]
- 45.Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther. 2002;72(6):702-710 [DOI] [PubMed] [Google Scholar]
- 46.Linder MW, Looney S, Adams JE, III, et al. Warfarin dose adjustments based on CYP2C9 genetic polymorphisms. J Thromb Thrombolysis 2002;14(3):227-232 [DOI] [PubMed] [Google Scholar]
- 47.Kamali F, Khan TI, King BP, et al. Contribution of age, body size, CYP2C9 genotype to anticoagulant response to warfarin. Clin Pharmacol Ther. 2004;75(3):204-212 [DOI] [PubMed] [Google Scholar]
- 48.Wadelius M, Sörlin K, Wallerman O, Moia M, Mannucci PM. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics 2004;4(1):40-48 [DOI] [PubMed] [Google Scholar]
- 49.Peyvandi F, Spreafico M, Siboni SM, et al. CYP2C9 genotypes and dose requirements during the induction phase of oral anticoagulant therapy. Clin Pharmacol Ther. 2004;75(3):198-203 [DOI] [PubMed] [Google Scholar]
- 50.Borgiani P, Cicacci C, Forte V, Romano S, Federici G, Novelli G. Allelic variants in CYP2C9 and VKORC1 loci and interindividual variability in anticoagulant dose effect of warfarin in Italians. Pharmacogenomics 2007;8(11):1545-1550 [DOI] [PubMed] [Google Scholar]
- 51.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008February;83(2):312-321 Epub 2007 Jul 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obayashi K, Nakamura K, Kawana J, et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clin Pharmacol Ther. 2006;80(2):169-178 [DOI] [PubMed] [Google Scholar]
- 53.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005;7(2):97-104 [DOI] [PubMed] [Google Scholar]
- 54.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008April15;111(8):4106-4112 Epub 2008 Feb 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009March;5(3)e1000433 Epub 2009 Mar 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood 2009January;113(4):784-792 Epub 2008 Jun 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature 2004;427(6974):541-544 [DOI] [PubMed] [Google Scholar]
- 58.D'Andrea G, D'Ambrosio RI, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood 2005January15;105(2):645-649 Epub 2004 Sep 9 [DOI] [PubMed] [Google Scholar]
- 59.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285-2293 [DOI] [PubMed] [Google Scholar]
- 60.Veenstra DL, You JH, Rieder MJ, et al. Association of vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics 2005;15(10):687-691 [DOI] [PubMed] [Google Scholar]
- 61.McClain MR, Palomaki GE, Piper M, Haddow JE. Rapid-ACCE review of CYP2C9 and VKORC1 alleles testing to inform warfarin dosing in adults at elevated risk for thrombotic events to avoid serious bleeding. Genet Med. 2008;10(2):89-98 [DOI] [PubMed] [Google Scholar]
- 62.Schwartz UI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;58(10):999-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood 2005October1;106(7):2329-2333 Epub 2005 Jun 9 [DOI] [PubMed] [Google Scholar]
- 64.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262-270 [DOI] [PubMed] [Google Scholar]
- 65.Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91(1):87-94 [DOI] [PubMed] [Google Scholar]
- 66.Bodin L, Verstuyft C, Tregouet DA, et al. Cytochrome P450 2C9 (CYP2C) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood 2005July1;106(1):135-140 Epub 2005 Mar 24 [DOI] [PubMed] [Google Scholar]
- 67.Voora D, Eby C, Linder MW, et al. Prospective dosing of warfarin based on cytochrome P450 2C9 genotype. Thromb Haemost. 2005;93(4):700-705 [DOI] [PubMed] [Google Scholar]
- 68.Hillman MA, Wilke RA, Yale SH, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res. 2005;3(3):137-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimura R, Miyashita K, Kokubo Y, et al. Genotypes of vitamin K epoxide reductase, gamma glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120(2):181-186 Epub 2006 Oct 17 [DOI] [PubMed] [Google Scholar]
- 70.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007April;7(2):99-111 Epub 2006 Sep 19 [DOI] [PubMed] [Google Scholar]
- 71.Anderson JL, Horne BD, Stevens SM, et al. Couma-Gen Investigators Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation 2007November27;116(22):2563-2570 Epub 2007 Nov 7 [DOI] [PubMed] [Google Scholar]
- 72.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin [published correction appears in Clin Pharmacol Ther 2008;84(3):430] Clin Pharmacol Ther. 2008September;84(3):326-331 Epub 2008 Feb 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008March;83(3):460-470 Epub 2007 Sep 12 [DOI] [PubMed] [Google Scholar]
- 74.International Warfarin Pharmacogenetics Consortium Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(80):753-764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007March;121(1):23-34 Epub 2006 Oct 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wen MS, Lee MT, Chen JJ, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther. 2008July;84(1):83-89 Epub 2008 Jan 9 [DOI] [PubMed] [Google Scholar]
- 77.Kimes M. Medco's big bio bet: the company that manages your drug benefit also wants to get into your genes. Fortune 500 October24, 2008. http://money.cnn.com/2008/10/24/news/companies/kimes_medco.fortune/index.htm Accessed September 4, 2009 [PubMed]
- 78.Harvard's warfarin dose study will analyze cost/benefit of using gene-based dosing. Genomeweb Pharmacogenomics Reporter Web site http://www.genomeweb.com/dxpgx/harvards-warfarin-dose-study-will-analyze-costbenefit-using-gene-based-dosing Accessed September 4, 2009
- 79.Could genetics improve warfarin dosing? New research says yes for now. National Insitute of General Medical Sciences Web site http://www.nigms.nih.gov/News/Results/Warfarin02182009.htm Accessed September 4, 2009