Abstract
Constitutive activity, or ligand-independent activity, of mutant G protein-coupled receptors (GPCRs) has been described extensively and implicated in the pathology of many diseases. Using the corticotropin-releasing factor (CRF) receptor and the thrombin receptor as a model, we present a ligand-dependent constitutive activation of a GPCR. A chimera in which the N-terminal domain of the CRF receptor is replaced by the amino-terminal 16 residues of CRF displays significant levels of constitutive activation. The activity, as measured by intracellular levels of cAMP, is blocked in a dose-dependent manner by the nonpeptide antagonist antalarmin. These results support a propinquity effect in CRF receptor activation, in which the amino-terminal portion of the CRF peptide is presented to the body of the receptor in the proper proximity for activation. This form of ligand-dependent constitutive activation may be of general applicability for the creation of constitutively activated GPCRs that are regulated by peptide ligands such as CRF. These chimeras may prove useful in analyzing mechanisms of receptor regulation and in the structural analysis of ligandactivated receptors.
Corticotropin-releasing factor (CRF) is a 41-residue C-terminally amidated neuropeptide, which was first isolated and characterized from ovine hypothalamic extracts (1). CRF is a key regulator of stress responses and mediates its physiological actions by activating G protein-coupled receptors (GPCRs). The cloning of the human CRF type 1 receptor (R1) (2) indicated that this receptor belonged to the secretin-like family of GPCRs, also designated the class 2 or class B receptor family. The secretin-like family of receptors includes receptors for secretin, calcitonin, gastric inhibitory peptide, growth hormone-releasing hormone, glucagon, glucagon-like peptide I, parathyroid hormone (PTH), and vasoactive intestinal polypeptide (3). These peptides all stimulate cAMP formation upon binding their respective receptors.
Constitutively active secretin-like receptors have been described (4, 5). The proposed involvement of constitutively active PTH receptors in Jansen-type metaphyseal chondrodysplasia revealed two positions at which mutations can induce ligand-independent activity. The mutations that conferred constitutive activity in the human PTH receptor were His-223–Arg and Thr-410–Pro at the beginning of transmembrane helices 2 and 6, respectively (5). These positions are highly conserved in the secretin-like receptor family. Position 223 in the PTH receptor is one helical turn above the conserved arginine, which, based on computer modeling, is proposed to correspond to the conserved arginine in the DRY sequence of the rhodopsin-like receptors (6). The arginine, which substitutes for His-223, may compete with the conserved arginine of the PTH receptor for a polar pocket in the receptor and may shift the conserved arginine out of this pocket and toward the cytosol and the G proteins. The switching between different side-chain conformations of the conserved arginine has been proposed to be the mechanism by which the rhodopsin-like receptors activate G proteins (7). That lysine is the only other substitution for His-223 that produces constitutive activity in the human PTH receptor (8) lends support to this explanation.
Point mutations at the cytoplasmic end of transmembrane helix 6 are known to induce ligand-independent activity in several rhodopsin-like receptors (9–11). In the α1 adrenergic receptor, mutation of position 293 by any other amino acid induces constitutive activity (12). Thus, it generally is believed that this region of the receptor plays a critical role in constraining the receptor in an inactive conformation. In the human PTH receptor, numerous mutations of the conserved Pro-410 induce constitutive activity (8). Therefore, this area may be similarly important for constraining the human PTH receptor in the inactive conformation.
Introduction of the His-223–Arg and Thr-410–Pro mutations at equivalent positions in the other secretin-like receptors results in a constitutively active phenotype for only the glucagon and vasoactive intestinal polypeptide receptors (8, 13). The comparable mutant versions of the receptors for glucagon-like peptide I, gastric inhibitory peptide, calcitonin, secretin, growth hormone-releasing hormone, as well as for CRF fail to show ligand-independent activity (8). Even more surprisingly, the Thr-410–Pro point mutant in the rat PTH receptor also fails to produce ligand-independent activity (8).
Here, we present a strategy to obtain constitutively activated receptors based on the thrombin receptor system. Structure–activity relationship studies on CRF (14–16), the proposed endogenous ligand for R1, imply that the peptide determinant involved in activation is localized in the amino-terminal portion of CRF. For example, amino-terminally truncated analogs such as the CRF(12–41) peptide bind to the receptor without activating it, thereby acting as competitive antagonists (16). Astressin (16), a high-affinity peptide antagonist, developed by using CRF(12–41) as a template, binds to the N-terminal domain of R1 with high affinity (17), supporting a model in which the carboxyl-terminal portion of CRF binds to the N-terminal domain of the receptor. This binding event then may position the amino-terminal portion of CRF in proximity to other regions of the receptor responsible for activation. To obtain constitutive activation of CRF receptors, we designed a chimera in which we replaced the N-terminal domain of R1 with the activating portion of CRF in a manner reminiscent of the activated thrombin receptor (18). Tethering the endogenous ligand to the receptor may offer a general strategy for obtaining constitutively activated receptors and avoiding the complex phenotypes of receptors with point mutations.
Experimental Protocol
The cDNA encoding the human R1 (2) was subcloned into the pCI vector (Promega), positioning an optimized Kozak sequence upstream of the initiation codon (19). Silent MluI and BspEI restriction sites were created at positions 285 and 450, respectively, and the endogenous BspEI site at position 1121 was removed. All constructs were made by a modified overlap-extension PCR (20) by using flanking primers with a 5′ add-on sequence (21). All PCR products were cloned as EcoRI/BspEI fragments into the pCI vector containing R1 except for the CRF(1–16)/R1 chimera, which was cloned as EcoRI/MluI. For R1ΔN, residues 1–111 were replaced by the hemagglutinin (HA)-signal peptide (22) followed by the FLAG epitope. The chimeras CRF(1–16)/R1ΔN and CRF(17–41)/R1ΔN have residues 1–111 replaced by the HA-signal peptide (22) followed by the indicated portions of rat/human CRF (23). Chimera CRF(1–16)/R1 has the c-myc epitope and a glycine residue separating it from CRF(1–16) (EQKLISEEDLGSEEPPISLDLTFHLLR) inserted between residues 28 and 29 of R1. All chimeras were identified by restriction enzyme digestion and verified by automatic sequencing. For the functional assay, 1 day before transfection, 2 × 106 COS M6 cells were plated in a 10-cm dish (Falcon) and transfected by using the DEAE–dextran method (17). Approximately 20 h later, the cells were trypsinized (Sigma) and seeded into 48-well (Costar) plates at a density of 2 × 104 cells per well. The cells were assayed approximately 40–48 h after transfection. Cells were washed twice with DMEM containing 0.1% FCS and incubated for 2 h in 200 μl of this medium in a 37°C humidified atmosphere containing 5% CO2. Cells subsequently were preincubated for 15 min with antagonist before 3-isobutyl-1-methylxanthine was added (final concentration, 0.4 mM). Ten minutes later, agonists were added, and the assay was stopped 15 min after that by aspiration of medium and addition of 0.5 ml of ice-cold ethanol containing 0.1 M HCl. The cAMP content was determined by RIA (Biomedical Technologies).
Results and Discussion
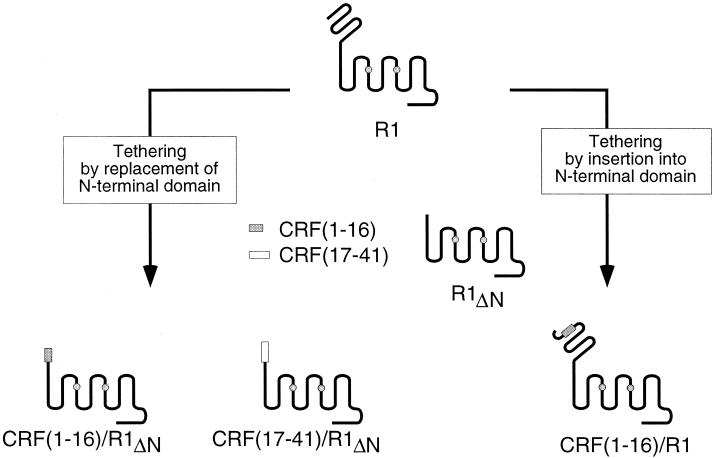
We designed chimeras between R1 and the amino-terminal residues (1–16) or the carboxyl-terminal residues (17–41) of CRF and positioned the peptide portion in place of the N-terminal domain of the receptor (Fig. 1). Because these chimeras lack the receptor's signal peptide (24), they are expressed by using the HA-signal peptide derived from influenza HA at their amino termini (22). The introduction of the HA-signal peptide ensures proper membrane targeting of the expressed constructs. Furthermore, the HA-signal peptide is cleaved by the expressing cells (22), leaving the peptide tethered to the body of the receptor (i.e., the transmembrane region including loops) with a free amino terminus.
Figure 1.
Schematic outline of R1 and related constructs. Based on alignment and structural modeling of the transmembrane region of secretin-like receptors (6), the transmembrane segment 1 starts at position 124 in the R1. Constructs in the lower left of the figure all have the N-terminal domain corresponding to residues 1–111 of the receptor replaced with the indicated portion of CRF and are expressed with the HA-signal peptide placed upstream of the peptide portion. The construct in the lower right has residues 1–16 of CRF inserted into the N-terminal domain of R1 between residues 28 and 29. The shaded circles in transmembrane segments 3 and 5 indicate residues important for binding of an R1-specific, nonpeptide antagonist (27).
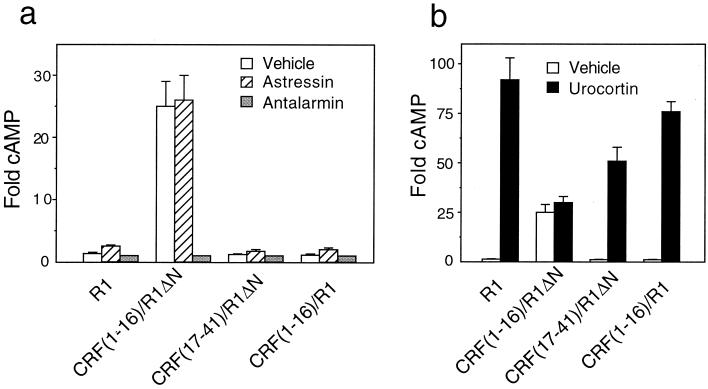
Transient transfection of COS-M6 cells with the peptide/receptor chimera in which the first 16 residues of CRF replace the receptor's N-terminal domain [CRF(1–16)/R1ΔN] results in high levels of receptor activity as measured by an increase in intracellular cAMP (Fig. 2a). The activity of this chimera is 25-fold (Fig. 2a and Table 1) higher than the activity observed in the presence of 10 μM antalarmin (25) (an R1-specific, nonpeptide antagonist) and is blocked in a dose-dependent manner by it (Fig. 3a). By contrast, the constitutive activation is not inhibited by 10 μM astressin (16), a peptide antagonist (Fig. 2a and Table 1). The body of this chimera is not responsible for the constitutive activation because the N-terminally truncated receptor, R1ΔN (Fig. 1), does not display constitutive activity (data not shown). This observation indicates that the N-terminal domain of R1 does not constrain the body of the receptor in an inactive conformation as has been proposed for the thyrotropin receptor (26). Urocortin, another mammalian member of the CRF-like family of peptides (23), activates R1ΔN with an EC50 ≅ 0.1 μM (data not shown), which is a 500-fold reduction in potency compared with that of the native receptor. This reduction in potency probably reflects the distinct, yet overlapping, receptor regions that contribute to the urocortin–receptor interaction. The N-terminal receptor domain is involved in relatively high-affinity binding (17), whereas the body of the receptor appears to display a lower-affinity interaction. Urocortin treatment of cells expressing the chimera CRF(1–16)/R1ΔN results in a minor, not statistically significant, further stimulation of cAMP production (Figs. 2b and 3a and Table 1).
Figure 2.
Constitutive activation and stimulation of R1 and peptide/R1 chimeras. (a) Level of cAMP in the absence (open bars) and presence of 10 μM astressin (hatched bars) or 10 μM antalarmin (shaded bars). (b) Level of cAMP in the absence (open bars) and presence of 10 μM urocortin (solid bars). The cAMP level is normalized in each experiment to that observed in the presence of 10 μM antalarmin. Data are presented as mean ± SEM from 4–8 independent experiments, each performed in triplicate. The absolute level of cAMP in the presence of 10 μM antalarmin is similar for all constructs.
Table 1.
Activities and potencies of peptide ligands on R1 and peptide/R1 chimeras
| Vehicle, -fold | Astressin, -fold | Urocortin
|
||
|---|---|---|---|---|
| -fold | EC60, nM | |||
| RI (wild type) | 1.3 ± 0.2 | 2.5 ± 0.2 | 92 ± 11 | 0.21 ± 0.04 |
| CRF(1–16)/R1ΔN | 25 ± 4 | 26 ± 4 | 30 ± 3 | — |
| CRF(17–41)/R1ΔN | 1.2 ± 0.1 | 1.7 ± 0.3 | 51 ± 7 | 110 ± 20 |
| CRF(1–16)/R1 | 1.1 ± 0.2 | 2.0 ± 0.3 | 76 ± 5 | 0.20 ± 0.1 |
Data are presented as mean ± SEM from 4–15 independent experiments, each performed in triplicate. The cAMP level is normalized in each experiment to that observed in the presence of 10 μM antalarmin. Because of the low level of urocortin stimulation of CRF(1–16)/R1ΔN, the EC50 could not be determined.
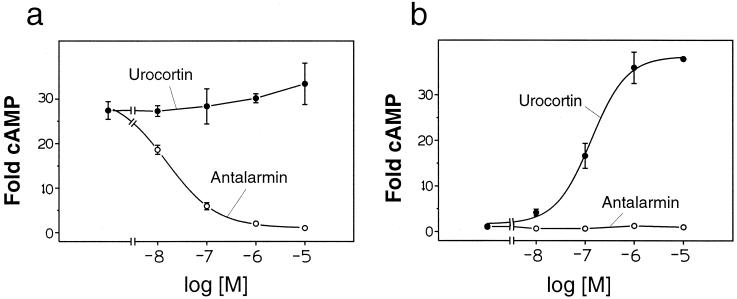
Figure 3.
Dose response with urocortin and antalarmin on CRF(1–16)/R1ΔN and CRF(1–16[L8A])/R1ΔN. (a) CRF(1–16)/R1ΔN. (b) CRF(1–16[L8A])/R1ΔN. Data are presented as mean ± SD from triplicate determinations and are representative of three independent assays performed in parallel. For CRF(1–16)/R1ΔN, the IC50 for inhibition of constitutive activation by antalarmin is 14 ± 1 nM. For CRF(1–16[L8A])/R1ΔN, the EC50 for stimulation by urocortin is 140 ± 20 nM.
Using an ELISA with antibodies raised against CRF(1–21) as well as against CRF(1–41), high levels of peptide epitope were detected on cells transfected with CRF(1–16)/R1ΔN (data not shown). Therefore, the low level of stimulation of CRF(1–16)/R1ΔN by urocortin is not caused by an absence of membrane-localized constructs.
Mutational analysis of R1 has revealed two residues, namely, His-199 and Met-276, in the transmembrane segments 3 and 5, respectively, which affect the binding of NBI 27914, another nonpeptide antagonist (27) that is functionally similar to antalarmin. It is likely that similar receptor segments are involved in the binding of antalarmin. That antalarmin is able to inhibit the constitutive activation of CRF(1–16)/R1ΔN is consistent with this assumption and supports the view derived from numerous studies on rhodopsin-like receptors (28) that nonpeptide antagonists bind in the transmembrane domains of the receptors.
The chimera in which the carboxyl-terminal (17–41) residues of CRF replace the N-terminal domain of R1, [CRF(17–41)/R1ΔN], is deficient in constitutive activation (Fig. 2a and Table 1), yet produces an ≈50-fold stimulation of intracellular cAMP in response to 10 μM urocortin (Fig. 2b and Table 1). These results are consistent with the amino-terminal portion of the CRF peptide being required for activation.
The significance of the proximity between CRF(1–16) and the body of the receptor was examined by inserting CRF(1–16) into the N-terminal domain of R1 between residues 28 and 29 within the intact receptor (Fig. 1). This chimera, CRF(1–16)/R1, does not display constitutive activation (Fig. 2a and Table 1), but shows a large response to urocortin, with similar potency (EC50 ∼ 0.2 nM) to that of R1 (Fig. 2b and Table 1). An anti-myc antibody as well as the aforementioned CRF antibodies detect high levels of chimera expression on cells transfected with CRF(1–16)/R1 (data not shown), indicating that the 16 residues of CRF are present in CRF(1–16)/R1. Therefore, the lack of constitutive activation of this chimera is not caused by loss of the peptide during expression. The lack of constitutive activation of this construct possibly may be a result of the ability of the N-terminal domain of the receptor to function as a spacer and, thus, diminish the proximity between the active portion of CRF and the body of the receptor.
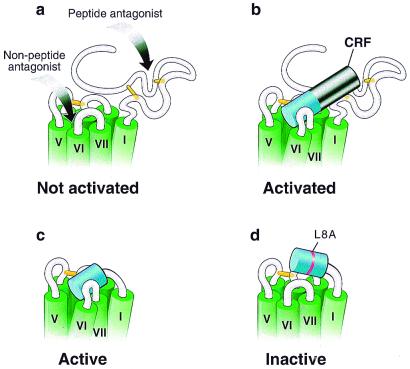
An analog of CRF in which Leu-8 is replaced by Ala retains full intrinsic activity but has a 300-fold decrease in the relative potency compared with that of CRF as measured by in vitro corticotropin release from cultured rat anterior pituitary cells (14). Similar results for cAMP production were found for R1 (data not shown). The corresponding modification in the peptide portion of the chimera, CRF(1–16[L8A])/R1ΔN, abolishes the constitutive activation (Fig. 3b). This chimera, however, can be stimulated by urocortin (Fig. 3b). Using an ELISA with antibodies raised against CRF(1–21) and CRF(1–41), the level of expression of CRF(1–16[L8A])/R1ΔN was found to be similar to that of CRF(1–16)/R1ΔN (data not shown). These results support the hypothesis that the constitutive activation of the CRF(1–16)/R1ΔN chimera is produced by specific interactions between the tethered amino-terminal part of CRF and the body of the receptor and that the specificity is reminiscent of that between CRF and native R1 (Fig. 4).
Figure 4.
Propinquity model for R1 and CRF(1–16)/R1ΔN activation. Schematic representations of the upper portion of R1. The transmembrane segments are indicated in green and are based on models of GPCRs (6, 44). The putative disulfide bridges important for binding are indicated in yellow (45). (a) R1. The putative regions involved in binding of antagonists are indicated by arrows. The peptide antagonist is presumed to bind to the N-terminal domain (17), whereas the nonpeptide antagonists bind in the transmembrane region (27). The receptor in the absence of agonist is represented in an inactivated state. (b) R1 activated by CRF. The first 16 residues of CRF are shown in blue, whereas the remaining (17–41) residues are shown in black. The receptor is represented in an activated state. (c) Constitutively activated chimera. The figure shows the chimera in which the first 16 residues of CRF are tethered to the receptor in place of its N-terminal domain. The tethered peptide stabilizes the chimera in an active state. (d) Inactive chimera. The figure shows the chimera in which the CRF peptide portion contains the mutation L8A and is tethered to the receptor in place of its N-terminal domain. This point mutation prevents the tethered peptide from stabilizing the chimera in an active state.
Recently, GPCR genes have been cloned that display homology to the transmembrane region of secretin-like receptors (29–33). However, these genes encode receptors with N termini of up to 950 residues with distinct structures compared with that of secretin-like receptors. The present study sheds light on how this subclass of secretin-like receptors is activated. The main roles of the various N termini presumably are to provide sufficient binding energy and specificity for presenting only a part of the ligand to the body of the receptor in the proper proximity for activation. In support of this model of activation, chimeric peptides consisting of the amino-terminal part of PTH and the carboxyl-terminal part of calcitonin were able to fully activate a chimeric receptor in which the N-terminal domain of the calcitonin receptor replaced that of the PTH receptor (34). Full stimulation also was achieved for the reciprocal peptide and receptor chimera (34). Tethering a ligand to the body of a receptor may be a general method for obtaining ligand-dependent constitutive activation of receptors, as well as being useful for delineating regions of the ligand involved in receptor activation.
A picture of GPCRs with multiple, yet distinct, activated states currently is emerging for secretin-like receptors (35) as well as rhodopsin-like receptors (36–40). These discrete, activated states have been shown to be stabilized by specific agonists (35–37) or induced by point mutations in the receptor (38, 40). The present approach of tethering a ligand to the receptor presumably will restrict the chimera to the same signaling pathways as the native receptor in responding to the ligand, thus making these chimeras appropriate for analysis of mechanisms underlying receptor desensitization and regulation. These constitutively activated chimeras are also suitable for transgenic mice by using tissue-specific conditional expression (41) for the study of the pathophysiology associated with a specific receptor system. The constitutively activated chimera CRF(1–16)/R1ΔN is a new type of RASSL (receptor activated solely by a synthetic ligand) (42). This chimera does not respond to endogenous levels of agonist like a RASSL. However, in contrast to a RASSL, which is activated only by administering a synthetic agonist, the CRF(1–16)/R1ΔN chimera will be constitutively activated upon induced expression. The activity of this chimera can be pharmacologically inhibited by an orally active, R1-specific, nonpeptide antagonist such as antalarmin (25). This type of engineered chimera may be designated as a RISSL (receptor inhibited solely by a synthetic ligand).
Mutations causing constitutive or ligand-independent activity in GPCRs generally are believed to destabilize the receptor (43), making the constitutively active receptor mutants difficult to handle for structural purposes. However, agonists as well as antagonists protect against receptor denaturation (43). Tethering the active portion of CRF in close proximity of the body of R1 creates a chimera in which the peptide part presumably binds and stabilizes the body of the receptor in an active conformation. Thus, the present approach may be a novel way of obtaining biochemically more stable, constitutively activated receptor proteins suitable for structural analysis.
In summary, the present results show that the body of the CRF type 1 receptor can be activated by a thrombin receptor-like mechanism. Tethering the amino-terminal part of CRF to the receptor in place of its N-terminal domain results in a constitutively activated chimera. It is likely that capture of an agonist by the N-terminal domain of secretin-like receptors, in this case the CRF type 1 receptor, spatially constrains the activating residues of the agonist relative to the body of the receptor in the proper proximity for receptor activation. This spatial constraint may be mimicked by tethering the activating residues of the agonist to the body of the receptor as presented here for the CRF(1–16)/R1ΔN chimera.
Acknowledgments
We thank Dr. T. Schwartz for introducing us to the concept of tethered-peptide receptor chimeras. We recognize R. Kaiser, K. Kunitake, J. Norton, and J. Vaughan for technical assistance and D. Dalton, S. Guerra, J. Simon, and Dr. C. Elling for manuscript and figure preparation. In addition, we thank Drs. G. Gaudriault, P. Gray, S. Koerber, C. Perrin, and S. Choe for fruitful discussions and Drs. J. Noel and A. Hsueh for critical reading of the manuscript. We acknowledge the generous gifts of antalarmin and peptides from Drs. G. Chrousos and J. Rivier, respectively. This work was supported by National Institutes of Health Program Project Grant DK26741 and the Foundation for Research. W.W.V. is a Foundation for Research Senior Investigator.
Abbreviations
- CRF
corticotropin-releasing factor
- GPCR
G protein-coupled receptor
- R1
human CRF type 1 receptor
- PTH
parathyroid hormone
- HA
hemagglutinin
References
- 1.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Lewis K A, Perrin M H, Vale W W. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laburthe M, Couvineau A, Gaudin P, Maoret J J, Rouyer-Fessard C, Nicole P. Ann NY Acad Sci. 1996;805:94–109. doi: 10.1111/j.1749-6632.1996.tb17476.x. , 110–111. [DOI] [PubMed] [Google Scholar]
- 4.Schipani E, Kruse K, Juppner H. Science. 1995;268:98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- 5.Schipani E, Langman C B, Parfitt A M, Jensen G S, Kikuchi S, Kooh S W, Cole W G, Juppner H. N Engl J Med. 1996;335:708–714. doi: 10.1056/NEJM199609053351004. [DOI] [PubMed] [Google Scholar]
- 6.Frimurer T M, Bywater R P. Proteins. 1999;35:375–386. [PubMed] [Google Scholar]
- 7.Oliveira L, Paiva A C, Sander C, Vriend G. Trends Pharmacol Sci. 1994;15:170–172. doi: 10.1016/0165-6147(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 8.Schipani E, Jensen G S, Pincus J, Nissenson R A, Gardella T J, Juppner H. Mol Endocrinol. 1997;11:851–858. doi: 10.1210/mend.11.7.9934. [DOI] [PubMed] [Google Scholar]
- 9.Cotecchia S, Exum S, Caron M G, Lefkowitz R J. Proc Natl Acad Sci USA. 1990;87:2896–2900. doi: 10.1073/pnas.87.8.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samama P, Cotecchia S, Costa T, Lefkowitz R J. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- 11.Egan C T, Herrick-Davis K, Teitler M. J Pharmacol Exp Ther. 1998;286:85–90. [PubMed] [Google Scholar]
- 12.Kjelsberg M A, Cotecchia S, Ostrowski J, Caron M G, Lefkowitz R J. J Biol Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- 13.Hjorth S A, Orskov C, Schwartz T W. Mol Endocrinol. 1998;12:78–86. doi: 10.1210/mend.12.1.0045. [DOI] [PubMed] [Google Scholar]
- 14.Kornreich W D, Galyean R, Hernandez J F, Craig A G, Donaldson C J, Yamamoto G, Rivier C, Vale W, Rivier J. J Med Chem. 1992;35:1870–1876. doi: 10.1021/jm00088a024. [DOI] [PubMed] [Google Scholar]
- 15.Rivier J, Rivier C, Vale W. Science. 1984;224:889–891. doi: 10.1126/science.6326264. [DOI] [PubMed] [Google Scholar]
- 16.Gulyas J, Rivier C, Perrin M, Koerber S C, Sutton S, Corrigan A, Lahrichi S L, Craig A G, Vale W, Rivier J. Proc Natl Acad Sci USA. 1995;92:10575–10579. doi: 10.1073/pnas.92.23.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrin M H, Sutton S, Bain D L, Berggren W T, Vale W W. Endocrinology. 1998;139:566–570. doi: 10.1210/endo.139.2.5757. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Ishii M, Wang L, Ishii K, Coughlin S R. J Biol Chem. 1994;269:16041–16045. [PubMed] [Google Scholar]
- 19.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 21.Stappert J, Wirsching J, Kemler R. Nucleic Acids Res. 1992;20:624. doi: 10.1093/nar/20.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan X M, Kobilka T S, Kobilka B K. J Biol Chem. 1992;267:21995–21998. [PubMed] [Google Scholar]
- 23.Vaughan J, Donaldson C, Bittencourt J, Perrin M H, Lewis K, Sutton S, Chan R, Turnbull A V, Lovejoy D, Rivier C. Nature (London) 1995;378:287–292. doi: 10.1038/378287a0. [DOI] [PubMed] [Google Scholar]
- 24.Liaw C W, Lovenberg T W, Barry G, Oltersdorf T, Grigoriadis D E, de Souza E B. Endocrinology. 1996;137:72–77. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- 25.Webster E L, Lewis D B, Torpy D J, Zachman E K, Rice K C, Chrousos G P. Endocrinology. 1996;137:5747–5750. doi: 10.1210/endo.137.12.8940412. [DOI] [PubMed] [Google Scholar]
- 26.Duprez L, Parma J, Costagliola S, Hermans J, Van Sande J, Dumont J E, Vassart G. FEBS Lett. 1997;409:469–474. doi: 10.1016/s0014-5793(97)00532-2. [DOI] [PubMed] [Google Scholar]
- 27.Liaw C W, Grigoriadis D E, Lorang M T, de Souza E B, Maki R A. Mol Endocrinol. 1997;11:2048–2053. doi: 10.1210/mend.11.13.0034. [DOI] [PubMed] [Google Scholar]
- 28.Holst B, Zoffmann S, Elling C E, Hjorth S A, Schwartz T W. Mol Pharmacol. 1998;53:166–175. doi: 10.1124/mol.53.1.166. [DOI] [PubMed] [Google Scholar]
- 29.Hamann J, Eichler W, Hamann D, Kerstens H M, Poddighe P J, Hoovers J M, Hartmann E, Strauss M, van Lier R A. J Immunol. 1995;155:1942–1950. [PubMed] [Google Scholar]
- 30.Baud V, Chissoe S L, Viegas-Pequignot E, Diriong S, N′Guyen V C, Roe B A, Lipinski M. Genomics. 1995;26:334–344. doi: 10.1016/0888-7543(95)80218-b. [DOI] [PubMed] [Google Scholar]
- 31.Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, Yoshida S, Ono M, Kuwano M, Nakamura Y, et al. Oncogene. 1997;15:2145–2150. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- 32.Osterhoff C, Ivell R, Kirchhoff C. DNA Cell Biol. 1997;16:379–389. doi: 10.1089/dna.1997.16.379. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Parker R M, Darby K, Eyre H J, Copeland N G, Crawford J, Gilbert D J, Sutherland G R, Jenkins N A, Herzog H. Genomics. 1999;55:296–305. doi: 10.1006/geno.1998.5644. [DOI] [PubMed] [Google Scholar]
- 34.Bergwitz C, Gardella T J, Flannery M R, Potts J T J, Kronenberg H M, Goldring S R, Juppner H. J Biol Chem. 1996;271:26469–26472. doi: 10.1074/jbc.271.43.26469. [DOI] [PubMed] [Google Scholar]
- 35.Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg P H, Journot L. Nature (London) 1993;365:170–175. doi: 10.1038/365170a0. [DOI] [PubMed] [Google Scholar]
- 36.Robb S, Cheek T R, Hannan F L, Hall L M, Midgley J M, Evans P D. EMBO J. 1994;13:1325–1330. doi: 10.1002/j.1460-2075.1994.tb06385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keith D E, Murray S R, Zaki P A, Chu P C, Lissin D V, Kang L, Evans C J, von Zastrow M. J Biol Chem. 1996;271:19021–19024. doi: 10.1074/jbc.271.32.19021. [DOI] [PubMed] [Google Scholar]
- 38.Perez D M, Hwa J, Gaivin R, Mathur M, Brown F, Graham R M. Mol Pharmacol. 1996;49:112–122. [PubMed] [Google Scholar]
- 39.Seifert R, Gether U, Wenzel-Seifert K, Kobilka B K. Mol Pharmacol. 1999;56:348–358. doi: 10.1124/mol.56.2.348. [DOI] [PubMed] [Google Scholar]
- 40.Zuscik M J, Porter J E, Gaivin R, Perez D M. J Biol Chem. 1998;273:3401–3407. doi: 10.1074/jbc.273.6.3401. [DOI] [PubMed] [Google Scholar]
- 41.Redfern C H, Coward P, Degtyarev M Y, Lee E K, Kwa A T, Hennighausen L, Bujard H, Fishman G I, Conklin B R. Nat Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- 42.Coward P, Wada H G, Falk M S, Chan S D, Meng F, Akil H, Conklin B R. Proc Natl Acad Sci USA. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gether U, Ballesteros J A, Seifert R, Sanders-Bush E, Weinstein H, Kobilka B K. J Biol Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 44.Elling C E, Raffetseder U, Nielsen S M, Schwartz T W. Biochemistry. 2000;39:667–675. doi: 10.1021/bi991777b. [DOI] [PubMed] [Google Scholar]
- 45.Qi L J, Leung A T, Xiong Y, Marx K A, Abou-Samra A B. Biochemistry. 1997;36:12442–12448. doi: 10.1021/bi970997r. [DOI] [PubMed] [Google Scholar]