Abstract
BACKGROUND
The molecular cause of inflammatory bowel disease is largely unknown.
METHODS
We performed genetic-linkage analysis and candidate-gene sequencing on samples from two unrelated consanguineous families with children who were affected by early-onset inflammatory bowel disease. We screened six additional patients with early-onset colitis for mutations in two candidate genes and carried out functional assays in patients’ peripheral-blood mononuclear cells. We performed an allogeneic hematopoietic stem-cell transplantation in one patient.
RESULTS
In four of nine patients with early-onset colitis, we identified three distinct homozygous mutations in genes IL10RA and IL10RB, encoding the IL10R1 and IL10R2 proteins, respectively, which form a heterotetramer to make up the interleukin-10 receptor. The mutations abrogate interleukin-10–induced signaling, as shown by deficient STAT3 (signal transducer and activator of transcription 3) phosphorylation on stimulation with interleukin-10. Consistent with this observation was the increased secretion of tumor necrosis factor α and other proinflammatory cytokines from peripheral-blood mononuclear cells from patients who were deficient in IL10R subunit proteins, suggesting that interleukin-10–dependent “negative feedback” regulation is disrupted in these cells. The allogeneic stem-cell transplantation performed in one patient was successful.
CONCLUSIONS
Mutations in genes encoding the IL10R subunit proteins were found in patients with early-onset enterocolitis, involving hyperinflammatory immune responses in the intestine. Allogeneic stem-cell transplantation resulted in disease remission in one patient.
Inflammatory bowel disease is a heterogeneous group of disorders, classified as Crohn’s disease, ulcerative colitis, and indeterminate colitis.1,2 In most patients, these disorders are manifested in adolescence or adulthood; however, they may present in infancy and may be inherited as an autosomal recessive trait.3–6
The genetic causes of inflammatory bowel disease are only partly understood. Studies in transgenic murine models7 and genomewide genetic-linkage and association studies have provided insights into the genetic complexity underlying these inflammatory conditions.8 Investigators using these approaches have implicated several genes in the pathogenesis of inflammatory bowel disease; the identity of these genes suggests that disruption of the innate and adaptive arms of the immune system,9–11 the process of autophagy,12,13 epithelial barrier function,14 and activation of the endoplasmic reticulum stress response15 may cause susceptibility. However, the functional relevance of most of these susceptibility genes is unclear. An alternative approach to identifying disease-causing genes is to study families in which inflammatory bowel disease is inherited as a potentially monogenic trait and to identify the relevant gene by positional cloning.
Interleukin-10 restricts excessive immune responses.16 Initially, it was described as a soluble factor that is released by type 2 helper T cells and that inhibits the secretion of type 1 helper T cytokines, such as interleukin-2 and interferon-γ.17 Interleukin-10 is secreted by a wide variety of cells and has pleiotropic effects on T cells, B cells, myeloid cells, and other cell types.16 In particular, interleukin-10 limits the secretion of proinflammatory cytokines, such as tumor necrosis factor α (TNF-α) and interleukin-12.18 The receptor for interleukin-10 consists of two alpha molecules (IL10R1) and two beta molecules (IL10R2).19 Although IL10R1 is specific to the interleukin-10 receptor, IL10R2 is a subunit of the receptors for several additional cytokines (e.g., interleukin-22 and interleukin-26).20 The assembly of this heterotetrameric interleukin-10 receptor results in the activation of the receptor-associated Janus tyrosine kinases, JAK1 and Tyk2, resulting in the phosphorylation of STAT3 (signal transducer and activator of transcription 3) and the induction of STAT3-dependent genes.21–25
Mice that are deficient in either interleukin-10 or interleukin-10 receptor 2 have been shown to have severe enterocolitis,26–28 a finding that underscores the pivotal role of interleukin-10 in the mediation of signaling that controls inflammation in the gut. These studies in mice have also proffered genes that encode proteins in the interleukin-10–signaling pathway as candidates for association with inflammatory bowel disease.
METHODS
PATIENTS
In Family A, the index patient (Patient II-3), who was of Turkish origin and born of parents who were first-degree cousins, presented at the age of 3 months with proctitis and abscesses in the peri-anal region, which required multiple surgical interventions. Protective colostomy was performed because of impaired wound healing. Multiple enterocutaneous fistulas originating from the small intestine required further partial bowel resections and ileostomy. In addition, he had early-onset cutaneous folliculitis. Recurrent infections such as otitis media, bronchitis, and two episodes of purulent gonarthritis were potentially linked to immunosuppressive therapy. He had normal numbers and functioning of T cells and B cells. Increased serum levels of IgG (21.8 g per liter; normal range, 5.2 to 12.9), IgA (7.66 g per liter; normal range, 0.47 to 2.1), and IgM (2.98 g per liter; normal range, 0.47 to 1.90) were interpreted as being associated with chronic ileocolitis. The neutrophil function was normal with respect to NADPH oxidase activity (data not shown). Histologic analysis of ileal- and colonic-biopsy samples that were obtained from the sites of inflammation showed multifocal ulcerations and agglomerations of polymorphic infiltrates (Fig. 1A and 1B in the Supplementary Appendix, available with the full text of this article at NEJM.org). The patient was treated with a wide spectrum of anti-inflammatory agents, including corticosteroids, methotrexate, thalidomide, and anti–TNF-α monoclonal antibodies. None of these therapies induced remission or long-term improvement.
In his affected sister, Patient II-4, proctitis and rectovaginal fistula developed in the first year of life. She had folliculitis, pneumonia, and one episode of renal abscess caused by Escherichia coli. She underwent several surgical interventions, including colonic resection and colostomies. The results of the immune workup were normal. The clinical course of the colitis was slightly milder in Patient II-4 than it was in her affected brother.
In Family B, the index patient (Patient II-5), who was of Lebanese descent, presented in her first year of life with severe enterocolitis (Fig. 1A) associated with enteric fistulas, perianal abscesses, and chronic folliculitis, consistent with a diagnosis of Crohn’s disease (Fig. 1B). She required multiple surgical interventions, including colectomy and ileostomy (Fig. 1C). The histopathological analysis of the specimen from the colon obtained during resection revealed circumscribed ulcerations of the mucosa, from which pear-shaped intramural abscesses extended into the submucosa and muscularis propria (Fig. 1D, 1E, and 1F). Inflammatory infiltrates were also seen in the small intestine, albeit to a lesser extent. Immunologic analyses showed normal numbers and function of T cells, B cells, and natural killer cells (data not shown). Serum immunoglobulin levels were normal or increased. Neutrophil granulocytes showed regular oxidative burst capacity (data not shown). Despite multiple therapies, including treatment with corticosteroids and azathioprine and total colectomy, a sustained, long-term remission could not be achieved.
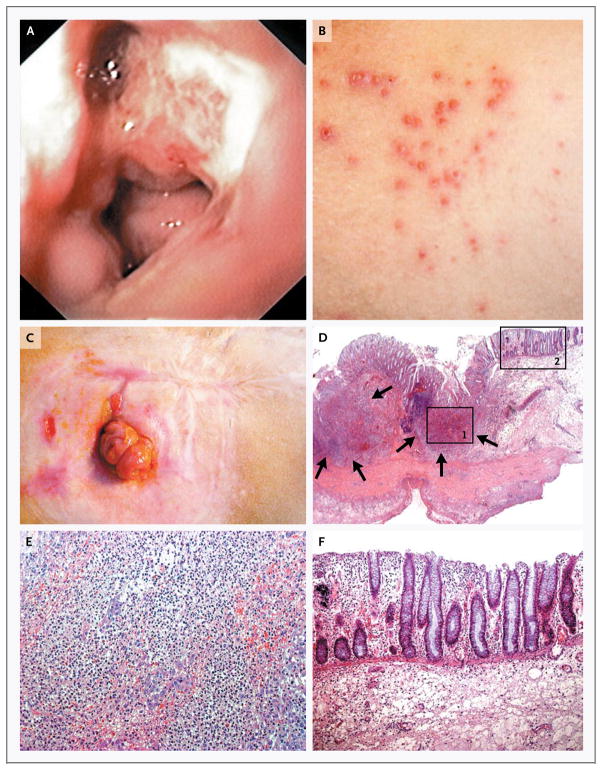
Figure 1. Clinical Phenotype in a Patient with a Deficiency in the Interleukin-10 Receptor.
The index patient in Family B, Patient II-5, had evidence of erosive lesions on colonoscopy, shown in Panel A, and of skin folliculitis, shown in Panel B. Panel C shows the patient’s status after ileostomy. Panel D shows the histopathological findings in a specimen from the colon obtained during resection in the patient, revealing circumscribed ulcerations of the mucosa from which pear-shaped intramural abscesses (arrows) extend into the submucosa and muscularis propria. Panel E shows a higher magnification of inset 1 in Panel D, revealing intramural microabscesses. Panel F shows a higher magnification of inset 2 in Panel D, revealing neighboring mucosa with scarce, superficial lymphoplasmocytic infiltrates without glandular distortion. There is minimal intramural lymphocytic hyperplasia (as seen in the dark area just to the left of inset 1 in Panel D) and no thickening of the intestinal wall as a result of fibrosis or granulomas.
We also performed studies in six patients with an onset of severe colitis during the first year of life.
We obtained written informed consent from adult patients and from the parents of children who participated in the study. In addition, the affected children provided their assent. The study was approved by the institutional review board at each study center (see the Supplementary Appendix).
GENETIC ANALYSES AND ASSAYS OF MOLECULAR FUNCTION
We analyzed the genotypes of each family to look for markers or intervals that showed perfect segregation with the disease phenotype. For each pedigree, we defined “perfect segregation” as meaning that the affected child or children in each family had the same homozygous genotype or haplotype. In addition, if both parents were heterozygous, then the unaffected children each needed to have at least one genotype or one haplotype that differed from those of the affected sibling or siblings.
Once we had identified perfectly segregating intervals, we chose genes therein to sequence for mutations on the basis of previous studies of the genes. We then performed gene-specific and protein-specific assays to determine the functional effects of the identified mutations on interleukin-10 signaling. The functional experiments included the transduction of cell lines and mutant cells with retroviral vectors encoding wild-type IL10RA and IL10RB complementary DNA, respectively, to show that the defect in interleukin-10 signaling was corrected by replacing the mutant interleukin receptor with a normal receptor. For more details on the genetic analyses and assays of molecular function, see the Supplementary Appendix.
RESULTS
GENETIC ANALYSES
We genotyped 253 microsatellite markers that spanned all 22 autosomes in members of Family A (Fig. 2A). Five markers showed perfect segregation with phenotype (corresponding to a logarithm of the odds [LOD] score of approximately 2.0) and were located on chromosomes 2, 7, 14, 19, and 21. Fine mapping with additional micro-satellites showed that three of these markers were within intervals that perfectly segregated with disease phenotype and that spanned multiple megabases. Table S1 in the Supplementary Appendix summarizes the extent of these intervals; the minimal interval was defined by the two most distant flanking, perfectly segregating markers, and the maximal interval was defined by extending, in each direction, to the first marker that did not perfectly segregate with disease phenotype. Genes that were located within these intervals and the genes we sequenced (one gene on chromosome 7 and four genes on chromosome 21) are listed in Table S2 in the Supplementary Appendix.
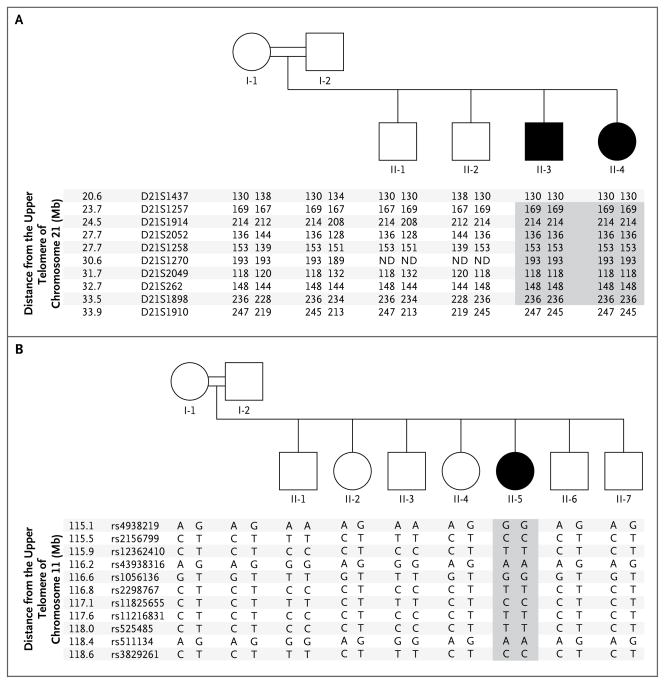
Figure 2. Haplotypes in Families A and B.
Panel A shows the pedigree of consanguineous Family A, which had two children with early-onset inflammatory bowel disease (Patients II-3 and II-4) and allelic segregation on chromosome 21q. Panel B shows the pedigree of a second consanguineous family, Family B, which had one affected child (Patient II-5) and allelic segregation on chromosome 11q. Squares indicate male family members, and circles female family members. Shading indicates the series of homozygous markers in the affected patients.
We focused on the chromosome 21 interval, since it contained more genes than the other two intervals combined and more functional candidate genes, including a family of interferon-related genes. We used all suitably positioned microsatellite markers on the Marshfield29 and deCode30 maps to reduce the uncertainty of the region most distal to the upper telomere to approximately 350 kb (Fig. 2A). The candidate gene, IL10RB, was the first gene (starting at 33.56 Mb) that was below and close to the perfect marker D21S1898. IL10RB is the only one of four consecutive immune-related genes (including the better- positioned IFNAR2) with an orthologous gene in the mouse that when “knocked out” results in colitis.28
We sequenced IL10RB and identified a homozygous point mutation in exon 4 in both affected siblings, resulting in a premature stop codon (c.G477A, p.Trp159X) (Fig. 2A in the Supplementary Appendix). Both healthy parents and the two healthy siblings carried a single heterozygous mutation (W159X) in IL10RB (data not shown). This mutation was absent in 180 unaffected German subjects of European descent, in 70 unaffected subjects of Turkish ancestry, and in 30 subjects of Iranian ancestry. We sequenced IL10RB in 90 patients with adult-onset inflammatory bowel disease: 45 patients with Crohn’s disease and 45 with ulcerative colitis. None of the unaffected subjects or the patients with adult-onset inflammatory bowel disease carried the mutation or any other sequence variations.
We used single-nucleotide–polymorphism arrays (Affymetrix) to map homozygous regions segregating with disease in Family B (in which the parents were second-degree cousins) (Fig. 2B). We identified eight regions larger than 1 Mb that were homozygous (with respect to haplotype) in the index patient and that “housed” in each unaffected family member a haplotype that was not present in the patient (Table S3 in the Supplementary Appendix). These regions yielded a peak LOD score of 2.5 (see the Supplementary Appendix). We observed that IL10RA was located in one of these regions (on chromosome 11q) and identified a homozygous missense mutation in exon 4 (c.G421A, p.Gly141Arg) in the index patient (Fig. 2B in the Supplementary Appendix). All the other members of Family B carried at least one IL10RA wild-type allele and did not have any inflammatory bowel disease. The mutation was absent in 100 unaffected Arabic subjects and 30 unaffected Iranian subjects.
We sequenced IL10RA and IL10RB in six additional patients who had an onset of severe colitis in the first year of life and identified a homozygous missense mutation in exon 3 of IL10RA (c.C325T, p.Thr84Ile) in one German patient of European ancestry (Fig. 2C in the Supplementary Appendix). This patient, who was 8 months of age, had received a diagnosis of severe pancolitis at the age of 3 months (Fig. 2D, 2E, and 2F in the Supplementary Appendix). This mutation was absent in 100 healthy white subjects of European ancestry. Furthermore, we observed no mutations in either IL10RA or IL10RB in 32 children with inflammatory bowel disease who had an onset of symptoms at more than 12 months of age.
EFFECTS OF MUTATIONS ON INTERLEUKIN-10–RECEPTOR PATHWAY FUNCTIONS
We studied the functional effects of the implicated mutations. Fluorescence-activated cell sorting (FACS) analysis disclosed no expression of IL10R2 in Epstein–Barr virus (EBV)–transformed B cells obtained from Patient II-3 in Family A, who carried the homozygous W159X mutation in IL10RB, in contrast to robust IL10R2 expression in cells from an unaffected subject (Fig. 3A).31 Figure 3B shows the localization of the W159X mutation in the IL10R2 protein.
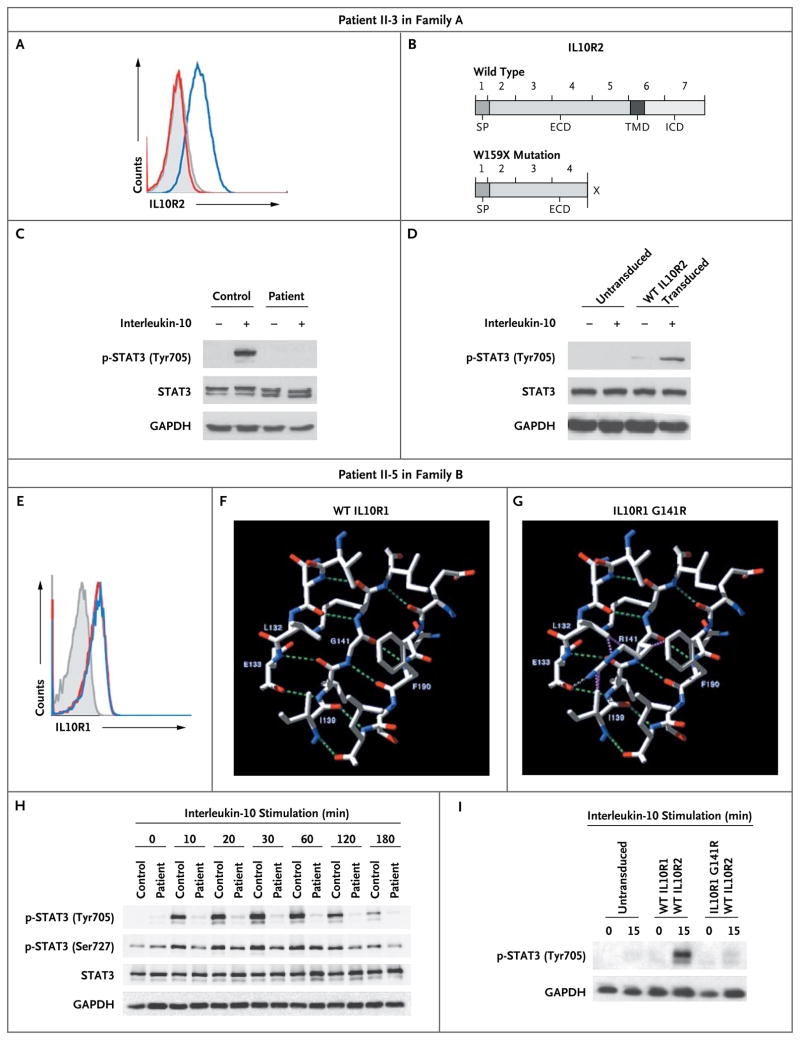
Figure 3. Structure and Functional Analysis of Mutations in the Interleukin-10 Receptor in Two Patients.
Panel A shows a representative analysis by fluorescence-activated cell sorting (FACS) that illustrates the lack of expression of IL10R2 protein in the index patient in Family A (Patient II-3), who carried an IL10RB W159X mutation. An isotype control sample is indicated by gray shading, a sample from a control subject by a blue line, and the sample from Patient II-3 by a red line. Panel B shows a schematic drawing of the IL10R2 protein, indicating known protein domains and localization of the nonsense W159X mutation, as compared with a wild-type sample. ECD denotes extracellular domain, ICD intracellular domain, SP signaling peptide, and TMD transmembrane domain. Panel C shows defective STAT3 (signal transducer and activator of transcription 3) phosphorylation (p-STAT3) at the residue tyrosine 705 on stimulation with interleukin-10 in peripheral-blood mononuclear cells (PBMCs) from Patient II-3, as compared with intact phosphorylation in a control sample. Panel D shows the reconstitution of STAT3 phosphorylation on transduction of cell lines from Patient II-3, with a lentiviral vector encoding wild-type (WT) IL10R2. Panel E shows normal expression levels of IL10R1 on FACS analysis in the index patient in Family B (Patient II-5), who carried an IL10RA G141R mutation. An isotype control sample is indicated by gray shading, a sample from a control subject by a blue line, and the sample from Patient II-5 by a red line. Panel F shows a structural model of wild-type IL10R1. Residues Leu132, Glu133, Ile139, Gly141, Lys142, and Phe190 are labeled. Also shown are nearby residues that participate with Gly141 or its neighbors in a beta-sheet conformation. The residues that are illustrated are those that are reported to have contact with Gly141 in wild-type IL10R1 (according to RankViaContact31) or with Arg141 in the G141R mutation and that lie within 6 Å. Panel G shows a structural model of the G141R mutation in IL10R1 (from Protein Data Bank structure 1Y6K, chain R). Putative hydrogen bonds are shown as green dashed lines in Panels F and G, and steric clashes are shown as purple dashed lines in Panel G. Panel H shows a Western blot analysis of STAT3 in PBMCs from Patient II-5 after stimulation for various periods with interleukin-10, as compared with a control sample. Panel I shows a Western blot analysis of phosphorylation of STAT3 at tyrosine 705 in interleukin-10–stimulated HeLa cells that were retrovirally transduced with wild-type IL10R2 along with either wild-type IL10R1 or IL10R1 with mutant G141R. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control for the experiments in Panels C, D, H, and I.
When interleukin-10 binds to its receptor, it signals predominantly through STAT3 to mediate its antiinflammatory effects.25,32 To examine the integrity of this pathway, we stimulated peripheral-blood mononuclear cells (PBMCs) or EBV-transformed B cells from Patient II-3 in Family A and those from an unaffected subject with interleukin-10 and assessed them for STAT3 phosphorylation at tyrosine 705, using Western blot analysis. Interleukin-10 induced STAT3 phosphorylation at tyrosine 705 in wild-type IL10R2 cells but not in cells with the W159X mutation (Fig. 3C, and Fig. 1C in the Supplementary Appendix). To reconstitute the function of IL10R, which has both IL10R1 and IL10R2 subunits, we analyzed EBV-transformed B cells from Patient II-3 in Family A that were transduced either with a lentiviral vector encoding IL10R2 along with green fluorescent protein (GFP) as a marker gene or with a control vector encoding GFP only (Fig. 1D in the Supplementary Appendix). In contrast to IL10R2-negative cells, IL10R2-reconstituted cells showed intact STAT3 phosphorylation at tyrosine 705 (Fig. 3D, and Fig. 1E in the Supplementary Appendix). We concluded that the W159X mutation abrogated the phosphorylation of STAT3 mediated by interleukin-10.
The mutations we observed in IL10RA were missense variants; analyses with the use of FACS and Western blot analysis indicated that the G141R mutation in the IL10R1 protein was expressed at levels similar to those in control cells (Fig. 3E, and Fig. 2G in the Supplementary Appendix). When interleukin-10 binds to the cell surface, it first interacts with IL10R1 and subsequently binds IL10R2. We used a structure of the intermediate interleukin-10–IL10R1 state (identifier 1Y6K in the Protein Data Bank) to model possible effects of missense changes,33 using computer algorithms34,35 (for details, see the Supplementary Appendix). We concluded that the IL10R1 G141R mutation, which was localized in a region binding to interleukin-10,36 probably has a substantial effect on the structure of the interleukin-10–receptor complex (Fig. 3F and 3G). The IL10R1 T84I variant was similarly predicted to be deleterious on the basis of computer modeling and because threonine 84 is highly conserved in homologous proteins (Fig. 2H and 2I in the Supplementary Appendix).
To assess the functional capacity of the interleukin-10 receptor, we stimulated PBMCs from Patient II-5 in Family B and those from an unaffected subject with recombinant human interleukin-10 and then measured the extent of phosphorylation of STAT3 at tyrosine 705 and serine 727. As expected, phosphorylation of STAT3 was abrogated at tyrosine 705 but was unaffected at serine 727 in the cells from the patient, as compared with those from the control subject (Fig. 3H). To validate these findings, we coexpressed either wild-type IL10R1 protein or that with the G141R mutation, along with wild-type IL10R2, in IL10R-negative HeLa cells. Coexpression of wild-type IL10R1 and wild-type IL10R2 resulted in STAT3 phosphorylation on interleukin-10 signaling. In contrast, coexpression of mutant IL10R1 G141R and wild-type IL10R2 did not result in STAT3 phosphorylation on exposure to interleukin-10 (Fig. 3I).
EFFECTS ON INTERLEUKIN-10 SIGNALING
Interleukin-10 is a pleiotropic cytokine with effects on T cells, B cells, monocytes, and other cell types.16 We hypothesized that the pathophysiology of a deficiency in the interleukin-10 receptor involves undue and prolonged activation of mononuclear cells on exposure to bacterial particles, resulting in an augmented efflux of inflammatory cytokines (e.g., TNF- α) and damage to the intestinal mucosa. To test this idea, we analyzed TNF- α secretion of monocytes and monocyte-derived macrophages on exposure to lipopolysaccharide (LPS) or LPS plus interleukin-10 in samples from Patient II-3 in Family A, Patient II-5 in Family B, the unrelated patient who was homozygous for the T84I mutation, and five control subjects. Interleukin-10 substantially reduced the release of TNF- α in cells from the control subjects. This inhibitory effect was absent in cells from Patient II-3, who carried the IL10RB W159X mutation (Fig. 4A); Patient II-5, who carried the IL10RA G141R mutation (Fig. 4B and 4C); and the unrelated additional patient, who carried the IL10RA T84I mutation (Fig. 2K in the Supplementary Appendix).
Figure 4. Defective Down-Regulation of Proinflammatory Cytokines Mediated by Interleukin-10 in Mononuclear Cells with a Mutation in the Interleukin-10 Receptor.
Panel A shows defective inhibition of the release of tumor necrosis factor α (TNF- α) by costimulation with interleukin-10 in lipopolysaccharide (LPS)–stimulated macrophages from the index patient in Family A (Patient II-3), as compared with the mean value from samples obtained from five control subjects, as measured by enzyme-linked immunosorbent assay (ELISA). Panel B shows a similar ELISA measuring TNF- α secretion in LPS-stimulated peripheral-blood mononuclear cells (PBMCs) from the index patient in Family B (Patient II-5) and three control subjects. Panel C shows defective interleukin-10–mediated suppression of TNF- α secretion in LPS-stimulated cells from Patient II-5 in Family B. Panel D shows increased secretion of proinflammatory cytokines upon stimulation of cells with LPS in Patient II-5, as compared with a healthy control subject. These cytokines include MIP-1α and MIP-1β (macrophage inflammatory proteins 1α and 1β), MCP1 (monocyte chemoattractant protein), and RANTES (regulated on activation, normal T expressed and secreted protein). Panel E shows the abrogated effect of interleukin-10 on the release of proinflammatory cytokines in cells from the patient that were costimulated with LPS and interleukin-10, as compared with a healthy control subject. Cells from Patient II-5 show increased secretion of inflammatory cytokines, which could not be counteracted by costimulation with exogenous interleukin-10. Panel F shows defective interleukin-10–mediated induction of messenger RNA expression in the suppressor of cytokine signaling 3 gene (SOCS3) in Patient II-3 in Family A; induction was more than four times as great in a control sample. Expression levels were measured relative to β-actin as a housekeeping gene. The I bars indicate standard errors.
To assess whether LPS induced sustained secretion of other proinflammatory cytokines in IL10R1-deficient PBMCs, we used protein array analysis to measure supernatants of LPS-stimulated PBMCs. As compared with control cells, the cells carrying the IL10RA G141R mutation secreted increased levels of TNF- α; TGF-β1; interleukin-1 α, -1β, -2, and -6; soluble receptor of interleukin-6; RANTES (regulated on activation, normal T expressed and secreted protein); MCP1 (monocyte chemoattractant protein 1); and MIP-1α and MIP-1β (macrophage inflammatory proteins 1α and 1β). None of these proteins were down-regulated by interleukin-10 (Fig. 4D and 4E, and Fig. 3A and 3B in the Supplementary Appendix). Similar results were seen in PBMCs from the patient with the IL10R1 T84I mutation (Fig. 3C to 3F in the Supplementary Appendix).
Suppressor of cytokine signaling 3 (SOCS3) is a downstream target gene of STAT3 that is induced by interleukin-10.25,31 We exposed PBMCs from Patient II-3 in Family A and from a healthy control subject to interleukin-10 and then assayed the messenger RNA (mRNA) expression of SOCS3, using a real-time polymerase-chain-reaction assay. PBMCs from the control subject showed an increase in the up-regulation of SOCS3 mRNA by a factor of four, as compared with that in unstimulated control cells. In contrast, SOCS3 mRNA levels in PBMCs from Patient II-3 did not change after incubation with interleukin-10, indicating a lack of interleukin-10 signaling (Fig. 4F).
ALLOGENEIC STEM-CELL TRANSPLANTATION
Bone marrow transplantation ameliorates disease in mice with colitis and interleukin-10 deficiency.37 In view of our discovery that a nonsense mutation in the IL10RB gene is probably the genetic cause of inflammatory bowel disease in the affected patients in Family A and given the severity of their disease, we considered allogeneic hematopoietic stem-cell transplantation as treatment. Patient II-3 had an unaffected, HLA-matched sibling who could serve as the donor for such transplantation. After written informed consent was obtained, the patient underwent conditioning with the use of alemtuzumab (1 mg per kilogram of body weight), fludarabine (180 mg per square meter of body-surface area), treosulfan (42 mg per square meter), and thiotepa (10 mg per kilogram). Strict gut decolonization was performed with the use of colistin and total parenteral nutrition during the peritransplantation period. Engraftment of donor cells was documented 13 days after transplantation. Grade III skin graft-versus-host disease subsequently developed, for which prednisone was administered. More than a year after transplantation, full chimerism without evidence of graft-versus-host disease was documented; no further adverse side effects were reported as of October 2009. Both cutaneous folliculitis and inflammatory anal fistulas resolved shortly after transplantation (Fig. 5A and 5B). The patient has remained in continuous remission from ileocolitis more than a year after stem-cell transplantation. He gained weight and had no further episodes of intestinal pseudo-obstruction.
Figure 5. Success of Allogeneic Hematopoietic Stem-Cell Transplantation.
In Patient II-3 from Family A, who had severe anocutaneous fistulas that were resistant to therapy (Panel A), allogeneic stem-cell transplantation resulted in clinical amelioration of all effects of disease (Panel B).
DISCUSSION
We have shown that loss-of-function mutations in either IL10RA or IL10RB can be found in children with severe, early-onset enterocolitis, findings that are consistent with the idea that a lack of negative-feedback signaling mediated by interleukin-10 perturbs homeostasis of the intestinal immune system. Since IL10R1 is expressed on many cells of the innate and adaptive immune system, further studies are needed to determine which types of cells are primarily responsible for the altered intestinal immunity. In contrast, IL10R2 is expressed not only on cells of the immune system but also on a wide range of nonimmune cells, such as epithelial cells and keratinocytes.38 Because IL10R2 is a component of the receptors for interleukin-10, -22, -26, -28A, -28B, and -29,38,39 defective signaling of any of these cytokines as a result of IL10R2 deficiency may have additive or synergistic effects. The presence of severe inflammatory bowel disease is the most prominent phenotype in patients with IL10R1 or IL10R2 deficiency. We therefore infer that a lack of interleukin-10 signaling is the principal malfunction and is a likely cause of inflammatory bowel disease in patients with IL10R2 deficiency. Nevertheless, interleukin-22 and interleukin-26 regulate immunity in the skin,40,41 suggesting that chronic folliculitis in IL10R2-deficient patients may be caused by irregular or diminished signaling by either interleukin-22 or interleukin-26.
Our findings are consistent with those regarding severe colitis in mice lacking either Il10 or Il10rb.27,28 Expression of the murine gene encoding interleukin-22 in the appropriate cell types provides protection against colitis and is associated with the resolution of colitis in two distinct murine models,42,43 suggesting that some IL10R2-related functions may be independent of interleukin-10. Moreover, interleukin-22 induces the antimicrobial proteins REGIIIβ and REGIIIγ and enhances mucus production in colonic epithelial cells, thereby maintaining the epithelial barrier function and preventing bacterial infections.42,44
We speculate that in the absence of an interleukin-10–mediated antiinflammatory response, the presence of intestinal commensal bacteria leads to activation of a fulminant immune response, resulting in a hyperinflammatory response with associated tissue damage. This may facilitate increased transmigration of intestinal bacteria and result in chronic intestinal lymphadenopathy or even organ-related abscesses.
Our limited search for mutations in IL10RA and IL10RB in patients with inflammatory bowel disease indicated that loss-of-function mutations may be confined to very severe cases with an onset in infancy. A polymorphism in IL10 has been associated with the risk of colitis in a genomewide association study,11 and this finding has been replicated,45 suggesting that milder genetic variants affecting interleukin-10–dependent pathways may be involved in the pathophysiology of inflammatory bowel disease.
Our study provides an example of the power of molecular medicine to go from the bedside (for diagnosis) to the bench (for the discovery of mutations) and back to the bedside (for treatment). Because no conventional therapeutic approach was successful in our patients and given the role of interleukin-10 signaling in cells of the hematopoietic system, we attempted a curative approach by means of allogeneic stem-cell transplantation, which would have been ethically difficult to justify without knowledge of the monogenic cause. The sustained remission after stem-cell transplantation in the patient suggests that interleukin-10 signaling in hematopoietic cells rather than signaling through a pathway associated with interleukin-22, interleukin-26, or interferon-λ in nonhematopoietic cells was critical for the therapeutic effect.
In summary, mutations in the genes encoding the two polypeptide chains of the interleukin-10 receptor abrogate interleukin-10–mediated immunomodulatory signaling and are strongly associated with hyperinflammation of the intestine.
Supplementary Material
Acknowledgments
Supported by grants from the European Commission Marie Curie Excellence program (MEXT-CT-2006-042316, to Dr. Grimbacher), Deutsche Forschungsgemeinschaft (SFB621, to Dr. Klein), and the German Federal Ministry of Education and Research (PID-NET), to Drs. Klein and Grimbacher; by the Intramural Research Program of the National Institutes of Health; and by fellowships from Deutsche José Carreras Leukämie-Stiftung (to Dr. Kotlarz) and Else-Kröner-Fresenius-Stiftung (to Dr. Boztug).
We thank the clinical and nursing staff at Hannover Medical School for their support, Jessica Pfannstiel for technical assistance, David Escors for providing an experimental reagent and technical advice, and Mahdad Noursadeghi and Lynn Williams for technical advice, suggestions, and discussions.
APPENDIX
The authors’ affiliations are as follows: the Department of Immunology, Royal Free Hospital and University College London (E.-O.G., M.P., C.W., B.G.), the Department of Paediatric Gastroenterology, Great Ormond Street Hospital, University College London (N.S.), and the Department of Medicine, University College London (A.W.S.) — all in London; the Departments of Pediatric Hematology/ Oncology (D.K., K.B., F.N., J.D., A.A., D.M., N.H., K.-W.S., M.S., C.K.), Pathology (H.K.), Pediatric Surgery (R.N.), and Pediatric Pulmonology (U.B.), Hannover Medical School, Hannover, Germany; the National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD (E.M.G., A.A.S.); the Department of Hematology/Oncology, Core Facility II Genomics, Freiburg University Medical Center (D.P.), and the Department of Rheumatology and Clinical Immunology, University Hospital Freiburg (U.S.) — both in Freiburg; Dr. von Hauner’sches Kinderspital, Ludwig-Maximilian University, Munich (M.L., S.K.); the Department of Pediatrics, HELIOS Hospital Erfurt, Erfurt (A.S.); and the Department of Pediatrics, St.-Marien-Hospital Bonn, Bonn (S.B.) — all in Germany; and Massachusetts General Hospital and Harvard Medical School — both in Boston (S.B.S.).
Footnotes
Dr. Snapper reports receiving consulting fees from Viamet and Cytokine PharmaSciences and lecture fees from UCB. No other potential conflict of interest relevant to this article was reported.
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Fried K, Vure E. A lethal autosomal recessive enterocolitis of early infancy. Clin Genet. 1974;6:195–6. doi: 10.1111/j.1399-0004.1974.tb00650.x. [DOI] [PubMed] [Google Scholar]
- 4.Mégarbané A, Sayad R. Early lethal autosomal recessive enterocolitis: report of a second family. Clin Genet. 2007;71:89–90. doi: 10.1111/j.1399-0004.2007.00735.x. [DOI] [PubMed] [Google Scholar]
- 5.Satsangi J, Silverberg MS, Vermeire S, Colombel J-F. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–53. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernier-Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn’s disease: a population-based cohort study. Gastroenterology. 2008;135:1106–13. doi: 10.1053/j.gastro.2008.06.079. [DOI] [PubMed] [Google Scholar]
- 7.Mizoguchi A, Mizoguchi E. Inflammatory bowel disease, past, present and future: lessons from animal models. J Gastroenterol. 2008;43:1–17. doi: 10.1007/s00535-007-2111-3. [DOI] [PubMed] [Google Scholar]
- 8.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 9.Hugot J-P, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 10.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke A, Balschun T, Karlsen TH, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 12.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of non-synonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 13.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoll M, Corneliussen B, Costello CM, et al. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476–80. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- 15.Kaser A, Lee A-H, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 17.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–95. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiorentino DF, Zlotnik A, Vieira P, et al. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol. 1991;146:3444–51. [PubMed] [Google Scholar]
- 19.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–79. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 20.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108–11. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19:563–73. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- 22.Finbloom DS, Winestock KD. IL-10 induces tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–90. [PubMed] [Google Scholar]
- 23.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894–903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber-Nordt RM, Riley JK, Greenlund AC, Moore KW, Darnell JE, Schreiber RD. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J Biol Chem. 1996;271:27954–61. doi: 10.1074/jbc.271.44.27954. [DOI] [PubMed] [Google Scholar]
- 25.Williams L, Bradley L, Smith A, Foxwell B. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J Immunol. 2004;172:567–76. doi: 10.4049/jimmunol.172.1.567. [DOI] [PubMed] [Google Scholar]
- 26.Berg DJ, Kühn R, Rajewsky K, et al. Interleukin-10 is a central regulator of the response to LPS in murine models of endotoxic shock and the Shwartzman reaction but not endotoxin tolerance. J Clin Invest. 1995;96:2339–47. doi: 10.1172/JCI118290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kühn R, Lohler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 28.Spencer SD, Di Marco F, Hooley J, et al. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–8. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–9. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 31.Shen B, Vihinen M. RankViaContact: ranking and visualization of amino acid contacts. Bioinformatics. 2003;19:2161–2. doi: 10.1093/bioinformatics/btg293. [DOI] [PubMed] [Google Scholar]
- 32.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–87. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon SI, Jones BC, Logsdon NJ, Walter MR. Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure. 2005;13:551–64. doi: 10.1016/j.str.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Salzer U, Bacchelli C, Buckridge S, et al. Relevance of biallelic versus mono-allelic TNFRSF13B mutations in distinguishing disease-causing from risk increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113:1967–76. doi: 10.1182/blood-2008-02-141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thusberg J, Vihinen M. Bioinformatic analysis of protein structure–function relationships: case study of leukocyte elastase (ELA2) missense mutations. Hum Mutat. 2006;27:1230–43. doi: 10.1002/humu.20407. [DOI] [PubMed] [Google Scholar]
- 36.Josephson K, Logsdon NJ, Walter MR. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity. 2001;15:35–46. doi: 10.1016/s1074-7613(01)00169-8. [DOI] [PubMed] [Google Scholar]
- 37.Bamba S, Lee C-Y, Brittan M, et al. Bone marrow transplantation ameliorates pathology in interleukin-10 knockout colitic mice. J Pathol. 2006;209:265–73. doi: 10.1002/path.1967. [DOI] [PubMed] [Google Scholar]
- 38.Wolk K, Sabat R. Interleukin-22: a novel T- and NK-cell derived cytokine that regulates the biology of tissue cells. Cytokine Growth Factor Rev. 2006;17:367–80. doi: 10.1016/j.cytogfr.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004;76:314–21. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- 40.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–54. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Wolk K, Witte E, Wallace E, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36:1309–23. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto K, Ogawa A, Mizoguchi E, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–44. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–57. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 45.Amre DK, Mack DR, Morgan K, et al. Interleukin 10 (IL-10) gene variants and susceptibility for pediatric onset Crohn’s disease. Aliment Pharmacol Ther. 2009;29:1025–31. doi: 10.1111/j.1365-2036.2009.03953.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.