Abstract
Development of drugs targeting lipid kinases has been delayed by the lack of robust screening assays. Methods are needed that can accommodate the presentation of different acceptor substrates in the optimal lipid environment. The Trancreener™ ADP Assay relies on homogenous immunodetection of ADP, using either fluorescence polarization (FP) or time-resolved fluorescence resonance energy transfer (TR-FRET) as a signal output. Detection of ADP - the invariant product of all kinase reactions - provides complete flexibility for varying lipid substrate parameters. We used this assay to optimize dispersal methods for C8 and C16 phosphatidylinositol 4,5 bisphosphate substrates and to assess the effects of chain length on the activity and inhibition of phosphoinositide-3-kinase (PI3K) isoforms. The non-physiological C8 substrate supported the highest activity. Known inhibitors were profiled using both the FP and TR-FRET based assays and there was excellent concordance (r2 = 0.93) in the IC50 values. The overall rank order of inhibitors was the same using the C8 and C16 substrates, except for minor deviations. ATP hydrolysis in the absence of substrate was detected with the PI3Kα isoform, and inhibitors affected PI3Kα intrinsic ATP hydrolysis activity similarly to lipid phosphorylation.
Keywords: ADP detection, phosphoinositide 3-kinase, fluorescence polarization, time-resolved fluorescence resonance energy transfer, intrinsic ATPase activity
INTRODUCTION
Phosphoinositide 3-kinases catalyze the phosphorylation of phosphatidylinositol (PIP) and its various phosphorylated forms at the 3′-hydroxyl of the inositol ring, generating the second messengers PI(3)P, PI(3,4)P2 and PI(3,4,5)P3 [1,2]. These lipid signaling molecules control diverse cellular activities including proliferation, apoptosis, motility, and morphology. For instance, Class I PI3-kinases transduce signals from activated receptor tyrosine kinases that ultimately result in stimulation of AKT-mediated anti-apoptotic and pro-survival phosphorylation cascades [3]. Moreover, the PI3K/AKT signaling pathway and its components are subject to more genetic aberrations than any other pathway in human cancer, including amplification and gain of function mutations of the PI3K catalytic and regulatory subunits [3]. Not surprisingly, PI3-kinases have come under intense focus as therapeutic targets for a broad range of diseases, most notably cancers [4,5] and inflammation [6].
There are several members in the PI3K family that differ in their substrate specificities, expression profiles, and regulation [7]. They share substantial homology in the ATP binding domains of their catalytic subunits, but have little similarity in other parts of the catalytic subunits or in their regulatory subunits [2]. PI3-kinases are divided into three classes on the basis of structural and functional homologies in the catalytic subunits. Class IA enzymes are heterodimers of one of three possible catalytic subunits, p110α, p110β, or p110δ, and one of five p85 or p55 regulatory subunits [2]. The Class IB enzymes are comprised of a p110γ catalytic subunit and a p101 or p84 regulatory subunit. The three Class II PI3Ks (CIIα, CIIβ, and CIIγ) are not known to have associated regulatory subunits. There is a single Class III PI3K comprised of unique catalytic and regulatory subunits.
The fungal metabolite Wortmannin and the synthetic compound LY 294002 have been widely used as pan inhibitors for PI3-kinases for several years; both compounds bind at the ATP-binding site [8]. Developing more selective inhibitors, with improved pharmacological and ADME properties is a compelling therapeutic strategy that is being pursued at several pharmaceutical and biotechnology companies to elucidate the functions of individual PI3K isoforms [9] and exploit the family for drug discovery [3].
Despite the high level of interest in the PI3K family, the development of robust high throughput screening (HTS) assay methods has lagged. This is partly because methods commonly used for monitoring protein kinase activity - such as phospho-peptide immunodetection that are not easily applied to lipid kinases. The most widely used methods take advantage of the selective binding of different phosphoinositide reaction products to the cognate pleckstrin homology (PH) domains of their intracellular targets. For example, TR-FRET competitive binding assays have been developed by attaching the donor fluorophore to the phophoinositide and the acceptor fluorophore to the PH domain [10,11]. The charge-dependent binding of phosphoinositides to immobilized metal ions has also been exploited to develop a direct binding assay based on fluorescence superquenching [12]. Traditional radioassays employing transfer of labeled phosphate from ATP have also been adapted to HTS using a flash-plate format [13]. These and related approaches based on detection of phosphoinositides require conjugation of the lipid substrates to detection components such as fluorophores [14] or biotin, which decreases the flexibility of the assay for accommodating different amounts and types of lipid substrates.
An additional factor complicating biochemical assay development is the requirement for three dimensional lipid aggregates, such as liposomes, to mimic cellular membrane surfaces [15]. Enzyme activity is affected by the type of lipid used, as well as its bulk and surface composition [16]. Optimization of these parameters can be time consuming, and is complicated by detection methods that rely on modified phosphoinositides as detection reagents.
In this study we extend validation of the homogenous fluorescent ADP detection assay, the Transcreener™ ADP Assay, as a flexible, robust assay method for detecting PI3K catalytic activity and its inhibition. The Transcreener™ ADP Assay employs a highly specific monoclonal antibody against ADP and a fluorescent ADP tracer for competitive immunodetection of ADP [17,18]. The assay has been formatted for both fluorescence polarization (FP) and time-resolved fluorescence resonance energy transfer (TR-FRET) and has been validated for HTS using protein kinases and other types of ATP-utilizing enzymes [17,19,20].
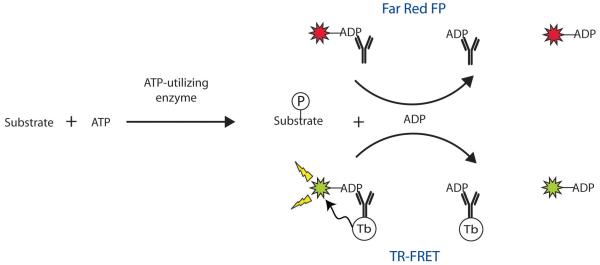
In both detection modes, enzymatically produced ADP displaces an ADP tracer from the antibody resulting in a change in the fluorescence signal (Fig. 1). In the case of the FP assay, enzyme activity results in decreased polarization values as a greater fraction of tracer is dissociated from the much larger antibody. The FP assay employs a far red tracer (670 nm emission), which minimizes interference from fluorescent compounds and light scattering [21,22]. The TR-FRET assay incorporates a terbium chelate-antennae complex (LanthaScreen™, Invitrogen, Carlsbad, CA) as the FRET donor, which is covalently attached to the ADP-specific monoclonal antibody; the acceptor is a fluorescein-ADP tracer. Displacement of tracer from antibody by enzymatically produced ADP disrupts luminescence energy transfer from the terbium to the tracer, resulting in decreased fluorescein intensity. The most advantageous feature of the TR-FRET format is the time-gated acquisition of the delayed signal, which reduces the prompt autofluorescence from test compounds. Both the FP and the TR-FRET detection modes provide robust signals (low % CV), because their self-correcting ratiometric measurements reduce well to well signal variation.
FIG. 1.
The Transcreener™ ADP Assays detect ADP, the invariant product of many ATP utilizing enzymes, including lipid kinases. ADP competes with the tracer for binding to the monoclonal ADP antibody. In fluorescence polarization (FP) detection, high polarization is observed when the ADP AlexaFluor® 633 tracer is bound to the ADP antibody. ADP displaces the far-red tracer resulting in low polarization. In time-resolved fluorescence resonance energy transfer (TR-FRET) detection, energy transfer from the terbium to the fluorescein (FAM) is observed when the ADP FAM tracer is bound to the ADP-antibody-Tb resulting in a high FAM520/Tb495 ratio. ADP disrupts TR-FRET by displacing the tracer from the antibody, resulting in a low ratio.
By using an assay that relies on detection of ADP rather than phosphoinositides, we were able to rapidly test different lipid substrates, their concentrations, and techniques for their preparation to arrive at conditions for optimal PI3K activity. We also examined the effects of lipid substrate chain length on the profiles of known inhibitors with three different PI3K isoforms in the IA and IB classes. Inhibitor pharmacology was shown to be essentially independent of the lipid substrate and equivalent using the TR-FRET and FP-based assays. Lastly, we demonstrated that the intrinsic ATP hydrolysis (ATPase) activity of PI3K in the absence of a lipid substrate can be used for identifying inhibitors and determining their potency. Detection and profiling of inhibitors by measuring kinase ATPase activity was recently examined in detail for a non-receptor protein tyrosine kinase from the Tec family, interleukin-2-inducible T cell kinase (ITK) [23] but to our knowledge, this is the first time this approach has been applied to lipid kinases.
MATERIALS AND METHODS
Materials
The monoclonal ADP antibody and Transcreener™ ADP Assays were developed at BellBrook Labs (Madison, WI) [17]. AlexaFluor633® succinimidyl ester and 5-carboxyfluorescein (FAM) succinimidyl ester were purchased from Molecular Probes (Eugene, OR) and were conjugated to ADP. LanthaScreen™ amine reactive terbium chelate was purchased from Invitrogen (Carlsbad, CA) and conjugated to the monoclonal ADP antibody. PI3 kinase isoforms were from Upstate (Dundee, Scotland). Lipids and lipid substrates were purchased from either Avanti Polar Lipids (Alabaster, AL) or Cellsignals (Columbus, OH). PI3 kinase inhibitors were from EMD Biosciences (La Jolla, CA). Basic buffer components were purchased from Sigma (St. Louis, MO) or Fisher (Hampton, NH).
Standard Conditions
The Transcreener™ ADP Assays follow a two-step ‘mix and read’ procedure. Lipid kinase reactions (10 μL) were performed in 384-well plates at 30°C, initiated with the addition of ATP. All PI3K isoform reactions were performed in 50 mM HEPES (pH 7.1), containing NaCl (100 mM), MgCl2 (4 mM), EGTA (2 mM), DTT (2 mM), DMSO (1%), ATP (30 μM), and either PI(4,5)P2 C8 lipid substrate (100 μM) or PI(4,5)P2 C16 lipid substrate (30 μM) unless otherwise noted. Enzyme concentration and reaction time were variable, but designed to produce 10% ATP conversion. Control reactions and standard curves contained all reaction components except enzyme. Enzyme reactions were stopped by the addition of ADP detection reagents (10 μL) containing EDTA. Reagent mixing was performed by orbital shaking for one minute, followed by incubation at the indicated times and temperatures. Detailed assay conditions are listed separately for FP and TR-FRET below. To stabilize signal, quenched reactions were allowed to equilibrate at room temperature for one hour prior to reading the plate. All assays were performed in Corning® (Corning, NY) white (#3673; for TR-FRET) or black (#3676; for FP) 384-well, round bottom, low volume, polystyrene, non-binding surface microtiter plates. Unless otherwise noted, the data were fit to a variable slope sigmoidal dose-response curve. IC50 values were calculated using Graphpad PRISM (GraphPad Software, San Diego, CA).
FP Detection
For FP detection, PI3K reactions were stopped by the addition of an equal volume (10 μL) of detection mix to yield final concentrations of: 50 mM HEPES (pH 7.5), 200 mM NaCl, 10 mM EDTA, 0.01% Brij-35, 2 nM ADP AlexaFluor® 633 tracer, and 15.5 μg/ml ADP antibody. The concentration of ADP antibody used was equal to the EC85 concentration in the presence of 30 μM ATP, the concentration of ATP used in all kinase reactions. Fluorescence polarization measurements were performed on a Tecan Ultra plate reader using the following filters and settings: 612 nm excitation filter (10 nm bandwidth), 670 nm emission filter (25 nm bandwidth), 10 flashes per well, 30°C, or on the Tecan Safire2™ plate reader using the following filters and settings: 635 nm excitation (LED), 670 nm emission (10 nm bandwidth), 10 flashes per well, 30°C. A free tracer reference was set to 20 mP, and the buffer (containing ADP antibody) was used as the buffer blank for both the sample and free tracer reference wells.
TR-FRET Detection
For TR-FRET detection, PI3K reactions were stopped by the addition of an equal volume (10 μL) of detection mix to yield final concentrations of: 50 mM HEPES (pH 7.5), 100 mM NaCl, 5 mM EDTA, 0.01% Brij-35, 2 nM ADP antibody-Tb, and 14 nM ADP FAM tracer. The concentration of ADP FAM tracer used was equal to the EC50 concentration in the presence of 30 μM ATP in the kinase enzyme reaction. TR-FRET measurements were performed on the Tecan Ultra plate reader (Durham, NC) using the following filters and settings: 340 nm excitation filter (35 nm bandwidth), 495 nm (10 nm bandwidth) and 520 nm (25 nm bandwidth) emission filters, 100 μsec delay, 100 μsec integration time, 10 flashes at 30°C.
Lipid Substrate Vesicle Preparation
Lipid vesicles were prepared by sonication, freeze/thaw, or a combination of the two methods. The phosphatidylinositol 4,5 bisphosphate (PI(4,5)P2) substrate with fatty acid side-chains of eight (C8) or sixteen (C16) carbons were suspended in water to a concentration of 1310 μM and 910 μM, respectively. In addition, an aliquot of the PI(4,5)P2 C16 sample was removed and an equimolar concentration of phosphatidylserine (PS) was added prior to sonication. Bath sonication was performed at 50/60 Hz/80 watts/117 volts for 1 hour at 27–33°C. In addition, aliquots from the sonicated PI(4,5)P2 C16 lipid substrate preparation were removed and frozen and thawed 5 times. The samples were frozen in an isopropanol/dry ice bath, with thawing in a water bath at 40°C and vigorous vortexing. Long chain fatty acids stick to plastic. Therefore, all manipulations of the PI(4,5)P2 C16 lipid substrate were performed in glass vials. Long term storage for lipid substrates was at −80°C.
ADP/ATP Standard Curve
12-point ADP/ATP standard curves designed to mimic an enzyme reaction were used to quantify ADP production in the PI3K enzyme reactions. Starting at 30 μM ATP - the concentration used in PI3K reactions - ATP was decreased and ADP increased proportionately, keeping the total adenosine concentration constant. The standard curves (n = 4) contained all of the components used in the authentic enzyme assays except enzyme, and were included on the same plates as the experimental reactions. Based on the standard curves for both TR-FRET and FP readouts, the concentration of ADP produced in the enzyme reactions was calculated using the Graphpad PRISM software using the four-parameter logistic regression curve fit. Because there are alternate ways to fit data to a non-linear standard curve, we validated the goodness of fit using the backcalculation method [24] and individual data points within an ADP/ATP standard curve. To minimize error propagation from the highest and lowest regions of the standard curves, enzyme reactions were designed so that the amount of ADP produced (in the absence of inhibitor) fell mostly within the middle region of the curves.
Inhibitor titrations
Dose dependency is shown for each inhibitor from a 20-point two-fold dilution in duplicate. Six PI3K inhibitors (Wortmannin, PI 103, PI3Kγ inhibitor, PI3KγII inhibitor, LY 294002, and Quercetin) were prepared as concentrated stocks in 100% DMSO. The inhibitor stocks were diluted into 4% DMSO and then serially diluted in 4% DMSO. The diluted inhibitor titration (2.5 μL) was transferred to the reaction plate prior to addition of the enzyme reaction mixture (5 μL). ATP (2.5 μL at 4X) was added to begin the enzyme reaction. All studies were performed using the same concentration of inhibitor dilutions.
Enzyme concentrations and reaction times were designed to achieve initial rates and were generally lowest and shortest, respectively, with the soluble C8-lipid substrate and TR-FRET readout. PI3Kα (0.4 or 2.8 nM) was incubated with either the PI(4,5)P2 C8 or PI(4,5)P2 C16 lipid substrate for 1.5 hr. More PI3Kα (28.2 nM) and longer incubation times (TR-FRET = 2 hr, FP = 4 hr) were required in the absence of substrate (intrinsic ATPase reaction). PI3Kβ (2.8–7.5 nM) was incubated with either the PI(4,5)P2 C8 or PI(4,5)P2 C16 lipid substrate for 2–2.5 hr, and 1.5 hr, respectively. The PI3Kγ (6.1–12.2 nM) was incubated with either PI(4,5)P2 C8 or PI(4,5)P2 C16 lipid substrate for 4 hr (TR-FRET) or 4–6 hr (FP), respectively. Z’ values (n = 16) were determined for each PI3K reaction without inhibitor, with the maximal signal set at or near initial velocity conditions, and the minimal signal from control reactions lacking enzyme. All enzyme studies were performed at or near initial rate conditions (between 1.1% and11% consumption of ATP). Observed enzyme rates were consistent with pilot experiments, indication that enzymes were stable in storage and during long incubations.
RESULTS
Enzyme and substrate dependent ADP production
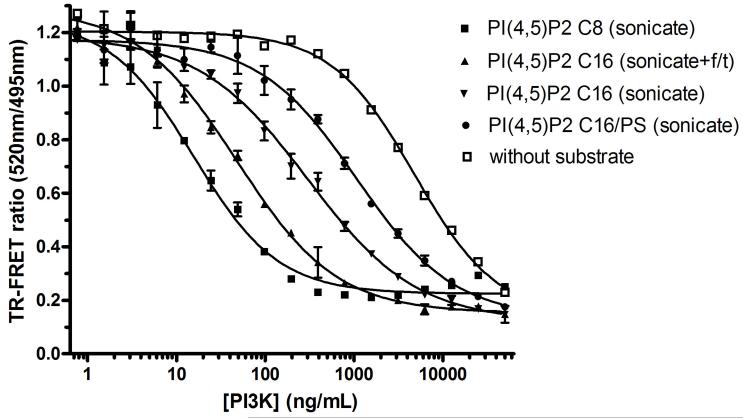
The effects of the fatty acid chain length and dispersion method of the lipid substrate on PI3K activity were investigated using the Transcreener™ ADP Assay with a TR-FRET readout (Fig. 2). Both the physiological PI(4,5)P2 C16 lipid substrate and the PI(4,5)P2 C8 lipid substrate were used at saturating concentrations based on literature Km values [10,25]. Therefore, the results should reflect differences in the Vmax that were dependent on the type of lipid substrate used and its method of preparation, and not its concentration. We tested a number of different lipid preparation methods drawn from the literature including sonication [16], a combined method of sonication and freeze/thaw, [26] and alteration of phospholipid composition [13,15,25-27]; all are treatments used to increase the dispersion and uniformity of substrate in lipid vesicles. These methods are generally used only for longer chain lipids, such as the PI(4,5)P2 C16, and are not as necessary for the more soluble C8 substrate. The data presented here represent a sampling of potential methods.
FIG. 2.
Effect of lipid substrate side chain length and preparation technique on PI3α kinase ADP production using time-resolved fluorescence resonance energy transfer (TR-FRET). Raw data for the two-fold serial dilution of PI3Kα (n = 2) is shown with error bars with PI(4,5)P2 C8 (30 μM), PI(4,5)P2 C16 (30 μM), and without substrate. The C16 lipid substrate was prepared using either sonication alone, sonication followed by freeze/thawing, or sonication in the presence of phosphatidylserine. Enzyme reactions were stopped after incubation for 1.5 hr at 30°C. EC50 values for the C8 lipid (sonication only method), C16 lipid (sonication and freeze/thaw method), C16 lipid (sonication only method), C16 lipid (sonication in presence of phosphatidylserine), and without substrate were 15.6, 47.8, 299, 1069, 4765 ng/mL, respectively.
ADP production is dependent upon enzyme concentration for both the physiological substrate (C16) and the shorter fatty acid chain substrate (C8) as indicated by a decrease in the ratio of acceptor fluorophore emission to donor fluorophore emission (Fig. 2). The length of the fatty acid chain of the lipid substrate had a moderate effect on ADP production; the EC50 value for the optimal preparation of C16 substrate (sonication plus freeze/thaw; EC50 = 47.8 ng/mL) was three-fold higher than the EC50 for the C8 substrate (EC50 = 15.6 ng/mL). However, this lipid chain length-dependence was not always observed in subsequent experiments using different C16 lipid dispensing protocols, and may have been caused by adherence of lipids to dispensing tips. The effects of different treatments used to disperse the less soluble C16 substrate were additive. The C16 substrate is improved as a substrate when the sonicated preparation is further processed through cycles of freezing and thawing. The EC50 values for C16 lipid sonication alone sample was 299 ng/mL. Interestingly, addition of equimolar phosphatidylserine (PS) to the C16 vesicles had a strong negative effect on kinase activity (EC50 = 1069 ng/mL). Others report the addition of PS to substrate showed an opposite effect when optimization of PS:PI(4,5)P2 C16 ratio was explored [15] however we did not pursue mixed micelle studies further. Indeed, ideal mixed micelle surface concentration is dependent not only upon enzyme class but also on isoform [15,16].
Interestingly, ADP was produced by PI3Kα in the absence of acceptor lipid substrate (Fig 2), although >300-fold more enzyme (EC50 = 4,765 ng/mL) was required, relative to reactions with the C8 substrate. Most if not all of this activity was likely due to non-productive ATP hydrolysis. Autophosphorylation is not an alternate explanation, because with the relatively small amount of enzyme used (30 nM) over 100 phosphorylation events would be required per enzyme molecule to produce the observed signal. PI3Kβ and PI3Kγ also exhibited intrinsic ATPase activity at very high enzyme concentration (data not shown). However, the lower turnover for β and γ isoforms and low enzyme stock concentrations prevented further studies.
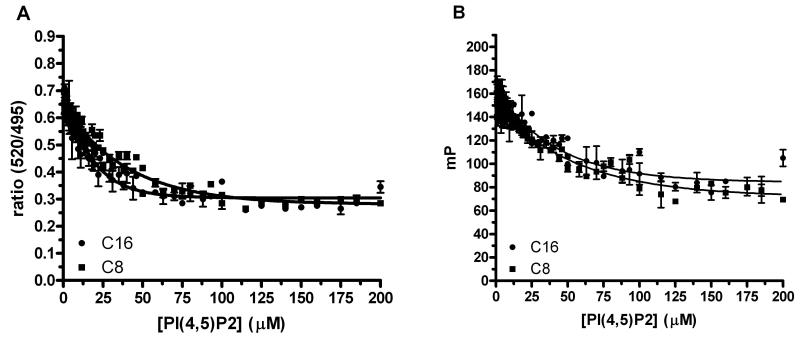
The dependence of ADP formation on the concentration of lipid substrate is shown with PI3Kα using freezing and thawing as the lipid preparation method and measured with both the TR-FRET (Fig. 3A) and FP (Fig. 3B) detection methods. These experiments confirmed the dependence of assay signal on the acceptor lipid and allowed determination of the optimal lipid concentrations to use for subsequent inhibitor studies. Note that these and inhibitor experiments were done using ATP at 30 μM, which is near the reported Km concentration [10,12], to reflect a typical kinase screening situation. Using the assay data at a one-hour time-point, the concentration of lipid that resulted in a half-maximal assay signal (S50) were as follows: S50 values were 30 μM and 39 μM for the C16 and C8 lipid substrates for the FP method, respectively and the S50 values for TR-FRET ADP detection were 12 μM and 28 μM. Based upon these results, we used C16 and C8 lipid substrate concentrations of 30 μM and 100 μM, respectively in the subsequent inhibitor studies .
FIG 3.
Influence on PI(4,5)P2 substrate concentration on PI3α kinase ADP production determined using (A) time-resolved fluorescence resonance energy transfer (TR-FRET) and (B) fluorescence polarization (FP). Raw data are shown with error bars for C8 and C16 lipid substrate (n = 2) titrations. The PI(4,5)P2 C8 and C16 lipid substrates were prepared using the freeze/thaw method and serially titrated (n = 2) before adding a constant enzyme concentration (2.4 nM).
Inhibitor Profiles for PI3 Kinase Activity
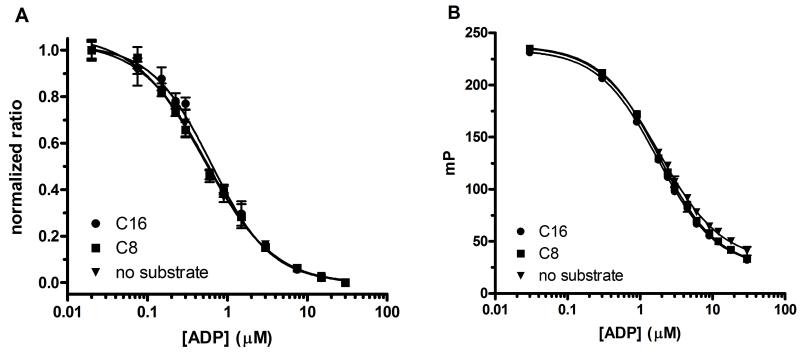
To enable determination of IC50 values based upon product formation instead of raw signal, standard curves were prepared mimicking the conversion of 30μM ATP to ADP. Importantly, the standard curves (Fig. 4) prepared in the presence and absence of lipids verify that neither the C8 nor the C16 lipid substrate (at 100 μM and 30 μM, respectively) had a significant effect on ADP immunodetection using TR-FRET (Fig. 4A) or FP (Fig, 4B). The data illustrate that using both TR-FRET and FP ADP detection, the change in assay signal is proportional to [ADP] and that a large assay signal is seen for ≤10% ATP conversion. Of the two ADP detection modes, TR-FRET is slightly (approximately 3-fold) more sensitive.
FIG 4.
Comparison of the effect of the presence and absence of lipid substrate on the ADP/ATP standard curves. Time-resolved fluorescence resonance energy transfer (TR-FRET; Fig. 4A) and fluorescence polarization (FP; Fig. 4B) data (n = 4) are shown with error bars for C8, C16 lipid substrate and without substrate. The data show that assay signal is proportional to concentration ADP. The PI(4,5)P2 C8 and C16 lipid substrates were prepared using the freeze/thaw method.
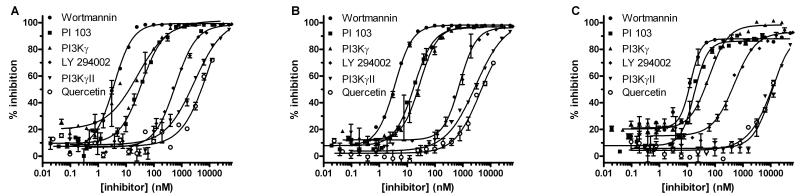
We examined inhibitor profiles for three PI3K isoforms performed in the presence of the C8 and C16 PI(4,5)P2 lipid substrates as well as with PI3Kα in the absence of substrate. Two Class IA isoforms, PI3Kα (p110α) and PI3Kβ (p110β) and the Class IB isoform PI3Kγ were tested with six well characterized inhibitors. The inhibitors (Wortmannin, PI 103, PI3Kγ inhibitor, PI3KγII inhibitor, LY 294002, and Quercetin) were chosen because they exhibit a wide range (1,000-fold) in IC50 values, and because their reported potency profiles are different amongst the three PI3K isoforms. Since this is an extensive matrix, representative inhibitor dose dependency curves are shown for PI3Kα with the following conditions: C16 substrate, TR-FRET detection (Fig. 5A); C16 substrate, FP detection (Fig. 5B); and without acceptor substrate, FP detection (Fig. 5C).
FIG. 5.
Comparison of inhibition curves between fluorescence polarization (FP) ADP detection and time-resolved fluorescence resonance energy transfer (TR-FRET) ADP detection for PI3Kα kinase and intrinsic ATPase activities. PI3Kα sensitivity to six known lipid kinase inhibitors (Wortmannin, PI 103, PI3Kγ inhibitor, LY 294002, PI3KγII inhibitor, and Quercetin) is shown with error bars for the 20-point dose dependency curves (n = 2). (A) Using TR-FRET, ADP detection in the PI3Kα (0.36 nM)/PI(4,5)P2 C16 lipid reaction progressed to 4.7% ATP conversion (Z’ = 0.82). (B) Using FP, ADP detection in the PI3Kα (1.0 nM)/PI(4,5)P2 C16 lipid reaction progressed to 10.3% ATP conversion (Z’ = 0.89). (C) Using FP, ADP detection in the PI3Kα (28.2 nM) intrinsic ATPase reaction progressed to 4.1% ATP conversion (Z’ = 0.82). Polarization values were converted to percent inhibition using an ATP/ADP standard curve and Graphpad PRISM software.
The ATP concentration was constant at 30 μM, which is equal to or less than the reported ATP Km values for all three PI3K isoforms [10,12,25]. The PI(4,5)P2 C8 and PI(4,5)P2 C16 substrates were used at concentrations (100 μM and 30 μM, respectively) either equal to or above their substrate Km values. A single preparation of the lipid substrate was used for the entire study. Reactions were designed so that ADP production was within initial velocity range and also fell mostly within the middle region of the standard curves, so that the propagation of error was minimized.
The complete inhibitor profiling data for all three isoforms is shown in Table 1 (FP) and Table 2 (TR-FRET); partial dose dependency curves over the inhibitor entire concentration range are set at >20 μM. Both the rank order and more importantly the IC50 values are in concordance with other studies [9,13,28-31]. Notably, the rank order of the inhibitors in these experiments was the same, with a few minor exceptions, regardless of the substrate or detection format. The covalent inhibitor Wortmannin [32] was the most potent PI3Kα inhibitor in the series with IC50 values of 4 nM and 2 nM observed using the C16 substrate with the TR-FRET and FP assay, respectively and somewhat higher values of 35 nM and 6 nM observed with the C8 substrate. Intermediate IC50 values of 18 nM and 13 nM were determined for Wortmannin by measuring the intrinsic ATPase activity of PI3Kα with the TR-FRET and FP assays, respectively. The patterns observed for the 5 remaining inhibitors with PI3Kα were similar; the IC50 values were relatively independent (less than a five-fold difference) of either the presence of the lipid acceptor, or of its chain length. The IC50 values determined using the two different detection methods - FP and TR-FRET - correlated even more closely, with less than a four-fold difference for all inhibitor/acceptor substrate combinations tested for PI3Kα, with the exception of Wortmannin with the C8 substrate, where IC50 values of 35 nM and 6 nM were obtained with the TR-FRET and FP assays, respectively. As expected, Quercetin was the weakest inhibitor of PI3Kα activity, with IC50 values of >20 μM, 12 μM, and 12 μM determined by FP using the C16 and C8 substrates and without substrate, and >20 μM, 11 μM, and 15 μM using TR-FRET detection. For comparison, literature IC50 values for Wortmannin with PI3Kα range from 1 nM to 5 nM and for Querectin from 1.4 μM to 3.8 μM [8,13,28,29,32-34]. Literature values were determined using various assay conditions.
Table 1. IC50 Values for Inhibition of PI3K Isoforms Using Fluorescence Polarization ADP Assay.
|
PI3Kα IC50 Values (μM) |
PI3Kβ IC50 Values (μM) |
PI3Kγ IC50 Values (μM) |
|||||
|---|---|---|---|---|---|---|---|
| Inhibitor | C16 | C8 | no substrate | C16 | C8 | C16 | C8 |
| Wortmannin | 0.002 | 0.006 | 0.013 | 0.019 | 0.009 | 0.003 | 0.003 |
| PI 103 | 0.018 | 0.024 | 0.018 | 0.040 | 0.048 | 0.16 | 0.31 |
| PI3Kγ inhibitor |
0.019 | 0.019 | 0.060 | 0.45 | 0.15 | 0.029 | 0.016 |
| LY 294002 | 0.77 | 0.45 | 0.49 | 1.3 | 0.68 | 2.0 | 2.8 |
| PI3KγII inhibitor |
>20 | 6.0 | 11 | >20 | >20 | 0.82 | 2.4 |
| Quercetin | >20 | 12 | 12 | >20 | >20 | 7.5 | 7.0 |
Six known PI3 kinase inhibitors were tested in 20-point dose-response curves using three PI3K isoforms, two phosphatidylinositol 4,5 bisphosphate lipid substrates (side chain length either 8 or 16 carbons) or without substrate (PI3Kα only). The data represent the results of a single experiment performed with duplicate wells for each inhibitor concentration. Between 2.7% and 11% ATP was consumed in all control PI3K reactions resulting in Z’ values of 0.64 to 0.89, except the PI3Kγ lipid (2.0%) reaction, which yielded a Z’ = 0.44.
Table 2. IC50 Values for Inhibition of PI3K Isoforms Using Time Resolved-Fluorescence Resonance Energy Transfer ADP Assay.
|
PI3Kα IC50 Values (μM) |
PI3Kβ IC50 Values (μM) |
PI3Kγ IC50 Values (μM) |
|||||
|---|---|---|---|---|---|---|---|
| Inhibitor | C16 | C8 | no substrate | C16 | C8 | C16 | C8 |
| Wortmannin | 0.004 | 0.035 | 0.018 | 0.027 | 0.009 | 0.006 | 0.003 |
| PI 103 | 0.036 | 0.023 | 0.017 | 0.058 | 0.038 | 0.18 | 0.34 |
| PI3Kγ inhibitor |
0.026 | 0.020 | 0.083 | 0.39 | 0.12 | 0.020 | 0.031 |
| LY 294002 | 0.59 | 0.14 | 0.42 | 1.1 | 0.67 | 2.3 | 3.6 |
| PI3KγII inhibitor |
6.3 | 9.1 | 10 | 17 | >20 | 1.1 | 3.2 |
| Quercetin | >20 | 11 | 15 | 7.8 | >20 | 6.7 | 13 |
Six known PI3 kinase inhibitors were tested in 20-point dose-response curves using three PI3K isoforms, two phosphatidylinositol 4,5 bisphosphate lipid substrates (side chain length either 8 or 16 carbons) or without substrate (PI3Kα only). The data represent the results of a single experiment performed with duplicate wells for each inhibitor concentration. Between 1.8% and 8.0% ATP was consumed in all control PI3K reactions resulting in Z’ values of 0.60 to 0.82, except the PI3Kβ/C16 lipid (1.1%) and the PI3Kγ/C16 lipid (1.6%) reaction, which yielded Z’ values of 0.45 and 0.35 respectively.
Similar rank orders of inhibitor potency were observed independently for the β and γ isoforms irrespective of the acceptor substrate or the detection method used. Wortmannin, the most potent inhibitor for all three isoforms showed no significant isoform selectivity with IC50 values ranging from 2 nM to 35 nM depending on the acceptor substrate and detection method used. PI 103 was the second most potent inhibitor for the α and β isoforms, with IC50 values ranging from 17 nM to 58 nM; however, in the case of the γ isoform, the PI3Kγ inhibitor - which was developed to be selective for the γ isoform - was more potent than PI 103. Interestingly, this is due both to lower IC50 values for the PI3Kγ inhibitor with the γ isoform and to higher IC50 values for PI 103 (160 nM to 340 nM) relative to the other two isoforms. IC50 values determined for the PI3Kγ inhibitor ranged from 19 nM to 83 nM for PI3Kα, from 120 nM to 450 nM for PI3Kβ, and from 16 nM to 31 nM for PI3Kγ, depending on the lipid substrate and detection method used. A similar trend is shown for the isoforms using the less effective PI3KγII inhibitor. Quercetin inhibited the γ isoform more effectively than the α and β isoforms, with IC50 values in of 7.0 μM to 7.5 μM using FP detection and 6.7 μM to 13 μM using TR-FRET detection.
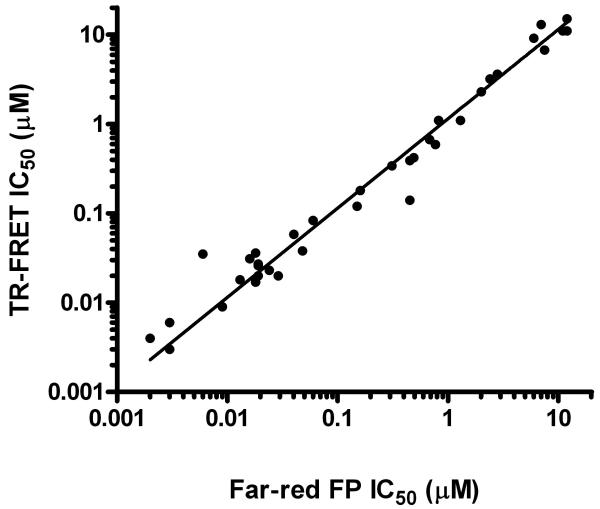
To compare overall concordance between the TR-FRET and FP-based assays we plotted all of the experimentally determined IC50 values, with the TR-FRET data on the Y-axis and the FP data on the X-axis (Fig. 6). This data includes a total of 36 IC50 values including 40 data points for each inhibitor titration. There is excellent correlation between TR-FRET and FP ADP detection (r2 = 0.93). The IC50 values determined using TR-FRET were on average 15% higher that those determined using FP, which is not statistically significant given the error in the measurements.
FIG. 6.
Correlation of IC50 values measured by time-resolved fluorescence resonance energy transfer (TR-FRET) and fluorescence polarization (FP) ADP detection. PI3K isoform inhibitor potency IC50 data for six inhibitors (Wortmannin, PI 103, PI3Kγ inhibitor, LY 294002, PI3KγII inhibitor, and Quercetin) determined using TR-FRET and FP detection methods was plotted in a scattergraph with the line equivalence shown using Graphpad PRISM software. A significant correlation (r2 = 0.93) was obtained for the comparison.
DISCUSSION
Identifying the lipid substrate and optimizing conditions for its preparation are important considerations in a lipid kinase HTS campaign. Decisions regarding the need to use the longer chain (C16) physiological substrate versus the shorter, more soluble lipids must take into account their relative ease of use, effects on enzyme activity and potential differences in inhibitor pharmacology. Examining these parameters in a systematic way requires a kinase enzyme assay method that is not affected by alterations to the lipid substrate. The assay methods most frequently used for lipid kinases do not meet these requirements because they rely on the use of modified lipid substrates - most commonly fluorescently labeled - which interact with detection reagents such as proteins or immobilized metal ions upon phosphorylation. Such assay methods place restrictions on the lipid composition, concentration, and preparation methods that can be used.
Detection of ADP or ATP provides alternative kinase assay methods that are well suited for examining the substrate requirements of lipid kinases. Though monitoring ADP formation - or ATP depletion - has long been used as a generic kinase assay method, the concept of direct immunodetection of ADP is novel, and it overcomes the key problems associated with prior methods. Earlier ADP detection methods relied on the use of coupling enzymes - as many as four - to produce a chromogenic or fluorescent product such as NADH or resorufin [35,36]. Such assays are prone to interference from test compounds and assay components, especially in light of the fact that one of the coupling enzymes is generally a glycolytic kinase, such as pyruvate kinase [37,38]. Luciferase-based ATP depletion assays rely on a single coupling enzyme that is relatively resistant to ATP-competitive inhibitors found in HTS kinase compound libraries [23], however this method suffers from the kinetic complications associated with any substrate depletion assay. That is, as much as 40–50% of the ATP must be consumed to observe a significant decrease in luminescence signal (assay window). This can cause non-linear kinetics - especially since ATP is often used at or near its Km - and thus results in deviations from the Michaelis-Menten assumptions commonly used to determine inhibitor potencies.
In contrast, the Transcreener™ method enables direct detection of ADP generated in a kinase reaction without the use of coupling enzymes. The use of a competitive binding immunoassay format allows optimization of assay conditions, most importantly antibody concentration, so that robust initial velocity measurements are possible over a broad range of initial ATP concentrations [17]. Thus in this study, all kinase reactions had between 1.8 and 11% ATP consumption and the Z’ values were significantly greater than 0.5, with the exception of 4 (of 14) enzyme reactions which progressed to ≤2% ATP conversion (Z’ = 0.17 to 0.45). Though the potential for binding of ATP-competitive ligands to the ADP antibody exists, the ability of the antibody to discriminate between ADP and related nucleotides such as ATP, AMP, and cAMP with ≥85-fold selectivity [17] make this an unlikely possibility.
The dependence of assay signal on enzyme concentration provides basic validation for ADP immunodetection as an assay method for PI3 kinases. The C16 phosphatidylinositol 4,5 bisphosphate is the physiological substrate for all of the PI3K isoforms used in this study, yet we observed a similar dose response using the C8 substrate (Fig. 2 and Fig. 3) in the case of PI3Kα. Because of its ease of preparation, the shorter lipid might be a more attractive choice for an automated HTS setting. However, to make this decision, one would want to be aware of any potential differences in inhibitor pharmacology that might be observed. Although our study was limited to six inhibitors, the results indicate that the length of the fatty acid side chain of the lipid did not affect the general pharmacology for PI3Kα, PI3Kβ or PI3Kγ. Using either substrate, there were only the expected minor deviations in the rank order of inhibitors for a given isoform, and the IC50 values varied less than five-fold for any specific inhibitor and isoform (Table 1 and Table 2). The largest variability in IC50 values is shown for the inhibitor Wortmannin. The IC50 values for this covalent inhibibor [31,32], are dependent on preincubation time with enzyme prior the addition of ATP to begin the reaction [30,39].
The ability to measure PI3K activity in the absence of a lipid substrate provides yet another option, though the large amounts of enzyme required - approximately 300-fold more than in the presence of the C8 substrate - would probably limit its use to cases where substrates are unknown or very difficult to obtain. It should be noted that the capacity of kinases to catalyze phosphoryl transfer to water in the absence of an acceptor is well described for protein kinases [23,40-42]. This inherent activity generally occurs at less than 10% of the rate observed for substrate phosphorylation [40,42] and is largely eliminated in the presence of substrate [41]. Moreover, in cases where inhibitor pharmacology has been investigated, similar results are obtained in the presence and absence of peptide acceptors [43]. Our extension of a similar observation to lipid kinases is the first reported to our knowledge.
The Transcreener™ FP ADP and TR-FRET ADP Assays show very good correlation to one another. The kinase activity data sets obtained using the FP and TR-FRET ADP detection modes were very similar, with an excellent correlation coefficient equal to 0.93 (Fig. 6). Both FP and TR-FRET methods are equally suitable for examining lipid kinase biochemistry or for inhibitor screening, with the decision dependent on compatibility with specific reagents and/or the compound classes being used. We have found that the FP assay generally gives less variability in control reactions (i.e., high and low mP values), is less susceptible to interference from exogenous agents, and has greater signal stability. The TR-FRET detection module is 3-fold more sensitive, and thus may be preferable in cases where enzyme is in short supply. The variability and sensitivity differences can be attributed reagent choices for antibody, tracer, and buffer components in the FP and TR-FRET detection mixtures.
ACKNOWLEDGMENTS
This work was supported by NIH SBIR grant CA110535-01A1.
Contributor Information
TONY A. KLINK, BellBrook Labs (608-227-4505; tony.klink@bellbrooklabs.com)
KAREN M. KLEMAN-LEYER, BellBrook Labs (608-227-4504; karen.kleman@bellbrooklabs.com)
ANDREW KOPP, BellBrook Labs (608-227-4511; andy.kopp@bellbrooklabs.com)
THANE A. WESTERMEYER, BellBrook Labs (608-227-4500; thane.westermeyer@bellbrooklabs.com)
ROBERT G. LOWERY, BellBrook Labs (608-227-4501; bob.lowery@bellbrooklabs.com)
REFERENCES
- 1.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 4.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 5.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 6.Ruckle T, Schwarz MK, Rommel C. PI3Kgamma inhibition: towards an ‘aspirin of the 21st century’? Nat Rev Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
- 7.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 8.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 9.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Li P, Feldberg L, Kim SC, Bowman M, Hollander I, Mallon R, Wolf SF. A directly labeled TR-FRET assay for monitoring phosphoinositide-3-kinase activity. Comb Chem High Throughput Screen. 2006;9:565–570. doi: 10.2174/138620706777935360. [DOI] [PubMed] [Google Scholar]
- 12.Stankewicz C, Rininsland FH. A Robust Screen for Inhibitors and Enhancers of Phosphoinositide-3 Kinase (PI3K) Activities by Ratiometric Fluorescence Superquenching. J Biomol Screen. 2006 doi: 10.1177/1087057106286402. [DOI] [PubMed] [Google Scholar]
- 13.Fuchikami K, Togame H, Sagara A, Satoh T, Gantner F, Bacon KB, Reinemer P. A versatile high-throughput screen for inhibitors of lipid kinase activity: development of an immobilized phospholipid plate assay for phosphoinositide 3-kinase gamma. J Biomol Screen. 2002;7:441–450. doi: 10.1177/108705702237676. [DOI] [PubMed] [Google Scholar]
- 14.Drees BE, Weipert A, Hudson H, Ferguson CG, Chakravarty L, Prestwich GD. Competitive fluorescence polarization assays for the detection of phosphoinositide kinase and phosphatase activity. Comb Chem High Throughput Screen. 2003;6:321–330. doi: 10.2174/138620703106298572. [DOI] [PubMed] [Google Scholar]
- 15.Meier TI, Cook JA, Thomas JE, Radding JA, Horn C, Lingaraj T, Smith MC. Cloning, expression, purification, and characterization of the human Class Ia phosphoinositide 3-kinase isoforms. Protein Expr Purif. 2004;35:218–224. doi: 10.1016/j.pep.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Deems RA. Interfacial enzyme kinetics at the phospholipid/water interface: practical considerations. Anal Biochem. 2000;287:1–16. doi: 10.1006/abio.2000.4766. [DOI] [PubMed] [Google Scholar]
- 17.Kleman-Leyer KM, Klink TA, Kopp AA, Westermeyer TA, Staeben MJ, van de Kar ST, Zaman GJR, Hornberg JJ, Lowery RG. Design and Characterization of the Transcreener ADP Detection Assay for Kinases and other ATP-utilizing Enzymes. In preparation for Assay and Drug Development Technologies. 2008 [Google Scholar]
- 18.Lowery RG, Kleman-Leyer K. Transcreener: screening enzymes involved in covalent regulation. Expert Opin Ther Targets. 2006;10:179–190. doi: 10.1517/14728222.10.1.179. [DOI] [PubMed] [Google Scholar]
- 19.Huss KL, Blonigen PE, Campbell RM. Development of a TranscreenerTM Kinase Assay for Protein Kinase A and Demonstration of Concordance of Data with a Filter-Binding Assay Format. J Biomol Screen. 2007 doi: 10.1177/1087057107300221. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Zalameda L, Kim KW, Wang M, McCarter JD. Discovery of acetyl-coenzyme A carboxylase 2 inhibitors: comparison of a fluorescence intensity-based phosphate assay and a fluorescence polarization-based ADP Assay for high-throughput screening. Assay Drug Dev Technol. 2007;5:225–235. doi: 10.1089/adt.2006.045. [DOI] [PubMed] [Google Scholar]
- 21.Vedvik KL, Eliason HC, Hoffman RL, Gibson JR, Kupcho KR, Somberg RL, Vogel KW. Overcoming compound interference in fluorescence polarization-based kinase assays using far-red tracers. Assay Drug Dev Technol. 2004;2:193–203. doi: 10.1089/154065804323056530. [DOI] [PubMed] [Google Scholar]
- 22.Turek-Etienne TC, Lei M, Terracciano JS, Langsdorf EF, Bryant RW, Hart RF, Horan AC. Use of red-shifted dyes in a fluorescence polarization AKT kinase assay for detection of biological activity in natural product extracts. J Biomol Screen. 2004;9:52–61. doi: 10.1177/1087057103259346. [DOI] [PubMed] [Google Scholar]
- 23.Kashem MA, Nelson RM, Yingling JD, Pullen SS, Prokopowicz AS, 3rd, Jones JW, Wolak JP, Rogers GR, Morelock MM, Snow RJ, Homon CA, Jakes S. Three mechanistically distinct kinase assays compared: Measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors. J Biomol Screen. 2007;12:70–83. doi: 10.1177/1087057106296047. [DOI] [PubMed] [Google Scholar]
- 24.Nix B, Wild D. Calibration curve-fitting. ed 2nd edition Nature Publishing Group; New York: 2001. [Google Scholar]
- 25.Carpenter CL, Duckworth BC, Auger KR, Cohen B, Schaffhausen BS, Cantley LC. Purification and characterization of phosphoinositide 3-kinase from rat liver. J Biol Chem. 1990;265:19704–19711. [PubMed] [Google Scholar]
- 26.Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol. 2003;170:2647–2654. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- 27.Kerchner KR, Clay RL, McCleery G, Watson N, McIntire WE, Myung CS, Garrison JC. Differential sensitivity of phosphatidylinositol 3-kinase p110gamma to isoforms of G protein betagamma dimers. J Biol Chem. 2004;279:44554–44562. doi: 10.1074/jbc.M406071200. [DOI] [PubMed] [Google Scholar]
- 28.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 29.Woscholski R, Kodaki T, McKinnon M, Waterfield MD, Parker PJ. A comparison of demethoxyviridin and wortmannin as inhibitors of phosphatidylinositol 3-kinase. FEBS Lett. 1994;342:109–114. doi: 10.1016/0014-5793(94)80482-6. [DOI] [PubMed] [Google Scholar]
- 30.Downing GJ, Kim S, Nakanishi S, Catt KJ, Balla T. Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin sensitivity of type III phosphatidylinositol kinases. Biochemistry. 1996;35:3587–3594. doi: 10.1021/bi9517493. [DOI] [PubMed] [Google Scholar]
- 31.Stoyanova S, Bulgarelli-Leva G, Kirsch C, Hanck T, Klinger R, Wetzker R, Wymann MP. Lipid kinase and protein kinase activities of G-protein-coupled phosphoinositide 3-kinase gamma: structure-activity analysis and interactions with wortmannin. Biochem J. 1997;324(Pt 2):489–495. doi: 10.1042/bj3240489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norman BH, Shih C, Toth JE, Ray JE, Dodge JA, Johnson DW, Rutherford PG, Schultz RM, Worzalla JF, Vlahos CJ. Studies on the mechanism of phosphatidylinositol 3-kinase inhibition by wortmannin and related analogs. J Med Chem. 1996;39:1106–1111. doi: 10.1021/jm950619p. [DOI] [PubMed] [Google Scholar]
- 34.Knight ZA, Chiang GG, Alaimo PJ, Kenski DM, Ho CB, Coan K, Abraham RT, Shokat KM. Isoform-specific phosphoinositide 3-kinase inhibitors from an arylmorpholine scaffold. Bioorg Med Chem. 2004;12:4749–4759. doi: 10.1016/j.bmc.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M. A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 2004;16:3304–3325. doi: 10.1105/tpc.104.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schirmer A, Kennedy J, Murli S, Reid R, Santi DV. Targeted covalent inactivation of protein kinases by resorcylic acid lactone polyketides. Proc Natl Acad Sci U S A. 2006;103:4234–4239. doi: 10.1073/pnas.0600445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monasterio O, Cardenas ML. Kinetic studies of rat liver hexokinase D (‘glucokinase’) in non-co-operative conditions show an ordered mechanism with MgADP as the last product to be released. Biochem J. 2003;371:29–38. doi: 10.1042/BJ20020728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart JM, Blakely JA. Long chain fatty acids inhibit and medium chain fatty acids activate mammalian cardiac hexokinase. Biochim Biophys Acta. 2000;1484:278–286. doi: 10.1016/s1388-1981(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 39.Reichling L, Lebakken C, Vedvik K, Kopp L, Robers M, Riddle S, Vogel K. A Homogenous Fluroescence-Based Assay for mTOR Kinase Activity; Poster presented at the Keystone Symposium; Sante Fe, NM. February 2007. [Google Scholar]
- 40.Ward NE, O’Brian CA. The intrinsic ATPase activity of protein kinase C is catalyzed at the active site of the enzyme. Biochemistry. 1992;31:5905–5911. doi: 10.1021/bi00140a029. [DOI] [PubMed] [Google Scholar]
- 41.Rominger CM, Schaber MD, Yang J, Gontarek RR, Weaver KL, Broderick T, Carter L, Copeland RA, May EW. An intrinsic ATPase activity of phospho-MEK-1 uncoupled from downstream ERK phosphorylation. Arch Biochem Biophys. 2007;464:130–137. doi: 10.1016/j.abb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Fox T, Fitzgibbon MJ, Fleming MA, Hsiao HM, Brummel CL, Su MS. Kinetic mechanism and ATP-binding site reactivity of p38gamma MAP kinase. FEBS Lett. 1999;461:323–328. doi: 10.1016/s0014-5793(99)01488-x. [DOI] [PubMed] [Google Scholar]
- 43.Chen G, Porter MD, Bristol JR, Fitzgibbon MJ, Pazhanisamy S. Kinetic mechanism of the p38-alpha MAP kinase: phosphoryl transfer to synthetic peptides. Biochemistry. 2000;39:2079–2087. doi: 10.1021/bi9919495. [DOI] [PubMed] [Google Scholar]