Abstract
Neuroglobin (Ngb) is a tissue globin specifically expressed in neurons. Our laboratory and others have shown that Ngb overexpression protects neurons against hypoxia/ischemia, but the underlying mechanisms remain poorly understood. Recent studies demonstrate that hypoxia/ischemia induces a multitude of spatially and temporally regulated responses in gene expression, and initial evidence suggested that Ngb might function in altering biological processes of gene expression. In this study, we asked how Ngb may help regulate genes responsive to hypoxia. Expression of hypoxic response genes following oxygen glucose deprivation (OGD) was examined using mRNA arrays in Ngb-overexpressing transgenic (Ngb-Tg) and wild type (WT) neurons. From a total of 113 genes on the micro-array, mRNA expression of 65 genes were detected. Under rest condition, 14 genes were downregulated in Ngb-Tg neurons compared to WT. In WT neurons, after 4-hr OGD followed by 4-hr reoxygenation (O4/R4), 20 genes were significantly downregulated, and only Fos mRNA was significantly increased. However, out of the 20 downregulated genes in WT neurons, 12 of them were no longer significantly changed in Ngb-Tg neurons: Add1, Arnt2, Camk2g, Cstb, Dr1, Epas1, Gna11, Hif1a, Il6st, Khsrp, Mars and Rara. Among these 12 genes, 8 (Add1, Camk2g, Cstb, Dr1, Epas1, Gna11, Hif1a, Khsrp) were already reduced in Ngb-Tg neurons compared to WT under rest conditions. Additionally, 3 genes that initially showed no changes in WT neurons (Ctgf, Egfr and Pea15) were downregulated after OGD in the Ngb-Tg neurons. These findings suggest that Ngb overexpression modulates mRNA expression of multiple hypoxic response genes in the early phase after O/R. Further studies on these gene networks and interactions may lead to better understanding of Ngb in signaling pathways that contribute to neuroprotection.
Keywords: Neuroglobin, Oxygen Glucose Deprivation, Hypoxic Response Genes, Oligo GEArray Hybridization
Neuroglobin (Ngb) is a recently discovered tissue globin with a high affinity for oxygen that is widely expressed in vertebral central and peripheral nerve systems as well as retina and endocrine tissues (Burmester and Hankeln 2004, Awenius et al. 2001, Zhang et al. 2001, Burmester et al. 2000, Garry and Mammen 2003). As a newly discovered member of globin family, Ngb has been considered the brain or nerve equivalent of tissue haemoglobin (Mammen et al. 2002). Previous studies demonstrated that overexpression of Ngb is neuroprotective against hypoxic/ischemic brain injuries (Khan et al. 2006, Sun et al. 2003, Wang et al. 2008, Liu et al. 2009).
However, how Ngb protects neurons against hypoxia/ischemia still remains unclear (Greenberg et al. 2008). In general, tissue globins mediate multiple cellular and molecular responses to hypoxic/ischemic insults. For example, myoglobin in cardiomyocytes and oxidative skeletal myofibers helps facilitate oxygen transport, maintain nitric oxide homeostasis, and scavenge reactive oxygen species (Garry and Mammen 2003). It is possible that Ngb has similar actions in brain. Importantly, Ngb expression is confined to metabolically most active, oxygen-consuming cell types (Burmester and Hankeln 2004). Those cells including neurons are more sensitive and vulnerable to hypoxic/ischemic insults. Importantly, emerging reports suggested that Ngb may function in regulating gene expression and signaling pathway against hypoxia/ischemia. For example, Ngb can act as a guanine nucleotide dissociation inhibitor (GDI) by inhibiting the exchange rate of GDP for GTP (Wakasugi et al. 2003). Ngb also interacts with Flotillin, a protein existing with lipid raft that are critical for signal transduction (Wakasugi et al. 2004). In the context of metabolic and hypoxic challenge, understanding how Ngb regulates hypoxic gene expression will be very helpful in better defining the neuroprotective roles and mechanisms of Ngb.
In this study we screened mRNA profiles of hypoxic response genes before and after OGD in cultured WT and Ngb-Tg cortical neurons with a “Mouse Hypoxia Signaling Pathway Microarray”, which contains 113 genes related to oxidative stress, apoptosis, protein metabolism, and transcription regulation. Our data showed that (1) stereotyped alterations in hypoxia-response gene pathways are triggered by OGD in neurons, (2) Ngb overexpression significantly alters these gene profiles even under resting baseline conditions, and (3) Ngb overexpression eliminated downregulation of mRNA expression of some genes related to neuronal survival and function after OGD.
Experimental procedures
Animals
All animal experiments were performed following protocols approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee in compliance with the NIH Guide for the Care and Use of Laboratory Animals. Wild type C57BL/6 mice were used as control in this study. Ngb over-expression transgenic (Ngb-Tg) mice were previously generated in our lab (Wang et al. 2008).
Primary cortical neuronal culture
Primary neuronal culture was prepared from the cortex of embryonic day 15 mouse. In brief, the cortical neurons were suspended in neuron-defined culture medium and plated onto poly-D-lysine-coated 35-mm dishes (3×105 cells per dish). Neural basal medium supplemented with 2% B27, 0.3 mM L-Glutamine and 1% Penicillin-Streptomycin was used. Half of the medium was replaced every 3 days.
Western blot
At 9 days of culture, cortical neurons were collected for protein extraction. Ngb protein levels were examined by western blot using rabbit anti-Ngb antibody (BioVendor) following the protocol as previously described (Wang et al. 2008). Relative Ngb protein levels were assessed by quantification of optical density of Ngb protein bands with NIH Image software.
Oxygen-glucose deprivation (OGD) and reoxygenation
The neuron cultures were used for experiments at 9 days after plating. Oxygen-glucose deprivation (OGD) experiments were performed using a specialized, humidified chamber (Heidolph, incubator 1000, Brinkmann instruments, Westbury, NY) kept at 37°C, which contained an anaerobic gas mixture (90% N2, 5% H2, and 5% CO2). To initiate OGD, culture medium was replaced with deoxygenated, glucose-free extracellular solution-Locke's medium (154 mM NaCl, 5.6 mM KCl, 2.3 mM CaCl2, 1.0 mM MgCl2, 3.6 mM NaHCO3, 5 mM HEPES, pH 7.2). After 4 hrs' challenge, cultures were removed from the anaerobic chamber, and the OGD solution in the cultures was replaced with maintenance medium. Cells were then allowed to recover for 20 hrs (for neurotoxicity assay) or 4 hrs (for Oligo GEArray hybridization) in a regular incubator.
Neurotoxicity
Lactate dehydrogenase (LDH) release assay was used to measure neurotoxicity. LDH release is an indicator of plasma membrane damage and commonly used for the determination of neurotoxicity as we previously described (Wang et al. 2002).
Oligo GEArray hybridization
“Oligo GEArray® Mouse Hypoxia Signaling Pathway Microarray” was obtained from SuperArray Bioscience Corporation (Fredrick, MD). Total RNA was isolated from four groups of primary neuron culture samples: wild type (WT) cells, WT after OGD treatment, Ngb-Tg cells, Ngb-Tg cells after OGD treatment. Amplification labeling of cRNA was conducted following the user’s manual of TrueLabeling-AMP 2.0 Kit. Hybridization was performed following the manual of Oligo GEArray® Reagent Kit and repeated three times using RNA samples isolated from 3 sets of cell cultures. Hybridization images were analyzed using the GEArray Expression Analysis Suite. Gene spots with signal intensity higher than the blank on the same membrane are considered detectable.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Quantitative RT-PCR was performed to validate the expression patterns of several randomly selected genes using total RNA similar as that used for Oligo GEArray hybridization. The primer sets of these genes, the RT2 first strand kit and the RT2 qPCR master mix were obtained from SuperArray Bioscience Corp. RT-PCR was performed following the instruction of “RT2 qPCR Primer Assays” kit. Data were analyzed according to the comparative threshold cycle (Ct) method with B2m (Beta-2 microglobulin) for sample normalization, which was also used for data normalization in Oligo GEArray hybridization.
Statistical analysis
We used ANOVA followed by Tukey-HSD posthoc tests in data analysis for Ngb protein level, LDH release and Oligo GEArray hybridization data. Data were given as mean±standard deviation (SD). P<0.05 was considered statistically significant. If the overall p value is lower than 0.05 by ANOVA for each gene’s mRNA levels under four conditions (WT, WTOGD, Ngb-Tg, NgbOGD), and additionally the pairwise Tukey-HSD posthoc result is also significant (P<0.05), then the change between that pair of conditions was considered statistically significant. Heat-map of Oligo array result was generated using the “Cluster” and “TreeView” programs from the Eisen Lab (Lawrence Berkeley National Lab, Berkeley, CA).
Results
Ngb protein levels in WT and Ngb-Tg neurons, under normal condition or after O4/R4
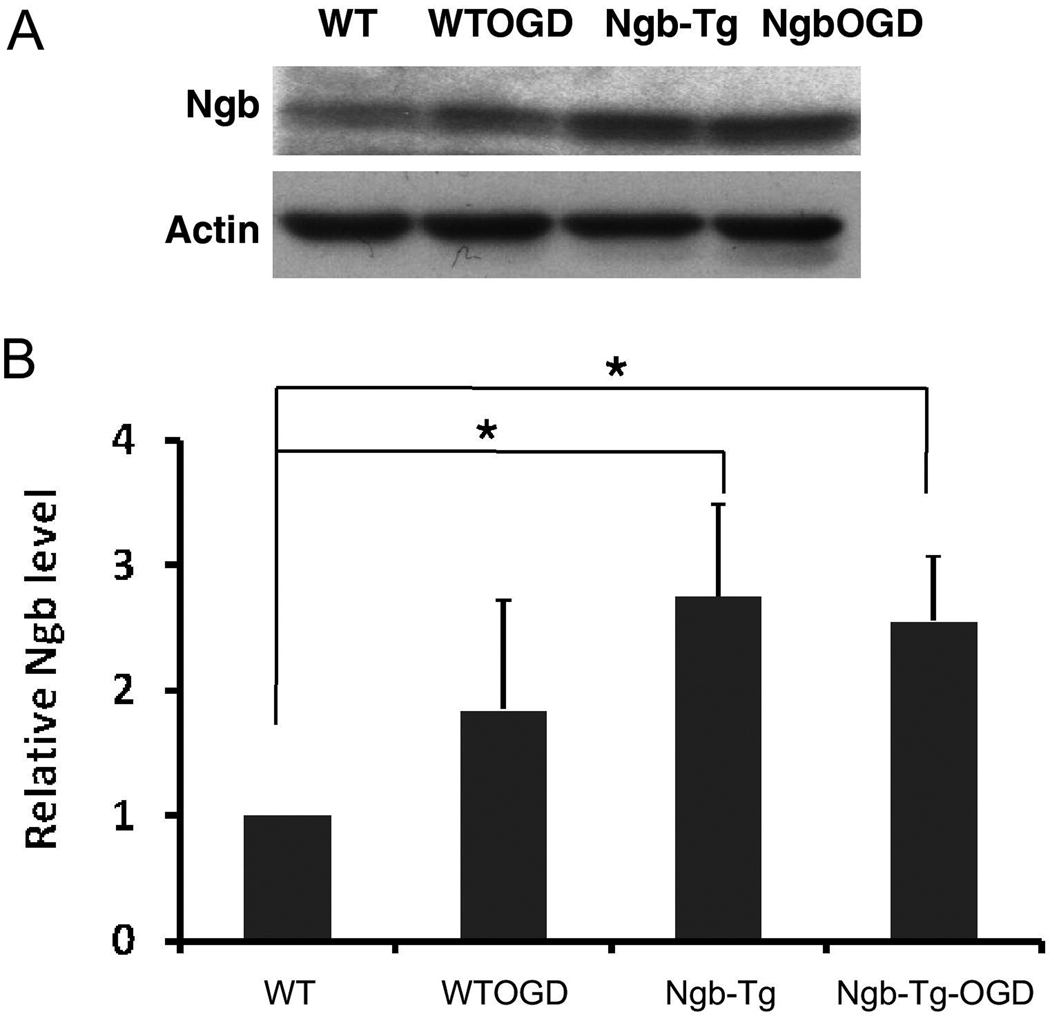
Total proteins were extracted from WT and Ngb-Tg neuron, either under normal condition, or after being treated by O4/R4. Western blotting was performed using rabbit anti-Ngb antibody (BioVendor) to examine Ngb protein levels as we previously described (Liu et al. 2009). Ngb protein level was slightly increased in WT neurons after O4/R4 (~1.8 fold), but was significantly increased in Ngb-Tg neurons, either under normal condition (~2.7 fold) or after O4/R4 (~2.5 fold) (Fig. 1).
Fig 1.
Ngb protein levels under four conditions: WT (wild type normal condition), WTOGD (wild type after O4/R4), Ngb-Tg (Ngb-transgenic normal condition), NgbOGD (Ngb-transgenic after O4/R4). A, Representative western blot showed Ngb level was slightly increased by O4/R4, and more significantly increased in Ngb-Tg under normal condition or after O4/R4. Actin served as equal loading controls. B, Relative Ngb protein levels were quantified by optical density of Ngb protein bands. Mean+SD, n=2 per group, *P<0.05.
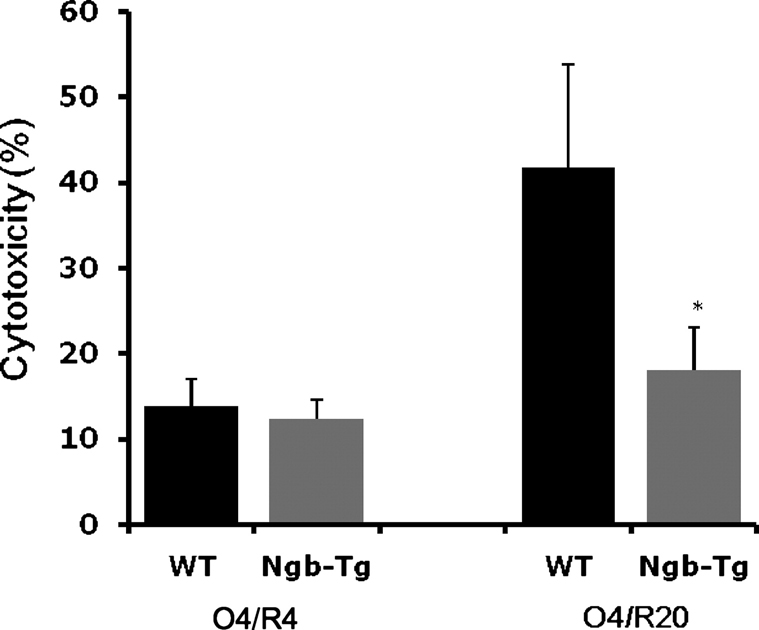
OGD-induced neurotoxicity was reduced in Ngb-Tg neurons compared to WT neurons
The neurotoxicity for WT or Ngb-Tg neurons was examined using a standard LDH release assay after 4-hr OGD followed by reoxygenation for 4 hrs or 20 hrs. Neurotoxicity after 4-hr OGD followed by 4-hr reoxygenation (O4/R4) was about 13% for both WT and Ngb-Tg neurons. In WT neurons, reoxygenation led to a continued evolution of neuron death, and neurotoxicity was increased to 40% by the 20 hr time point. However, Ngb overexpression clearly prevented the continued progression of neuron death (Fig. 2).
Fig 2.
Neurotoxicity of WT and Ngb-Tg neurons after OGD/reoxygenation. At 9 days of culture, primary cortical neurons were treated with 4-hr OGD followed by reoxygenation for 4 hrs (O4/R4) or 20 hrs (O4/R20). The neurotoxicity after OGD/reoxygenation was examined with LDH release assay. Mean±SD, n=5 per group, *P<0.05.
Oligo GEArray hybridization
We performed hybridization using “Oligo GEArray® Mouse Hypoxia Signaling Pathway Microarray” to study the effect of Ngb overexpression on 113 selected hypoxia response genes. Significant reduction of OGD-induced neurotoxicity was observed in Ngb-Tg neurons only after continued reoxygenation for 20 hrs. Thus, it may be reasonable to assume that protective effects of OGD-induced gene expression changes occur at an earlier time point. So we extracted total RNA from cultured neurons treated with 4-hr OGD plus 4-hr reoxygenation (O4/R4). Hybridization was performed using the RNA samples of the following four groups of primary neuron cultures: normal wild type neurons, wild type neurons after O4/R4, normal Ngb-Tg neurons, Ngb-Tg neurons after O4/R4 (Figure 3).
Fig 3.
Oligo GEArray hybridization images. (one set of images representative of three repeats) A. Wild type neurons under normal condition; B. Wild type neurons after O4/R4; C. Ngb-Tg neurons under normal condition; D. Ngb-Tg neurons after O4/R4.
Among the 113 genes included in the “Oligo GEArray® Mouse Hypoxia Signaling Pathway Microarray”, mRNA of 65 genes were detected in our primary neuron cultures (Table 1). The relative mRNA levels of these genes were normalized by internal control Beta-2 microglobulin (B2m) (Table 2). For each gene, three types of comparisons were analyzed: (I), Ngb-Tg normal baseline versus WT normal baseline; (II), WT O4/R4 versus WT normal control; (III), Ngb-Tg O4/R4 versus Ngb-Tg normal control. Statistical analysis was performed for these changes, and only those genes with at least one significant change in the three comparisons (p< 0.05) were listed in Table 2.
Table 1.
Gene symbols and descriptions
| Gene Symbols | Description |
|---|---|
| Cytoskeleton | |
| Add1 | Adducin 1a |
| Response to Stress | |
| Arnt2 | Aryl hydrocarbon receptor nuclear translocator 2 |
| Bax | Bcl2-associated X protein |
| Cat | Catalase |
| Cygb | Cytoglobin |
| Gpx1 | Glutathione peroxidase 1 |
| Kit | Kit oncogene |
| Mt3 | Metallothionein 3 |
| Sod2 | Superoxide dismutase 2, mitochondrial |
| Transcription Factors and Regulators: | |
| Dr1 | Down-regulator of transcription 1 |
| Epas1 | Endothelial PAS domain protein 1 |
| Fos | FBJ osteosarcoma oncogene |
| Hif1a | Hypoxia inducible factor 1, alpha subunit |
| Id1 | Inhibitor of DNA binding 1 |
| Id2 | Inhibitor of DNA binding 2 |
| Mybl2 | Myeloblastosis oncogene-like 2 |
| Mycn | V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| Notch1 | Notch gene homolog 1 (Drosophila) |
| Rara | Retinoic acid receptor, alpha |
| Signal Transduction | |
| Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) |
| Crebbp | CREB binding protein |
| Dctn2 | Dynactin 2 |
| Egfr | Epidermal growth factor receptor |
| Gna11 | Guanine nucleotide binding protein, alpha 11 |
| Il6st | Interleukin 6 signal transducer |
| Iqgap1 | IQ motif containing GTPase activating protein 1 |
| Psme2 | Proteasome (prosome, macropain) 28 subunit, beta |
| Extracellular Matrix (ECM)-Related Molecules | |
| Agtpbp1 | ATP/GTP binding protein 1 |
| Cstb | Cystatin B |
| Ctgf | Connective tissue growth factor |
| Mmp14 | Matrix metallopeptidase 14 (membrane-inserted) |
| Npy | Neuropeptide Y |
| Psmb3 | Proteasome (prosome, macropain) subunit, beta type 3 |
| Protein metabolism | |
| CamK2g | Calcium/calmodulin-dependent protein kinase II gamma |
| Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
| Eef1a1 | Eukaryotic translation elongation factor 1 alpha 1 |
| Mars | Methionine-tRNA synthetase |
| Ppp2cb | Protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| Rpl28 | Ribosomal protein L28 |
| Rpl32 | Ribosomal protein L32 |
| Rps37 | Ribosomal protein L37 |
| Rps2 | Ribosomal protein S2 |
| Rps7 | Ribosomal protein S7 |
| Rps8 | Ribosomal protein S8 |
| Spnb2 | Spectrin beta 2 |
| Sumo2 | SMT3 suppressor of mif two 3 homolog 2 (yeast) |
| Tuba3a | Tubulin, alpha 3A |
| Metabolism | |
| Gsto1 | Glutathione S-transferase omega 1 |
| Khsrp | KH-type splicing regulatory protein |
| Man2b1 | Mannosidase 2, alpha B1 |
| Mocs3 | Molybdenum cofactor synthesis 3 |
| Nudt2 | Nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| Sae1 | SUMO1 activating enzyme subunit 1 |
| Snrp70 | U1 small nuclear ribonucleoprotein polypeptide A |
| Cell growth | |
| Gap43 | Growth associated protein 43 |
| Gpi1 | Glucose phosphate isomerase 1 |
| Oxidoreductase | |
| Sdh1 (sord) | Sorbitol dehydrogenase |
| Apoptosis | |
| Pea15 | Phosphoprotein enriched in astrocytes 15 |
| Hemoglobin Complex Associated Genes | |
| Hbb | Hemoglobin Y, beta-like embryonic chain |
| Hmox1 | Heme oxygenase (decycling) 1 |
Table 2.
Relative gene mRNA levels of WT or Ngb-Tg neurons before and after O4/R4
| Gene Symbol |
WT | WT-OGD | Ngb-Tg | Ngb-OGD |
|---|---|---|---|---|
| Mean±SD | Mean± SD | Mean± SD | Mean± SD | |
| Gapdh | 8.15±0.79 | 7.23±0.62 | 7.66±0.14 | 8.07±0.57 |
| Add1 | 0.62±0.32 | 0.26±0.10 | 0.35±0.13 | 0.27±0.05 |
| Agtpbp1 | 0.83±0.09 | 0.42±0.32 | 0.68±0.25 | 0.73±0.13 |
| Arnt2 | 0.13±0.08 | 0.04±0.02 | 0.08±0.05 | 0.10±0.09 |
| Bax | 1.06±0.12 | 0.97±0.12 | 1.02±0.09 | 0.94±0.09 |
| Camk2g | 0.15±0.04 | 0.06±0.04 | 0.05±0.01 | 0.07±0.09 |
| Cat | 0.83±0.08 | 0.30±0.14 | 0.22±0.11 | 0.11±0.02 |
| Cdc42 | 6.09±0.48 | 4.04±0.30 | 5.77±0.44 | 3.03±0.27 |
| Crebbp | 0.33±0.19 | 0.52±0.17 | 0.37±0.24 | 0.29±0.15 |
| Cstb | 0.70±0.08 | 0.39±0.08 | 0.40±0.05 | 0.45±0.06 |
| Ctgf | 0.36±0.08 | 0.30±0.13 | 0.64±0.24 | 0.29±0.02 |
| Cygb | 0.79±0.15 | 0.48±0.10 | 0.50±0.15 | 0.42±0.07 |
| Dctn2 | 4.31±0.27 | 3.59±0.21 | 4.57±0.55 | 2.44±0.28 |
| Dnajb1 | 1.98±0.15 | 1.32±0.12 | 2.01±0.14 | 1.05±0.06 |
| Dr1 | 0.18±0.01 | 0.05±0.01 | 0.06±0.02 | 0.05±0.01 |
| Eef1a1 | 8.19±0.68 | 8.18±0.61 | 5.76±0.53 | 5.46±0.37 |
| Ece1 | 0.61±0.78 | 0.33±0.33 | 0.52±0.68 | 0.24±0.16 |
| Egfr | 0.21±0.03 | 0.16±0.04 | 0.21±0.01 | 0.13±0.04 |
| Egln2 | 3.29±0.27 | 1.75±0.21 | 2.87±0.24 | 2.04±0.14 |
| Eno1 | 0.42±0.48 | 0.16±0.15 | 0.19±0.22 | 0.13±0.13 |
| Epas1 | 0.40±0.03 | 0.17±0.06 | 0.16±0.06 | 0.11±0.01 |
| Fos | 0.17±0.07 | 1.20±0.11 | 0.55±0.38 | 1.03±0.09 |
| Gap43 | 5.14±0.37 | 4.66±0.41 | 4.05±0.44 | 2.38±0.19 |
| Gna11 | 0.16±0.01 | 0.11±0.05 | 0.08±0.03 | 0.07±0.04 |
| Gpi1 | 5.24±0.38 | 4.04±0.40 | 3.84±0.43 | 3.01±0.27 |
| Gpx1 | 1.07±0.06 | 0.93±0.01 | 0.77±0.17 | 0.62±0.09 |
| Gsto1 | 1.55±0.07 | 1.05±0.05 | 1.10±0.05 | 0.88±0.09 |
| Hbb-y | 4.38±0.08 | 4.64±0.11 | 4.03±0.13 | 2.15±0.08 |
| Hif1a | 0.18±0.06 | 0.06±0.02 | 0.05±0.01 | 0.04±0.01 |
| Hk2 | 0.45±0.58 | 0.20±0.21 | 0.19±0.23 | 0.11±0.10 |
| Hmox1 | 0.34±0.02 | 0.54±0.18 | 0.19±0.12 | 0.28±0.11 |
| Id1 | 0.29±0.08 | 0.28±0.17 | 0.11±0.07 | 0.07±0.03 |
| Id2 | 3.71±0.28 | 1.52±0.09 | 2.58±0.15 | 1.02±0.04 |
| Il6st | 0.92±0.06 | 0.44±0.17 | 0.79±0.21 | 0.68±0.13 |
| Iqgap1 | 0.19±0.10 | 0.16±0.09 | 0.15±0.08 | 0.10±0.04 |
| Khsrp | 0.93±0.08 | 0.54±0.10 | 0.54±0.16 | 0.28±0.12 |
| Kit | 0.20±0.07 | 0.03±0.01 | 0.11±0.04 | 0.04±0.01 |
| Man2b1 | 0.20±0.06 | 0.08±0.02 | 0.24±0.11 | 0.16±0.05 |
| Mars | 0.20±0.06 | 0.05±0.02 | 0.12±0.07 | 0.08±0.07 |
| Mmp14 | 0.59±0.16 | 0.08±0.02 | 0.20±0.07 | 0.02±0.01 |
| Mocs3 | 0.42±0.15 | 0.10±0.03 | 0.19±0.08 | 0.10±0.04 |
| Mt3 | 1.52±0.07 | 0.90±0.03 | 1.43±0.11 | 0.61±0.07 |
| Mybl2 | 0.23±0.02 | 0.18±0.07 | 0.16±0.03 | 0.13±0.01 |
| Nmyc1 | 2.73±0.16 | 1.25±0.29 | 1.18±0.07 | 0.29±0.02 |
| Notch1 | 0.72±0.21 | 0.36±0.18 | 0.61±0.31 | 0.13±0.06 |
| Npy | 2.70±0.17 | 2.11±0.09 | 3.51±0.34 | 1.50±0.12 |
| Nudt2 | 0.32±0.06 | 0.10±0.02 | 0.16±0.04 | 0.04±0.02 |
| Pea15 | 1.00±0.06 | 0.69±0.19 | 0.81±0.13 | 0.38±0.13 |
| Ppp2cb | 1.09±0.10 | 0.90±0.08 | 1.01±0.08 | 0.73±0.17 |
| Psmb3 | 4.32±0.27 | 3.44±0.31 | 4.61±0.53 | 1.90±0.14 |
| Psme2 | 0.96±0.15 | 0.34±0.23 | 0.75±0.50 | 0.70±0.11 |
| Rara | 0.14±0.01 | 0.07±0.01 | 0.09±0.03 | 0.11±0.05 |
| Rpl28 | 1.89±0.16 | 1.47±0.10 | 1.56±0.12 | 0.74±0.04 |
| Rpl32 | 5.96±0.37 | 6.72±0.72 | 6.22±0.84 | 3.95±0.27 |
| Rpl37 | 0.36±0.11 | 0.23±0.04 | 0.36±0.04 | 0.31±0.04 |
| Rps2 | 6.99±0.37 | 6.96±0.41 | 6.51±0.54 | 4.74±0.37 |
| Rps7 | 0.48±0.06 | 0.26±0.05 | 0.38±0.10 | 0.25±0.14 |
| Rps8 | 0.07±0.05 | 0.06±0.03 | 0.07±0.05 | 0.04±0.02 |
| Sdh1 | 1.07±0.04 | 1.10±0.02 | 0.89±0.14 | 0.95±0.03 |
| Snrp70 | 1.10±0.07 | 1.07±0.06 | 1.04±0.07 | 0.94±0.11 |
| Sod2 | 2.65±0.28 | 1.62±0.12 | 2.84±0.21 | 1.10±0.09 |
| Spnb2 | 0.11±0.07 | 0.09±0.06 | 0.15±0.07 | 0.08±0.03 |
| Sumo2 | 6.14±0.36 | 6.04±0.73 | 3.69±0.43 | 2.55±0.29 |
| Sae1a | 1.10±0.06 | 0.85±0.19 | 1.02±0.08 | 0.90±0.11 |
| Tuba1a | 5.78±0.48 | 6.06±0.59 | 5.49±0.74 | 3.31±0.27 |
| Tuba3a | 0.38±0.35 | 0.24±0.24 | 0.24±0.22 | 0.35±0.25 |
| Blank | 0.11±0.05 | 0.09±0.02 | 0.10±0.04 | 0.10±0.04 |
| Blank | 0.19±0.03 | 0.16±0.04 | 0.21±0.08 | 0.21±0.14 |
| Rps27a | 5.13±0.35 | 5.82±0.63 | 5.17±0.52 | 5.07±0.38 |
| B2m | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 | 1.00±0.00 |
| Hsp90ab1 | 6.16±0.38 | 6.26±0.41 | 6.18±0.45 | 6.79±0.48 |
| Ppia | 7.46±0.57 | 7.04±0.60 | 7.18±0.85 | 8.09±0.57 |
| BAS2C | 1.07±0.12 | 1.13±0.10 | 1.11±0.14 | 0.97±0.14 |
mRNA levels were normalized by internal control gene B2m. “WT” and “Ngb-Tg” refer to wild type and Ngb-Tg cells under normal rest condition, respectively; “WT-OGD” and “Ngb-OGD” refer to wild type and Ngb-Tg neurons after OGD, respectively. BAS2C is an artificial sequence serving as a detection control.
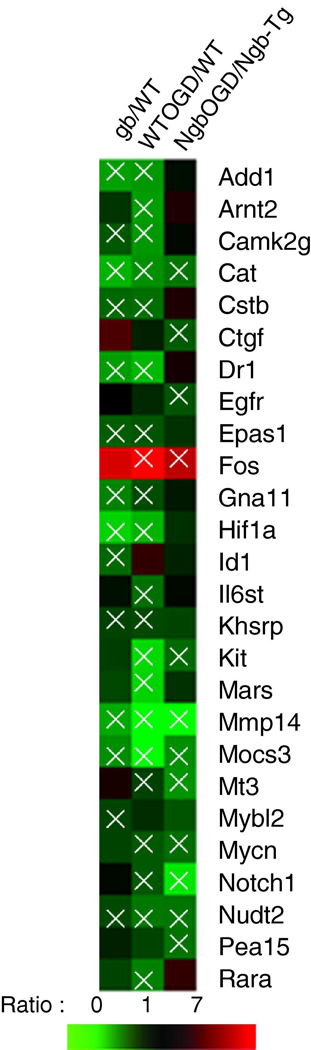
Gene mRNA profile was affected by Ngb overexpression under normal condition
Our results showed that under normal baseline conditions, mRNA levels of 14 genes in Ngb-Tg neurons were significantly reduced compared to WT neurons (p <0.05, Fig. 4, supplementary result). These genes include: Add1, Camk2g, Cat, Cstb, Dr1, Epas1, Gna11, Hif-1a , Id1, Khsrp, Mmp14, Mocs3, Mybl2, Nudt2.
Fig 4.
Heat-map of mRNA level comparisons. Green indicates reduced mRNA levels and red indicates elevated levels to each dedicated comparisons. For each gene, three comparisons of mRNA levels are made. Comparison I, mRNA levels in Ngb-Tg versus WT neurons under rest condition; Comparison II, mRNA levels in WT-OGD condition versus WT rest condition; Comparison III, mRNA levels in Ngb-Tg OGD condition versus Ngb-Tg rest condition. Significant mRNA level changes were marked by white crosses. The original relative data of mRNA expression were presented in Supplemental data, Table 2.
Gene mRNA profile was changed after O4/R4 in WT neurons
In WT neurons, mRNA levels of 20 genes were significantly reduced by OGD, and only Fos mRNA was significantly increased (Fig. 4, supplementary result). The downregulated genes include: Add1, Arnt2, Camk2g, Cat, Cstb, Dr1, Epas1, Gna11, Hif-1a, Il6st, Khsrp, Kit, Mars, Mmp14, Mocs3, Mt3, Mycn, Notch1, Nudt2, Rara.
Gene mRNA profile was changed after OGD in Ngb-Tg neurons
WT and Ngb-Tg neurons responded to the OGD challenge very differently. Overexpression of Ngb in the transgenic neurons seemed to decrease the number of genes that were downregulated by OGD; only 11 genes were significantly decreased: Cat, Ctgf, Egfr, Kit, Mmp14, Mocs3, Mt3, Mycn, Notch1, Nudt2, Pea15 (Fig. 4, supplementary result). But just as in WT neurons, Fos was still significantly upregulated (Table 2, comparison III).
Comparison of gene mRNA responses to OGD/reoxygenation between WT and Ngb-Tg neurons
The mRNA level changes of each gene in response to O4/R4 were compared in both WT and Ngb-Tg neurons (Fig. 4, supplementary result). Twelve of the genes that were downregulated by OGD in WT neurons were no longer significantly decreased in Ngb-Tg neurons: Add1, Arnt2, Camk2g, Cstb, Dr1, Epas1, Gna11, Hif1a, Il6st, Khsrp, Mars and Rara. On the other hand, mRNA levels of Ctgf, Egfr and Pea15 were downregulated in Ngb-Tg neurons, but these 3 genes were not significantly altered in WT neurons.
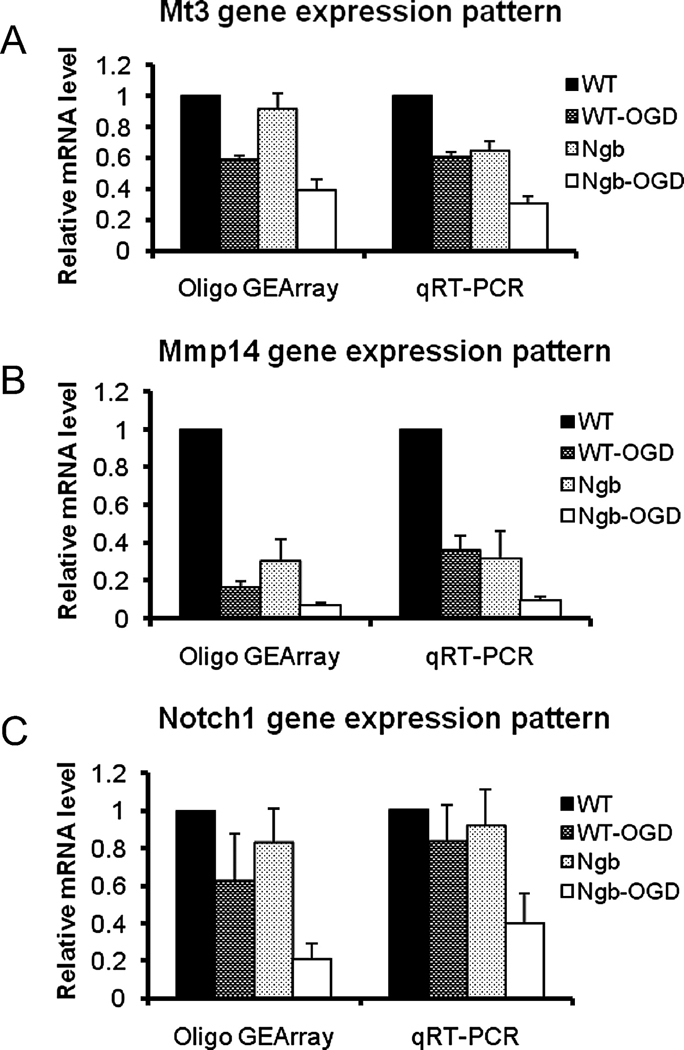
Validation of the microarray findings by quantitative RT-PCR
To validate the reliability of this Oligo GEArray approach, we performed quantitative RT-PCR with the same RNA samples for three representative genes: Mmp14, Mt3 and Notch1. The overall expression pattern of each gene revealed by RT-PCR is similar to that obtained by Oligo GEArray Hybridization (Fig. 5). Overexpression of Ngb significantly downregulated Mmp14, but not Mt3 or Notch1. For Mt3 and Notch1, OGD downregulated their expression in WT neurons, and this downregulation was enhanced in Ngb-Tg neurons.
Fig 5.
Gene expression patterns obtained from Oligo GEArray Hybridization compared with qRT-PCR. The same RNA samples used for Oligo GEArray hybridization were analyzed by qRT-PCR to four different neuron cultures: WT, WT-OGD (WT after O4/R4), Ngb (Ngb-Tg), Ngb-OGD (Ngb-Tg after O4/R4). Three randomly selected genes, Mmp14 (A), Mt3 (B) and Notch1 (C), were tested. Mean±SD, n=3 per group.
Discussion
Since its discovery in 2000 (Burmester et al. 2000), experimental studies have documented that Ngb overexpression is neuroprotective against hypoxia/ischemia (Garry and Mammen 2003, Khan et al. 2006, Sun et al. 2003, Wang et al. 2008) and direct oxidative stress (Li et al. 2008, Jin et al. 2008). However, the neuroprotective mechanisms remain mysterious and not fully elucidated (Nienhaus and Nienhaus 2007, Brunori and Vallone 2006, Brunori and Vallone 2007, Greenberg et al. 2008).
Ngb was initially considered to be an O2 storage protein. However, this function alone seems insufficient for protecting neurons against hypoxic/ischemic insults, in part due to the high P50 value of Ngb’s O2 binding (Fago et al. 2004) and low Ngb concentration in the brain (Burmester et al. 2000). It was also suggested that Ngb may function as a scavenger of NO and reactive oxygen species (ROS) (Herold et al. 2004, Liu et al. 2009). A recent study indicated that Ngb overexpression eliminated hypoxia-induced mitochondrial aggregation and neuron death (Khan et al. 2008), which is consistent with our previous findings that Ngb overexpression ameliorated mitochondria function disruption caused by hypoxia/reoxygenation (H/R) (Liu et al. 2009). Finally, accumulating evidence now indicate a crucial role of Ngb as a sensor molecule for signal transduction in response to hypoxia/ischemia (Wakasugi et al. 2003, Watanabe and Wakasugi 2008, Wakasugi et al. 2004).
In the context of hypoxia, neurons are known to have stereotyped responses of specific patterns of genes that contribute to overlapping events after injury, such as apoptotic cell death, inflammation, necrosis, and neurogenesis. In recent years, the development of multiplex gene analysis tools, such as DNA, mRNA and protein microarrays, have allowed us to assess larger-scale gene responses in brain after stroke and neurodegeneration (Lu et al. 2003, Kobori et al. 2002, Schmidt-Kastner et al. 2002). In this study, we asked whether Ngb confers neuroprotection against hypoxic/ischemic brain injury via modifying the expression of specific hypoxic/ischemia response genes.
As the first step, in this study we used Mouse Hypoxia Signaling Pathway Microarray to screen the mRNA profiles of total 113 hypoxic response genes under normal rest condition and after OGD/reoxygenation in cultured WT and Ngb-Tg cortical neurons. In this study, neuron death mostly occurred during the reoxygenation phase after OGD. During the early 4 hr time point, there were no clear differences in neurotoxicity between WT and Ngb-Tg neurons. But by the later 20 hr time point, WT neurons had continued to die up to 40%, whereas Ngb-Tg neurons had significantly lesser amounts of injury (20%). Hence, effects of Ngb on these early alterations in hypoxic response genes should at least partially contribute to the neuroprotective actions of Ngb.
From a total 113 genes, mRNA for 65 genes on this microarray were detectable. The overall response after OGD seemed to be dominated by a downregulation of many genes. In WT neurons, 20 genes were decreased, and only Fos mRNA was significantly increased. Out of the 20 downregulated genes in WT neurons, 12 of them were no longer altered by OGD in Ngb Tg neurons. Those 12 genes distributed to multiple gene groups, they were Add1, Arnt2, Camk2g, Cstb, Dr1, Epas1, Gna11, Hif1a, Il6st, Khsrp, Mars and Rara. However, 8 of these 12 genes (Add1, Camk2g, Cstb, Dr1, Epas1, Gna11, Hif1a, Khsrp) were already reduced under rest conditions in Ngb-Tg neurons compared to WT. These genes’ downregulation in WT after OGD may at least be partially ascribed to the OGD-induced Ngb level increase (Fig. 1). Above overlapping of certain genes’ down-regulation indicates Ngb-overexpression may have “pre-conditioning”-like effect. Among the 12 genes, except unidentified relations of genes Dr1, Il6st and Mars to neuron survival, all other genes have been suggested to play roles in neuronal function and survival, indicating that maintaining multiple cell survival signaling pathways might be one mechanism for Ngb’s neuroprotection (Rabenstein et al. 2005, Maltepe et al. 2000, Wayman et al. 2008, Kaasik et al. 2007, zur Nedden et al. 2008, Shinozaki et al. 2007). In contrast, levels of Ctgf, Egfr and Pea15 genes were downregulated by OGD in Ngb-Tg neurons whereas they were not initially altered in WT neurons, however roles of these three genes in neuronal function and survival have not been defined. Nevertheless, how these various genes relate to specific pathways of neuron death versus survival requires further dissection. But at least, this study provides a comprehensive database for these future studies.
There are several caveats in this study. First, the random integration of the Ngb overexpression construct during the transgenic process may cause constitutive mutant and adaptive compensations, and these might affect gene regulation in unknown ways. For example, we found that Ngb overexpression reduced the basal levels of several genes under resting basal conditions (Table 2). Since Ngb protein is primarily localized in cytoplasm of neurons (Geuens et al. 2003), it may not directly regulate gene expression by serving as a transcription factor. But it might indirectly interact with the regulation elements of these genes. Consequently, it is possible that altered gene expressions in Ngb-Tg neurons under normal rest condition may also play a role in further responses to hypoxia/ischemia and underlie its protective mechanisms. Second, we only tested gene mRNA level at one time point after OGD/R in this study, i.e, 4-hr OGD followed by 4-hr reoxygenation. This may not be the peak expression time point for all genes. It is therefore important to examine the expression change in a continuous time course for any specific gene in future investigations.
In summary, our results documented that Ngb overexpression alters the expression of a group of genes involved in hypoxia signaling pathways, suggesting this could at least partially contribute to mechanisms of Ngb’s neuroprotective actions against hypoxia/OGD. Further studies to dissect these pathways will help us understand how Ngb works in brain, and this may ultimately reveal novel therapeutic approaches for stroke and other neurodegenerative disorders.
Supplementary Material
Acknowledgments
This work was supported in part by a NIH grant R01-NS049476 and a Scientist Development Grant 0435087N from American Heart Association (to X.W.), and NIH grants R01-NS48422, R01-NS53560, and P01-NS55104 (to E.H.L).
References
- Awenius C, Hankeln T, Burmester T. Neuroglobins from the zebrafish Danio rerio and the pufferfish Tetraodon nigroviridis. Biochem Biophys Res Commun. 2001;287:418–421. doi: 10.1006/bbrc.2001.5614. [DOI] [PubMed] [Google Scholar]
- Brunori M, Vallone B. A globin for the brain. FASEB J. 2006;20:2192–2197. doi: 10.1096/fj.06-6643rev. [DOI] [PubMed] [Google Scholar]
- Brunori M, Vallone B. Neuroglobin, seven years after. Cell Mol Life Sci. 2007;64:1259–1268. doi: 10.1007/s00018-007-7090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester T, Hankeln T. Neuroglobin: a respiratory protein of the nervous system. News Physiol Sci. 2004;19:110–113. doi: 10.1152/nips.01513.2003. [DOI] [PubMed] [Google Scholar]
- Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, Weber RE. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J Biol Chem. 2004;279:44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Mammen PP. Neuroprotection and the role of neuroglobin. Lancet. 2003;362:342–343. doi: 10.1016/S0140-6736(03)14055-X. [DOI] [PubMed] [Google Scholar]
- Geuens E, Brouns I, Flamez D, Dewilde S, Timmermans JP, Moens L. A globin in the nucleus! J Biol Chem. 2003;278:30417–30420. doi: 10.1074/jbc.C300203200. [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Jin K, Khan AA. Neuroglobin: an endogenous neuroprotectant. Curr Opin Pharmacol. 2008;8:20–24. doi: 10.1016/j.coph.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Fago A, Weber RE, Dewilde S, Moens L. Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress. J Biol Chem. 2004;279:22841–22847. doi: 10.1074/jbc.M313732200. [DOI] [PubMed] [Google Scholar]
- Jin K, Mao XO, Xie L, Khan AA, Greenberg DA. Neuroglobin protects against nitric oxide toxicity. Neurosci Lett. 2008;430:135–137. doi: 10.1016/j.neulet.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik A, Kuum M, Aonurm A, Kalda A, Vaarmann A, Zharkovsky A. Seizures, ataxia, and neuronal loss in cystatin B heterozygous mice. Epilepsia. 2007;48:752–757. doi: 10.1111/j.1528-1167.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- Khan AA, Ou Mao X, Banwait S, Dermardirossian CM, Bokoch GM, Jin K, Greenberg DA. Regulation of hypoxic neuronal death signaling by neuroglobin. FASEB J. 2008 doi: 10.1096/fj.07-100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Wang Y, Sun Y, Mao XO, Xie L, Miles E, Graboski J, Chen S, Ellerby LM, Jin K, Greenberg DA. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17944–17948. doi: 10.1073/pnas.0607497103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori N, Clifton GL, Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res Mol Brain Res. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- Li RC, Morris MW, Lee SK, Pouranfar F, Wang Y, Gozal D. Neuroglobin protects PC12 cells against oxidative stress. Brain Res. 2008;1190:159–166. doi: 10.1016/j.brainres.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yu Z, Guo S, Lee SR, Xing C, Zhang C, Gao Y, Nicholls DG, Lo EH, Wang X. Effects of neuroglobin overexpression on mitochondrial function and oxidative stress following hypoxia/reoxygenation in cultured neurons. J Neurosci Res. 2009;87:164–170. doi: 10.1002/jnr.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Tang Y, Ran R, Clark JF, Aronow BJ, Sharp FR. Genomics of the periinfarction cortex after focal cerebral ischemia. J Cereb Blood Flow Metab. 2003;23:786–810. doi: 10.1097/01.WCB.0000062340.80057.06. [DOI] [PubMed] [Google Scholar]
- Maltepe E, Keith B, Arsham AM, Brorson JR, Simon MC. The role of ARNT2 in tumor angiogenesis and the neural response to hypoxia. Biochem Biophys Res Commun. 2000;273:231–238. doi: 10.1006/bbrc.2000.2928. [DOI] [PubMed] [Google Scholar]
- Mammen PP, Shelton JM, Goetsch SC, Williams SC, Richardson JA, Garry MG, Garry DJ. Neuroglobin, a novel member of the globin family, is expressed in focal regions of the brain. J Histochem Cytochem. 2002;50:1591–1598. doi: 10.1177/002215540205001203. [DOI] [PubMed] [Google Scholar]
- Nienhaus K, Nienhaus GU. Searching for neuroglobin's role in the brain. IUBMB Life. 2007;59:490–497. doi: 10.1080/15216540601188538. [DOI] [PubMed] [Google Scholar]
- Rabenstein RL, Addy NA, Caldarone BJ, Asaka Y, Gruenbaum LM, Peters LL, Gilligan DM, Fitzsimonds RM, Picciotto MR. Impaired synaptic plasticity and learning in mice lacking beta-adducin, an actin-regulating protein. J Neurosci. 2005;25:2138–2145. doi: 10.1523/JNEUROSCI.3530-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Zhang B, Belayev L, Khoutorova L, Amin R, Busto R, Ginsberg MD. DNA microarray analysis of cortical gene expression during early recirculation after focal brain ischemia in rat. Brain Res Mol Brain Res. 2002;108:81–93. doi: 10.1016/s0169-328x(02)00516-8. [DOI] [PubMed] [Google Scholar]
- Shinozaki Y, Sato Y, Koizumi S, Ohno Y, Nagao T, Inoue K. Retinoic acids acting through retinoid receptors protect hippocampal neurons from oxygen-glucose deprivation-mediated cell death by inhibition of c-jun-N-terminal kinase and p38 mitogen-activated protein kinase. Neuroscience. 2007;147:153–163. doi: 10.1016/j.neuroscience.2007.04.032. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jin K, Peel A, Mao XO, Xie L, Greenberg DA. Neuroglobin protects the brain from experimental stroke in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3497–3500. doi: 10.1073/pnas.0637726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Nakano T, Kitatsuji C, Morishima I. Human neuroglobin interacts with flotillin-1, a lipid raft microdomain-associated protein. Biochem Biophys Res Commun. 2004;318:453–460. doi: 10.1016/j.bbrc.2004.04.045. [DOI] [PubMed] [Google Scholar]
- Wakasugi K, Nakano T, Morishima I. Oxidized human neuroglobin acts as a heterotrimeric Galpha protein guanine nucleotide dissociation inhibitor. J Biol Chem. 2003;278:36505–36512. doi: 10.1074/jbc.M305519200. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu J, Zhu H, Tejima E, Tsuji K, Murata Y, Atochin DN, Huang PL, Zhang C, Lo EH. Effects of neuroglobin overexpression on acute brain injury and long-term outcomes after focal cerebral ischemia. Stroke. 2008;39:1869–1874. doi: 10.1161/STROKEAHA.107.506022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mori T, Sumii T, Lo EH. Hemoglobin-induced cytotoxicity in rat cerebral cortical neurons: caspase activation and oxidative stress. Stroke. 2002;33:1882–1888. doi: 10.1161/01.str.0000020121.41527.5d. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Wakasugi K. Neuroprotective function of human neuroglobin is correlated with its guanine nucleotide dissociation inhibitor activity. Biochem Biophys Res Commun. 2008;369:695–700. doi: 10.1016/j.bbrc.2008.02.089. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CG, Li L, Deng MY, Xie F, Wang CL, Zhou WQ, Wang HY, He FC. [Coding region cDNA sequence cloning of rat neuroglobin gene, its polymorphism feature and tissue expression profile analysis] Yi Chuan Xue Bao. 2001;28:997–1001. [PubMed] [Google Scholar]
- zur Nedden S, Tomaselli B, Baier-Bitterlich G. HIF-1 alpha is an essential effector for purine nucleoside-mediated neuroprotection against hypoxia in PC12 cells and primary cerebellar granule neurons. J Neurochem. 2008;105:1901–1914. doi: 10.1111/j.1471-4159.2008.05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.