Abstract
A career of following unplanned observations has serendipitously led to a deep appreciation of the capacity that bacterial cells have for restructuring their genomes in a biologically responsive manner. Routine characterization of spontaneous mutations in the gal operon guided the discovery that bacteria transpose DNA segments into new genome sites. A failed project to fuse λ sequences to a lacZ reporter ultimately made it possible to demonstrate how readily Escherichia coli generated rearrangements necessary for in vivo cloning of chromosomal fragments into phage genomes. Thinking about the molecular mechanism of IS1 and phage Mu transposition unexpectedly clarified how transposable elements mediate large-scale rearrangements of the bacterial genome. Following up on lab lore about long delays needed to obtain Mu-mediated lacZ protein fusions revealed a striking connection between physiological stress and activation of DNA rearrangement functions. Examining the fate of Mudlac DNA in sectored colonies showed that these same functions are subject to developmental control, like controlling elements in maize. All these experiences confirmed Barbara McClintock's view that cells frequently respond to stimuli by restructuring their genomes and provided novel insights into the natural genetic engineering processes involved in evolution.
THIS article is the reminiscence of a bacterial geneticist studying the processes of mutation and DNA rearrangements. I want to emphasize how my experience was full of surprises and unplanned discoveries that took me ever deeper into the mechanisms and regulation of natural genetic engineering by Escherichia coli cells.
For the benefit of younger molecular geneticists, there are at least three points to be made. First, you can find something truly novel only when you do not know exactly what you are looking for. If the experiment comes out just as you planned, you have not really learned anything you did not already know or suspect.
Second, routine characterization of your experimental material is critical because it will tell you where your understanding is incomplete—but only when the characterizations do not come out as you expect. In other words, it can be a good thing if an experimental result does not fit your expectations.
Third, science will inevitably lead us in the future to think about the subjects that we are studying in ways that we cannot currently predict. When I began my research, we thought we understood the basics of genome expression and mutation because we knew about DNA, RNA polymerase, and the triplet code for amino acids. The worlds of transcriptional regulation beyond simple repressor–operator models, signal transduction, chromatin formatting, transcript processing, protein modifications, and regulatory RNAs were all in the future. In my particular field, the molecular basis of genetic change, discoveries about mobile genetic elements, reverse transcription, programmed genome rearrangements, and other aspects of what I call “natural genetic engineering” were yet to be made.
The following account relates my own experimental journey into a new way of thinking about the molecular and cellular basis of genetic change. After detailing the journey, I will explain why and how I believe that this new mode of thought is likely to influence our ideas about evolution, the most basic of biological subjects.
I would be remiss if I did not acknowledge the powerful influence of Barbara McClintock on my thinking. After meeting her in 1976, I realized that she possessed an unmatched depth of experience about all aspects of biology, from natural history to the current status of molecular genetics. We engaged in a 16-year dialogue up to her death in 1992 (Shapiro 1992c). Only after many years did I finally come to appreciate the wisdom of her insistence that the ability of cells to sense and respond to “genome shock” was just as important in determining what happens to their genomes as are the biochemical mechanisms that they use in repairing and restructuring DNA molecules (McClintock 1984).
DISCOVERING DNA INSERTIONS
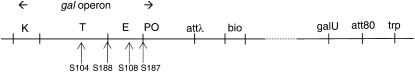
When I was a student at the University of Cambridge in the mid-1960s, I planned to study the effect of transcription on mutagenesis. Sydney Brenner at the MRC Laboratory for Molecular Biology suggested that a positive selection system for loss-of-function mutations would be the most sensitive experimental arrangement. The E. coli gal operon offered such a system in which the resulting mutations could easily be analyzed. A defect in the galU locus (Figure 1) made the bacteria sensitive to galactose (Gals phenotype). These galU mutants accumulated Gal-1-P, and the phosphorylated sugar inhibited growth. However, when the bacteria lost the galK-encoded galactokinase activity, they no longer accumulated Gal-1-P and shifted from a Gals to a Galr phenotype. Therefore, GalK− mutants could be selected as colonies on medium containing galactose and glycerol. Since galU was not linked to the galETK operon (Shapiro 1966), it was easy to separate the GalK-inactivating mutations from the original galU mutation for reversion studies and mapping.
Figure 1.—
The gal and trp regions of the E. coli chromosome. The dashed line indicates >400 kb between the two operons. This distance makes galU mutations easy to separate from gal operon mutations that inactivate galK expression. The positions of four strongly polar insertion mutations are indicated by the vertical arrows. The figure is based on the deletion mapping of Shapiro and Adhya (1969).
Following this plan, I isolated a series of spontaneous Galr mutants and proceeded to characterize the underlying gal operon mutations (Adhya and Shapiro 1969). Of almost 200 spontaneous GalK-inactivating mutations, 14 proved to be pleiotropic or polar and inactivated at least two of the gal operon's three cistrons. Of these, 10 failed to revert, and most of those were easy to interpret as deletions, which was later confirmed by mapping studies (Shapiro and Adhya 1969). But the pleiotropic gal mutations that reverted proved to be hard to understand using existing ideas about the mechanisms of genetic change. They reverted spontaneously and could be mapped to upstream regions of the galETK operon (Figure 1), but they were not traditional point mutations because reversion was not increased by either base substitution or frameshift mutagens. Other pleiotropic gal mutations isolated by the Lederberg and Starlinger groups behaved similarly (Morse 1967; Saedler and Starlinger 1967).
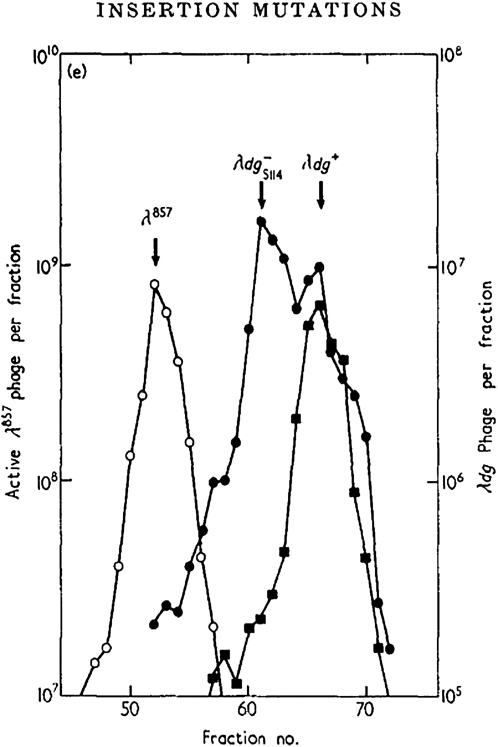
As I wrote my thesis in 1967, it occurred to me that the mysterious pleiotropic gal mutations might result from insertion of extra DNA into the operon (Shapiro 1967). For my postdoc, I went to the Institut Pasteur, where I learned to do CsCl density-gradient centrifugation so that I could confirm the insertion hypothesis using λdgal transducing phages: the λdgal particles carrying the mutations had extra DNA and were denser than the parental phage (Figure 2; Shapiro 1969; Cohen and Shapiro 1980). The Starlinger group quickly repeated these results (Jordan et al. 1968), and electron microscope heteroduplex studies subsequently showed that the same DNA segments inserted into various positions in the gal and lac operons (Fiandt et al. 1972; Hirsch et al. 1972). Thus, we learned that bacteria have the capacity to move specific segments of DNA through the genome, and these segments came to be called “insertion sequences” or IS elements (Starlinger and Saedler 1972). The world of transposable elements had moved from Barbara McClintock's maize cytogenetics (McClintock, 1950, 1953, 1987) to the molecular biology of E. coli (Cohen and Shapiro 1980).
Figure 2.—
The results of CsCl equilibrium density-gradient centrifugation of a mixture of λ-phages (λ 857, plaque-forming phage; λ dg+, phage carrying the wild-type gal operon; λ dgS114−, phage carrying an insertion mutation). The fractions on the left contain the densest particles. From Shapiro (1969) with permission.
CLONING lac INTO λ AND PURIFYING lac DNA WITHOUT RESTRICTION ENZYMES
After Paris, I moved to Jon Beckwith's laboratory at Harvard Medical School. Together with Ethan Signer, Jon had pioneered in vivo genetic manipulation of the lac operon, targeting insertions of Ftslac plasmids into the chromosome and using these insertions to isolate Φ80lac transducing phages (Beckwith et al. 1966). These in vivo genetic engineering methods complemented my own experience with spontaneous mutagenesis and specialized transducing phages. One of my early projects, together with Karen Ippen, was to try making λN-lacZ transcriptional fusions by adapting a double selection method from my thesis research to obtain deletions. The double selection depended upon simultaneous loss of lethal λ functions from a thermoinducible prophage (enabling growth at 42°) and loss of LacI repression (producing blue colonies on X-gal indicator plates). Although we never found the fusions that we sought, the effort ultimately paid off for both Karen and myself. The double selection was useful for isolating deletion mutations of the integrated F, which permitted Karen to map and analyze plasmid transfer functions (Ippen-Ihler et al. 1972). Double selection also allowed us to move the λ attachment site next to the transposed lac operon so that we could easily obtain λplac transducing phages that formed blue plaques on plates that contained a chromogenic β-galactosidase substrate (Ippen et al.1971). Effectively, we used a series of natural in vivo DNA rearrangements to clone defined segments of the lac operon into λ.
The λplac transducing phages provided a ready means of obtaining large quantities of lac operon DNA and proved to be important tools of in vitro genetic engineering in the 1970s. We already realized in 1969 that we had the potential to purify defined lac operon DNA on the basis of these phages. Garret Ihler came up with the clever idea of hybridizing DNA strands whose only complementarity was in lac sequences, so we could then digest away the unhybridized single strands. Fortunately, a Φ80plac transducing phage had the lac operon segment inserted in the opposite orientation to the one in λplac. The DNA isolations, strand separations, annealing, and nuclease digestions all worked the first time. Together with Lorne MacHattie's electron microscope expertise and Larry Eron's hybridization experiments, we were able to demonstrate the first purification of a genetically defined functional segment of DNA (Shapiro et al. 1969). To me, it was significant that all the genetic rearrangements necessary for lac purification had been accomplished by E. coli, not by humans. We simply selected the right products.
PLASMIDS, TRANSPOSONS, PHAGE Mu, AND THE MECHANISM OF TRANSPOSITIONAL DNA EXCHANGE
In 1975, my unrelated work on Pseudomonas hydrocarbon oxidation led me to the seminal ICN-UCLA Squaw Valley Symposium on Bacterial Plasmids. In a way that periodically happens in all fields of science, research from many labs converged at a key meeting to illuminate a general process. In this case, the topic was transposable elements, and the general process was DNA restructuring in bacteria. Great interest had been generated by the ability of plasmids to accumulate multiple antibiotic resistances. Realizing that DNA segments could insert at multiple genomic locations provided a critical part of the explanation. Naomi Datta and Fred Heffron spoke on the mobile β-lactamase element now known as Tn3 (Hedges and Jacob 1974; Heffron et al. 1975), Nancy Kleckner on the tetracycline-resistance transposon Tn10 (Kleckner et al. 1978), Doug Berg on the kanamycin-resistance transposon Tn5 (Berg et al. 1975), and Richard Deonier on the IS elements that integrate the F plasmid into the bacterial chromosome (Deonier and Davidson 1976).
The Squaw Valley meeting led me, my gal operon collaborator Sankar Adhya, and Mu colleague Ahmed Bukhari to organize a meeting on this exciting new field. In May 1976, the first meeting on DNA insertion elements, plasmids, and episomes took place at Cold Spring Harbor Laboratory, Ahmed's home institution (and also that of Barbara McClintock). We had guessed that we would be lucky if 50 people would want to come but were gratifyingly surprised when more than three times as many signed up from all over the world. The meeting led to the first book on mobile genetic elements (Bukhari et al. 1977). Talks on phages, bacteria, yeast, Drosophila, tissue culture cells, animal viruses, and plants made it abundantly clear that a general phenomenon of homology-independent DNA restructuring, previously disparaged as “illegitimate recombination,” was at work in virtually every living organism. “Illegitimate recombination” had suddenly become legitimate.
The mechanistic question of how transposable elements moved through the genome independently of DNA sequence homology became a critical issue. I approached this problem from the perspective of transposable elements as agents of genome restructuring. It had become clear to me that transposable elements not only move themselves around but also link other genomic segments together, as they did in Deonier's integrated F plasmids and in the compound transposon structures that are bounded on either end by IS elements (Tn5, Tn9, and Tn10). Arianne Toussaint and Michel Faelen had shown that the promiscuously inserting phage Mu can join unrelated DNA segments together and duplicate itself while doing so (Faelen and Toussaint 1976; Faelen et al. 1978). A similar duplication event appears as an intermediate in Tn3 transposition (Gill et al. 1978; Heffron et al. 1979), and Lorne MacHattie and I found that IS1 duplicated as it integrated phage λ into the bacterial chromosome (MacHattie and Shapiro 1978; Shapiro and MacHattie 1979). These observations fixed the association between transposable element duplication, DNA restructuring, and transposition in my mind.
Meanwhile, Nigel Grindley had discovered a different kind of duplication event: each newly inserted IS1 was flanked by a duplicated 9-bp sequence from the target site (Grindley 1978). Similar duplications of various sizes were soon found to surround insertions of other transposable elements. These target-site duplications (TSDs) indicated that target DNA was subjected to staggered interruptions of the two strands (9 bp apart in the case of IS1); such interruptions would allow the duplex to come apart during transposition, and the two single-strand overhangs could be copied to generate the TSD.
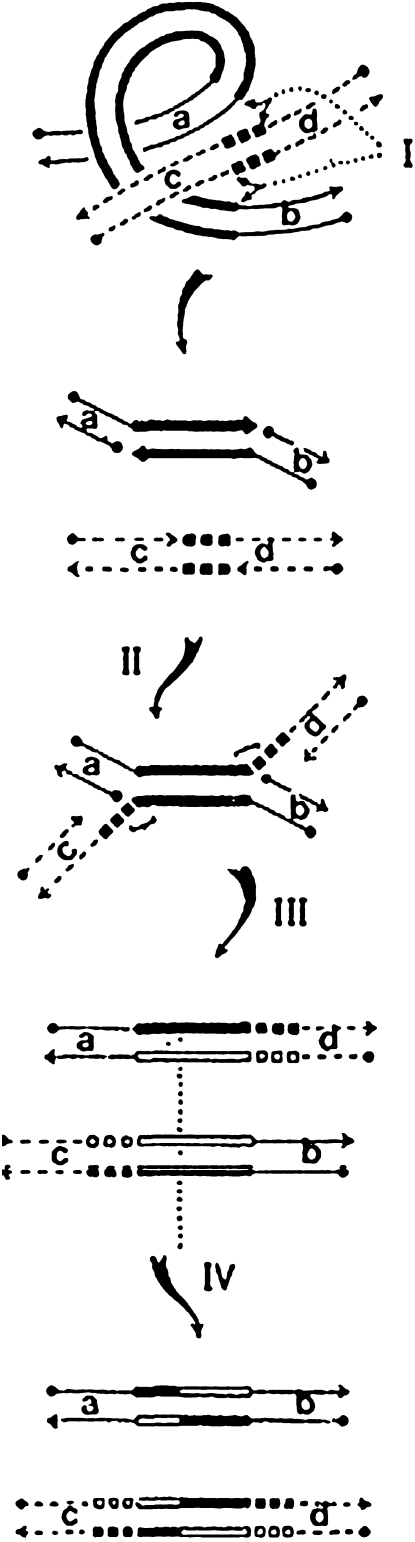
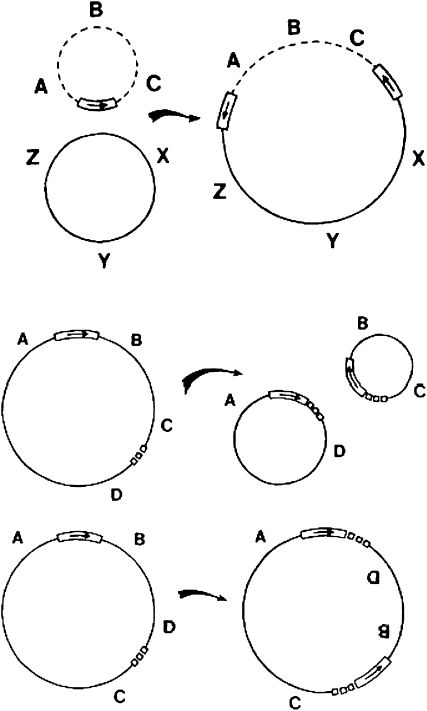
Having puzzled over all these observations for several months, I came home one night and asked myself what would happen if staggered interruptions also occurred at the ends of the transposable element (e.g., Tn3, IS1, or Mu). When I had worked out the consequences (paying close attention to keeping all my 5′ phosphates and 3′ hydroxyls straight so that phosphodiester bonds would open and reseal correctly), it was evident that the postulated sequence of events explained how a duplicated element could transpose from one site to another. The mechanism fortuitously provided an explanation for the puzzling finding that phage Mu could replicate its genome without excising from its original prophage site (Ljungquist and Bukhari 1977): excision was not necessary because the initial strand transfer event created a replication fork at each end of the unexcised Mu prophage (Figure 3). What finally convinced me that the model was likely to prove correct was its ability to explain a large number of other DNA rearrangements associated with IS elements, transposons, and phage Mu (Figure 3; Shapiro 1979). The rearrangements include fusions (or cointegrations) of circular molecules, deletions, and inversions (Figure 4).
Figure 3.—
A molecular model for IS element and phage Mu transposition and replication. The long rectangles indicate the two strands of the transposable element (TE). The sets of three squares indicate the strands of the duplicated target sequence. Arrowheads indicate 3′ hydroxyl ends, and solid circles indicate 5′ phosphate ends. Although the initial strand cleavages are indicated as occurring simultaneously before ligation at step I, we now know that TE termini are first cleaved in the transposasome complex to liberate 3′ hydroxyl groups, which then proceed to attack the target duplex and induce trans-esterification reactions to produce the strand-transfer product illustrated following step II. The open rectangles and boxes after step III represent newly replicated DNA. Many bacterial and the majority of eukaryotic transposons undergo a double-strand cleavage before attacking the target duplex to undergo nonreplicative transposition. This figure is reproduced from Shapiro (1979) with permission.
Figure 4.—
IS- and Mu-mediated DNA rearrangements. The panels illustrate replicon fusion or co-integration (top), deletion formation (middle), and inversion (bottom). These rearrangements occur when the final reciprocal exchange step IV of complete transposition illustrated in Figure 3 fails to occur. The open rectangle containing an arrow represents the transposable element. Note how it becomes duplicated in each process. In the bottom two panels, the small boxes represent the target sequence that is duplicated to form a TSD. Further rearrangements are possible after the ones illustrated here. For example, the BC circle excised in deletion formation can fuse with a target site to create an inserted segment flanked by direct repeats of the transposable element. This process would create a transposon carrying the BC segment. Reproduced and corrected from Shapiro (1979) with permission.
We now understand the molecular details of DNA strand exchange carried out by Mu and many other transposable elements. These elements include retroviruses such as HIV, whose integrase proteins function on the double-stranded products of reverse transcription in the same way as Mu transposase does on the Mu prophage (Mizuuchi and Craigie 1986; Lavoie and Chaconas 1996; Craig et al. 2002). Since novel junctions in all these processes arise by end-to-end joining of DNA strands, as illustrated in Figure 3, no sequence homology is required for their formation, and the locations of insertion events are determined by protein–DNA and protein–protein interactions. We now know that such interactions can serve to target transposable element insertions to quite specific DNA structures or genomic locations, as occurs during the integration of Tn7 into replication forks (Peters and Craig 2001) or of yeast retrotransposons into promoter regions (Kirchner et al. 1995) and silent chromatin (Xie et al. 2001). Thus, transposable elements display a double nonrandomness in their movement through the genome: (1) the same segment of DNA, comprising all its coding sequences and cis-acting signals, is repeatedly moved to new locations, and (2) those locations reflect the action of molecular recognition complexes.
REGULATING TRANSPOSABLE ELEMENT ACTIVITY: Mu ACTIVATION, ADAPTIVE MUTATION, AND COLONY DEVELOPMENT
My first graduate student, Spencer Benson, had as a postdoc made use of the bacteriophage Mu-based lacZ protein fusion method designed by Malcolm Casadaban (Casadaban 1975, 1976). Spencer told me that one had to use thick agar plates for the Casadaban technique because it took a long time for the colonies to appear. Intrigued, I urged Spencer to investigate this delay further, but he had too many other projects underway. So, having spent a lot of time thinking about Mu, I took up the project myself, using Casadaban's original strain for selecting araB-lacZ protein fusions. The results were eye-opening (Shapiro 1984a).
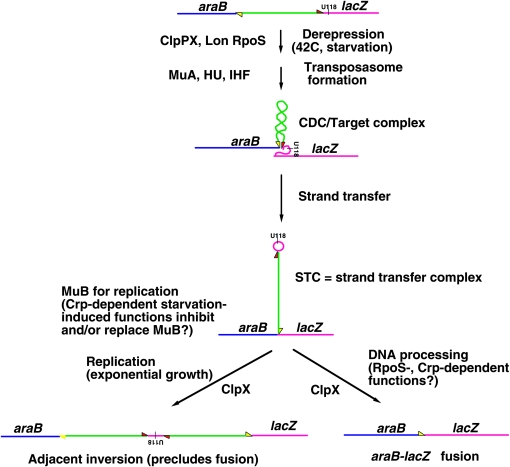
In Casadaban's prefusion strain, a Mu prophage separates the 5′-end of araB from a lacZ sequence that is blocked for transcription (it has no promoter) and translation (it has a chain termination triplet in the dispensable 5′ region) (Figure 5). Growth occurs on a medium with arabinose and lactose only if a deletion event fuses the araB cistron directly as a translational fusion to lacZ. I quickly confirmed what Spencer had told me, finding delays of between 5 and 19 days before the first fusion colonies appeared on selective plates. By reconstructing cultures seeded with a low proportion of preselected araB–lacZ fusion cells, I observed colonies appearing after 2 days of incubation. This control meant that no fusions could have formed during growth prior to plating because, if they had, they would have formed colonies 3 days earlier than I had observed in 187 experiments starting with a total of >3 × 1010 cells plated. I measured the increase in frequency of fusion formation per plated cell to be at least five orders of magnitude! Something interesting must have been happening in that initial prefusion delay after plating. Experiments showing that an additional Mu prophage inhibited colony appearance were an indication that “something” involved Mu derepression (Shapiro 1984a). The appearance of araB–lacZ fusions only (and at high frequency) following an induction process under selective conditions was the first clear demonstration of what later came to be called “adaptive mutation,” a term that I now understand in the sense that McClintock meant it: an increase in mutagenesis as adaptation to some biological challenge (McClintock 1984; Shapiro 1997).
Figure 5.—
Steps in the activation and execution of Mu-mediated araB–lacZ fusions. The process begins with Mu prophage derepression, which can occur either by aerobic starvation or by a temperature shift. Under starvation conditions, derepression involves ClpPX and Lon proteases and RpoS σ-factor. Expression of MuA transposase leads to formation of a super-twisted transposasome complex with the prophage termini. If this transposasome binds a site downstream of the U118 lacZ ochre mutation as illustrated, strand transfer will produce the branched strand transfer complex (STC) structure illustrated at the middle of the figure. Degradation of the bound transposase by ClpPX protease permits further processing of the STC. During active growth, this would produce a small inversion flanked by the replicated prophage. Under starvation conditions, an araB–lacZ translational fusion results, frequently containing at least one rearranged Mu terminus. Phosphodiester- and strand-specific details are in Shapiro and Leach (1990) and Maenhaut-Michel et al. (1997). Adapted from Shapiro (1997) with permission.
Experiments with a derivative of the prefusion strain MCS2 having a Mu A∷miniTn10 insertion blocking transposase expression yielded virtually no fusions (Shapiro and Leach 1990). This result showed that Mu transposition functions were necessary for fusions to form, and David Leach proposed how their origin could be explained as a variation of the process that would normally produce a Mu-mediated inversion (Figure 5). Sequence analysis of unexpected fusion structures that contained Mu fragments supported this model (Maenhaut-Michel et al. 1997).
Genevieve Maenhaut-Michel used indirect sib-selection methods (developed in the earliest days of bacterial genetics) to show that aerobic starvation triggered the necessary Mu activities independently of the selection substrates (Maenhaut-Michel and Shapiro 1994). Together with other colleagues, Genevieve and I were able to identify several regulatory factors involved in the starvation response (RpoS, ClpXP, Lon, HN-S, and Crp) (Figure 5; Gomez-Gomez et al. 1997; Lamrani et al. 1999). As has subsequently been discovered for other adaptive mutation systems in bacteria, aerobic starvation leads to signal transduction events and an increase in the genome restructuring activities of transposable elements as well as the SOS system and plasmid transfer functions (Peters and Benson 1995; Taddei et al. 1995; Hall 1999; Bjedov et al. 2003; Horak et al. 2004; Foster 2007; Galhardo et al. 2007).
A different facet of Mu regulation was revealed by studies of Mudlac behavior in bacterial colonies. While studying araB–lacZ fusion formation, I got in the habit of documenting the kinetics of daily colony appearance photographically (Shapiro 1984a). One day, I decided to photograph some X-Gal-stained Pseudomonas putida colonies carrying the MudII1681 translational lacZ fusion transposon (Castilho et al. 1984). When I developed the pictures, I was amazed to see that each colony looked like a flower, displaying the same kind of clonal (sectorial) and nonclonal (concentric) patterns that Barbara McClintock had documented in maize kernels (McClintock 1987; Shapiro 1984b,c, 1985, 1992b).
Together with Pat Higgins, I analyzed MudII1681 behavior in E. coli colonies that produced sectorial and concentric lacZ expression patterns by in situ colony hybridization with Mu-specific probes (Shapiro and Higgins 1988, 1989). We found a striking correlation between LacZ staining and MudII1681 DNA abundance. This result indicated that lacZ expression depended upon MudII1681 transposition and replication. A nontransposing, nonreplicating MudII1681 mutant did not produce visible β-galactosidase staining. In other words, when we saw expression patterns in the colonies on X-Gal medium, we were actually visualizing the transpositional activity of the mini-Mu construct creating new lacZ fusions, and that activity was subject to developmental regulation during colony morphogenesis. In a completely unexpected way, bacterial transposable elements revealed a control feature that McClintock had documented in maize decades earlier (McClintock 1950, 1953, 1987; Shapiro 1992b). This was quite a surprise observation in the 1980s, but it makes sense today because (1) we know that mobile genetic elements are subject to cellular regulatory networks (Shapiro 2009), and (2) there is a widespread recognition that bacteria use those regulatory networks to form organized multicellular populations, such as colonies and biofilms (Shapiro 1988, 1998).
THE FLUID GENOME, NATURAL GENETIC ENGINEERING AND A 21ST CENTURY VIEW OF EVOLUTION
My experience in the last four decades of the 20th century fits into the larger picture of how we have come to think about genome change in the 21st century. With the 1976 Cold Spring Harbor DNA insertion elements meeting, the era of the constant genome had come to an end, and the era of meetings dedicated to the fluid genome had begun. Over the next few decades, our picture of cellular systems that restructure the genome has grown to include various types of antigenic switching cassettes in prokaryotic and eukaryotic pathogens, retroviruses and retrotransposons, LINE and SINE elements, homing and retrohoming introns, immune system rearrangements, and a completely unanticipated diversity of mechanisms for DNA-based transposition (Craig et al. 2002; Wicker et al. 2007). Discoveries about genome restructuring have continued into the 21st century, with new classes of mobile elements and RNA-based mutagenesis mechanisms enriching our appreciation of cellular virtuosity in rewriting their DNA (Kapitonov and Jurka 2001; Medhekar and Miller 2007; Pritham et al. 2007).
As outlined above, my own experience with E. coli and its transposons taught me that living cells are prodigious genetic engineers. For the past 17 years, I have called mobile elements and the other biochemical complexes that restructure cellular DNA molecules “natural genetic engineering systems” and argued further that these systems fulfill major evolutionary functions (Shapiro 1992a, 1997, 1999, 2002, 2005, 2009; Shapiro and Sternberg 2005). Many researchers who study mobile elements share this view of evolution (e.g. Wessler et al. 1995; Brosius 1999; Wright and Finnegan 2001; Kidwell 2002; Khazazian 2004; Miller and Capy 2004; Bennetzen 2005; Hedges and Batzer 2005; Marino-Ramirez et al. 2005; Walsh 2006; Slotkin and Martienssen 2007; Böhne et al. 2008; Feschotte and Pritham 2007; Jurka 2008; Nishihara and Okada 2008). But the evolutionary biology community is resistant to accepting the fundamental importance of natural genetic engineering because biologically controlled genome restructuring does not fit with their assumptions about the random, accidental nature of hereditary variation. The use of the word “engineering” has generated further controversy because, some claim, it suggests the existence of an engineer and might thereby give comfort to the intelligent design community.
There are two ways to address reservations about the natural genetic engineering concept. The first way is to summarize what incontrovertible scientific evidence tells us about how cells use highly evolved biochemical systems to restructure their genomes. The wider literature deeply parallels and greatly extends my own experiences with E. coli:
Cells have biochemical activities that can do everything that human genetics engineers can accomplish with DNA (alter individual bases, cut it, splice it, and synthesize it from an RNA template or from no template at all).
Cells turn on and off their natural genetic engineering functions in response to a very wide range of stimuli. These stimuli range from physical and metabolic stresses to changes in the mating structure of populations. The adaptive benefits of this regulatory capacity are clearest in the cases of DNA changes integrated into the regular life cycles of organisms (our immune system is an example), but biological utility is also evident in the ability to stimulate variability under conditions where proliferation or survival are threatened.
Molecular mechanisms that include DNA–DNA, DNA–protein, protein–protein, RNA–protein, and RNA–DNA interactions can target changes within a genome (Shapiro 2005). This targeting is understood to be beneficial when it reduces disruption of coding sequences and favors establishment of novel transcription controls.
Cellular genetic engineering serves well-understood biological functions in cases such as phenotypic variation by pathogens, the spread and accumulation of antibiotic resistances, mating-type switching, telomere elongation, DNA damage repair, and adaptive immunity.
Sequenced genomes provide overwhelming evidence that mobile elements and DNA rearrangement functions have played a major role in episodes of evolutionary change. Examples include horizontal transfer of biochemical capabilities, protein evolution by exon shuffling and accretion, formation of novel transcriptional regulatory regions by insertional mutagenesis, construction of specialized chromatin domains, and chromosome elongation in organisms with large genomes. (Our own genomes, for example, have been molded by >2.8 million retrotransposition events.)
The second response to arguments against the natural genetic engineering concept is to reflect seriously on how living organisms are able to search effectively through the infinite space of possible genome configurations. The progenitors of extant organisms have had to make these searches during the course of repeated evolutionary challenges. So it should be no surprise that today's survivors possess evolved biochemical systems to facilitate the evolutionary rewriting of genomic information. These systems have the capacity to reduce the size of the genomic search space dramatically and to maximize the chances for success by using a combinatorial process based on existing functional components. New combinations of established coding sequences, transcriptional regulatory signals, and chromatin determinants are far more likely to prove effective than are a series of random changes altering individual genetic elements.
We know from genome sequences that evolution has followed the reliable engineering process of putting known pieces together in new arrangements. What we do not yet know is how far cells' abilities to regulate and target natural genetic engineering activities may contribute, as McClintock has suggested, to the generation of complex and useful evolutionary novelties. Until recently, investigation of this subject has not been feasible. Today, however, we can activate transposons and retrotransposons to search for functional multi-locus mutations. If we do indeed find out that regulatory and targeting mechanisms facilitate the kind of advantageous genome engineering documented in our databases, then there is no danger of falling into any epistemological trap with the natural genetic engineering concept. We will have identified the responsible “engineers” to be prokaryotic and eukaryotic cells that have evolved capacities to sense danger and rewrite their genomic memory storage systems as best they can (McClintock 1984). The research agenda will then be to analyze molecularly and computationally how those cellular control functions operate. My guess is that we will be amazed at what we find.
References
- Adhya, S., and J. A. Shapiro, 1969. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutants of the operon. Genetics 62 231–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith, J. R., E. R. Signer and W. Epstein, 1966. Transposition of the Lac region of E. coli. Cold Spring Harb. Symp. Quant. Biol. 31 393–401. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., 2005. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15 621–627. [DOI] [PubMed] [Google Scholar]
- Berg, D. E., J. Davies, B. Allet and J. D. Rochaix, 1975. Transposition of R factor genes to bacteriophage lambda. Proc. Natl. Acad. Sci. USA 72 3628–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov, I., O. Tenaillon, B. Gérard, V. Souza, E. Denamur et al., 2003. Stress-induced mutagenesis in bacteria. Science 300 1404–1409. [DOI] [PubMed] [Google Scholar]
- Böhne, A., F. Brunet, D. Galiana-Arnoux, C. Schultheis and J. N. Volff, 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16 203–215. [DOI] [PubMed] [Google Scholar]
- Brosius, J., 1999. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107 209–238. [PubMed] [Google Scholar]
- Bukhari, A. I., J. A. Shapiro and S. L. Adhya (Editors), 1977. DNA Insertion Elements, Plasmids and Episomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Casadaban, M. J., 1975. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc. Natl. Acad. Sci. USA 72 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban, M. J., 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophages lambda and Mu. J. Mol. Biol. 104 541–555. [DOI] [PubMed] [Google Scholar]
- Castilho, B. A., P. Olfson and M. Casdaban, 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. N., and J. A. Shapiro, 1980. Transposable genetic elements. Sci. Am. 242(2): 40–49. [DOI] [PubMed] [Google Scholar]
- Craig, N. L., R. Craigie, M. Gellert and A. M. Lambowitz (Editors), 2002. Mobile DNA II. American Society of Microbiology Press, Washington, DC.
- Deonier, R. C., and N. Davidson, 1976. The sequence organization of the integrated F plasmid in two Hfr strains of Escherichia coli. J. Mol. Biol. 107 207–222. [DOI] [PubMed] [Google Scholar]
- Faelen, M., and A. Toussaint, 1976. Bacteriophage Mu-1: a tool to transpose and to localize bacterial genes. J. Mol. Biol. 104 525–539. [DOI] [PubMed] [Google Scholar]
- Faelen, M., O. Huisman and A. Toussaint, 1978. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature 271 580–582. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., and E. J. Pritham, 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiandt, M., W. Szybalski and M. H. Malamy, 1972. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol. Gen. Genet. 119 223–231. [DOI] [PubMed] [Google Scholar]
- Foster, P. L., 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo, R. S., P. J. Hastings and S. M. Rosenberg, 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42 399–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, R., F. Heffron, G. Dougan and S. Falkow, 1978. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J. Bacteriol. 136 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez, J. M., J. Blásquez, F. Baquero and J. L. Martínez, 1997. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J. Bacteriol. 179 4620–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley, N. D., 1978. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell 13 419–426. [DOI] [PubMed] [Google Scholar]
- Hall, B. G., 1999. Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J. Bacteriol. 181 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, D. J., and M. A. Batzer, 2005. From the margins of the genome: mobile elements shape primate evolution. BioEssays 27 785–794. [DOI] [PubMed] [Google Scholar]
- Hedges, R. W., and A. E. Jacob, 1974. Transposition of ampicillin resistance from RP4 to other replicons. Mol. Gen. Genet. 132 31–40. [DOI] [PubMed] [Google Scholar]
- Heffron, F., C. Rubens and S. Falkow, 1975. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc. Natl. Acad. Sci. USA 72 3623–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron, F., B. J. McCarthy, H. Ohtsubo and E. Ohtsubo, 1979. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell 18 1153–1163. [DOI] [PubMed] [Google Scholar]
- Hirsch, H.J., P. Starlinger and P. Brachet, 1972. Two kinds of insertions in bacterial genes. Mol. Gen. Genet. 119 191–206. [DOI] [PubMed] [Google Scholar]
- Hõrak, R, H. Ilves, P. Pruunsild, M. Kuljus and M. Kivisaar, 2004. The ColR-ColS two-component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol. Microbiol. 54 795–807. [DOI] [PubMed] [Google Scholar]
- Ippen, K., J. A. Shapiro and J. R. Beckwith, 1971. Transposition of the lac region to the gal region of the Escherichia coli chromosome: isolation of lambda-lac transducing bacteriophages. J. Bacteriol. 108 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler, K., M. Achtman and N. Willetts, 1972. Deletion map of the Escherichia coli K-12 sex factor F: the order of eleven transfer cistrons. J. Bacteriol. 110 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, E., H. Saedler and P. Starlinger, 1968. O0 and strong-polar mutations in the gal operon are insertions. Mol. Gen. Genet. 102 353–363. [DOI] [PubMed] [Google Scholar]
- Jurka, J., 2008. Conserved eukaryotic transposable elements and the evolution of gene regulation. Cell. Mol. Life Sci. 65 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V. V., and J. Jurka, 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazazian, H. H., Jr., 2004. Mobile elements: drivers of genome evolution. Science 303 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115 49–63. [DOI] [PubMed] [Google Scholar]
- Kirchner, J., C. M. Connolly and S. B. Sandmeyer, 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267 1488–1491. [DOI] [PubMed] [Google Scholar]
- Kleckner, N., D. F. Barker, D. G. Ross and D. Botstein, 1978. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics 90 427–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamrani, S., C. Ranquet, M.-J. Gama, H. Nakai, J. A. Shapiro et al., 1999. Starvation-induced Mucts62-mediated coding sequence fusion: roles for ClpXP, Lon, RpoS and Crp. Mol. Microbiol. 32 327–343. [DOI] [PubMed] [Google Scholar]
- Lavoie, B. D., and G. Chaconas, 1996. Transposition of phage Mu DNA. Curr. Top. Microbiol. Immunol. 204 83–102. [DOI] [PubMed] [Google Scholar]
- Ljungquist, E., and A. I. Bukhari, 1977. State of prophage Mu DNA upon induction. Proc. Natl. Acad. Sci. USA 74 3143–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHattie, L., and J. A. Shapiro, 1978. Chromosomal integration of bacteriophage λ by means of a DNA insertion element. Proc. Natl. Acad. Sci. USA 75 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel, G., and J. A. Shapiro, 1994. The roles of starvation and selective substrates in the emergence of araB-lacZ fusion clones. EMBO J. 13 5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel, G., C. E. Blake, D. R. F. Leach and J. A. Shapiro, 1997. Different structures of selected and unselected araB-lacZ fusions. Mol. Microbiol. 23 1133–1146. [DOI] [PubMed] [Google Scholar]
- Marino-Ramirez, L., K. C. Lewis, D. Landsman and I. K. Jordan, 2005. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet. Genome Res. 110 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1953. Induction of instability at selected loci in maize. Genetics 38 579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1984. Significance of responses of the genome to challenge. Science 226 792–801. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1987. Discovery and Characterization of Transposable Elements: The Collected Papers of Barbara McClintock. Garland Press, New York.
- Medhekar, B., and J. F. Miller, 2007. Diversity-generating retroelements. Curr. Opin. Microbiol. 10 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W. J., and P. Capy, 2004. Mobile genetic elements as natural tools for genome evolution. Methods Mol. Biol. 260 1–20. [DOI] [PubMed] [Google Scholar]
- Mizuuchi, K., and R. Craigie, 1986. Mechanism of bacteriophage mu transposition. Annu. Rev. Genet. 20 385–429. [DOI] [PubMed] [Google Scholar]
- Morse, M. L., 1967. Reversion instability of an extreme polar mutant of the galactose operon. Genetics 56 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara, H., and N. Okada, 2008. Retroposons: genetic footprints on the evolutionary paths of life. Methods Mol. Biol. 422 201–225. [DOI] [PubMed] [Google Scholar]
- Peters, J. E., and S. A. Benson, 1995. Redundant transfer of F′ plasmids occurs between Escherichia coli cells during non-lethal selections. J. Bacteriol. 177 847–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J. E., and N. L. Craig, 2001. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 15 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham, E. J., T. Putliwala and C. Feschotte, 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390 3–17. [DOI] [PubMed] [Google Scholar]
- Saedler, H., and P. Starlinger, 1967. 0° mutations in the galactose operon in E. coli. I. Genetic characterization. Mol. Gen. Genetics 100 178–189. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1966. The chromosomal location of the gene determining uridine diphosphoglucose formation in Escherichia coli K-12. J. Bacteriol. 92 518–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., 1967. The structure of the galactose operon in Escherichia coli K-12. Ph.D. Thesis, University of Cambridge, Cambridge, UK.
- Shapiro, J. A., 1969. Mutations caused by the insertion of genetic material into the galactose operon of Escherichia coli. J. Mol. Biol. 40 93–105. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1979. A molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl. Acad. Sci. USA 76 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., 1984. a Observations on the formation of clones containing araB-lacZ cistron fusions. Mol. Gen. Genet. 194 79–90. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1984. b Transposable elements, genome reorganization and cellular differentiation in Gram-negative bacteria. Symp. Soc. Gen. Microbiol. 36(Part 2): 169–193. [Google Scholar]
- Shapiro, J. A., 1984. c The use of Mudlac transposons as tools for vital staining to visualize clonal and non-clonal patterns of organization in bacterial growth on agar surfaces. J. Gen. Microbiol. 130 1169–1181. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1985. Photographing bacterial colonies. ASM News 51 62–69. [Google Scholar]
- Shapiro, J. A., 1988. Bacteria as multicellular organisms. Sci. Am. 256(6): 82–89. [Google Scholar]
- Shapiro, J. A., 1992. a Natural genetic engineering in evolution. Genetica 86 99–111. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1992. b Kernels and colonies: the challenge of pattern, pp. 213–221 in The Dynamic Genome, edited by N. Federoff and D. Botstein. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shapiro, J. A., 1992. c Barbara McClintock, 1902–1992. BioEssays 14 791–792. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1997. Genome organization, natural genetic engineering, and adaptive mutation. Trends Genet. 13 98–104. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52 81–104. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 1999. Genome system architecture and natural genetic engineering in evolution. Ann. NY Acad. Sci. 870 23–35. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 2002. Genome organization and reorganization in evolution: formatting for computation and function. Ann. NY Acad. Sci. 981 111–134. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 2005. A 21st century view of evolution: genome system architecture, repetitive DNA, and natural genetic engineering. Gene 345 91–100. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., 2009. Revisiting the central dogma in the 21st century. Ann. NY Acad. Sci. 1178 6–28. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., and S. Adhya, 1969. The galactose operon of E. coli K-12. II. A deletion analysis of operon structure and polarity. Genetics 62 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., and N. P. Higgins, 1988. Variation of beta-galactosidase expression from Mudlac elements during the development of E. coli colonies. Ann. Inst. Pasteur 139 79–103. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., and N. P. Higgins, 1989. Differential activity of a transposable element in E. coli colonies. J. Bacteriol. 171 5975–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J. A., and D. Leach, 1990. Action of a transposable element in coding sequence fusions. Genetics 126 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, J., and L. MacHattie, 1979. Integration and excision of λ prophage mediated by the IS1 element. Cold Spring Harb. Symp. Quant. Biol. 43 1135–1142. [DOI] [PubMed] [Google Scholar]
- Shapiro, J. A., and R. V. Sternberg, 2005. Why repetitive DNA is essential to genome function. Biol. Rev. Camb. Philos. Soc. 80 227–250. [DOI] [PubMed] [Google Scholar]
- Shapiro, J., L. Machattie, L. Eron, G. Ihler, K. Ippen et al., 1969. Isolation of pure lac operon DNA. Nature 224 768–774. [DOI] [PubMed] [Google Scholar]
- Slotkin, R. K., and R. Martienssen, 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8 272–285. [DOI] [PubMed] [Google Scholar]
- Starlinger, P., and H. Saedler, 1972. Insertion mutations in microorganisms. Biochimie 54 177–185. [DOI] [PubMed] [Google Scholar]
- Taddei, F., I. Matic and M. Radman, 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92 11736–11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T. R., 2006. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 9 476–482. [DOI] [PubMed] [Google Scholar]
- Wessler, S. R., T. E. Bureau and S. E. White, 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5 814–821. [DOI] [PubMed] [Google Scholar]
- Wicker, T., F. Sabot, A. Hua-Van, J. L. Bennetzen, P. Capy et al., 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8 973–982. [DOI] [PubMed] [Google Scholar]
- Wright, S., and D. Finnegan, 2001. Genome evolution: sex and the transposable element. Curr. Biol. 11 R296–R299. [DOI] [PubMed] [Google Scholar]
- Xie, W., X. Gai, Y. Zhu, D. C. Zappulla, R. Sternglanz et al., 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]