Abstract
Digestion–ligation–amplification (DLA), a novel adaptor-mediated PCR-based method that uses a single-stranded oligo as the adaptor, was developed to overcome difficulties of amplifying unknown sequences flanking known DNA sequences in large genomes. DLA specifically overcomes the problems associated with existing methods for amplifying genomic sequences flanking Mu transposons, including high levels of nonspecific amplification. Two DLA-based strategies, MuClone and DLA-454, were developed to isolate Mu-tagged alleles. MuClone allows for the amplification of subsets of the numerous Mu transposons in the genome, using unique three-nucleotide tags at the 3′ ends of primers, simplifying the identification of flanking sequences that cosegregate with mutant phenotypes caused by Mu insertions. DLA-454, which combines DLA with 454 pyrosequencing, permits the efficient cloning of genes for which multiple independent insertion alleles are available without the need to develop segregating populations. The utility of each approach was validated by independently cloning the gl4 (glossy4) gene. Mutants of gl4 lack the normal accumulation of epicuticular waxes. The gl4 gene is a homolog of the Arabidopsis CUT1 gene, which encodes a condensing enzyme involved in the synthesis of very-long-chain fatty acids, which are precursors of epicuticular waxes.
INSERTIONAL mutagenesis is widely used in functional genomics. For example, insertion mutants obtained via T-DNA in Arabidopsis (Alonso et al. 2003) and rice (Sallaud et al. 2004) and via transposons in maize (Brutnell 2002; Brutnell and Conrad 2003; May et al. 2003; McCarty et al. 2005; Settles et al. 2007), rice (Kolesnik et al. 2004; Miyao et al. 2003; Kumar et al. 2005), and Arabidopsis (Speulman et al. 1999) have been used for both forward and reverse genetics. In both situations it is necessary to identify sequences flanking the insertional mutagen. For example, the availability of sequence-indexed collections of T-DNA insertion mutants (Alonso et al. 2003) has greatly facilitated the functional analysis of Arabidopsis. Such reverse genetic resources are generated by creating large numbers of independent insertion events and then identifying and sequencing the DNA flanking the insertional mutagen. To be cost effective such flanking sequences are typically amplified using one of several available “genome-walking” strategies (Shyamala and Ames 1989; Alonso et al. 2003; O'Malley et al. 2007; Vandenbussche et al. 2008; Uren et al. 2009).
Similarly, once mutant phenotypes have been identified following forward genetic screens, the challenge in cloning the affected gene is to identify the specific genic sequences that flank causative insertions. Insertional mutagensis is typically more productive if multiple copies of the insertional mutagens are present. The Mutator (Mu) transposon of maize has been widely used for forward genetics because of its high copy number and transposition activity (Benito and Walbot 1997). This high copy number can, however, complicate the identification of the specific insertion responsible for a mutant phenotype. Traditionally, identifying a gene sequence that had been tagged by an insertion involved genomic DNA blotting using multiple wild-type and mutant siblings to identify a DNA fragment that contained the insertion and that cosegregated with the mutant phenotype (James et al. 1995). However, both DNA blotting and subsequent postblotting gene isolation steps were laborious, time-consuming, and often unpredictable.
Here, we report two strategies, MuClone and DLA-454, for cloning mutant alleles derived from insertional mutagenesis. Both strategies are based on an adaptation of a novel highly specific and efficient genome-walking method, digestion–ligation–amplification (DLA) that uses a single-stranded oligo as the adaptor instead of the partially double-stranded adaptors used in other methods. MuClone, a cost-efficient strategy, adds unique three-nucleotide tags to the 3′ ends of the common adaptor primer so subsets of high-copy Mu transposons can be separately amplified in a manner analogous to AFLP technology (Yunis et al. 1991). It is then possible to identify which copy of the transposon cosegregates with the mutant allele in the cosegregating population. DLA-454 combines DLA with 454 pyrosequencing to amplify and sequence multiple independent alleles of a gene to be cloned. Analysis of the resulting Mu flanking sequences (MFSs) identifies the target gene. To illustrate the applicability of the MuClone and DLA-454 strategies, each was used to independently clone the glossy4 (gl4) gene. The maize gl4 is a homolog of the Arabidopsis CUT1 gene involved in epicuticular wax accumulation.
MATERIALS AND METHODS
Genetic stocks:
A total of 16 Mu-induced alleles of gl4a (gl4, to distinguish from its paralog, gl4b) were available for analysis. One (gl4a-92-1178-64) was previously isolated and the remaining 15 alleles were generated via direct tagging experiments as illustrated in supporting information, Figure S1. The su1 gene was used as a visual marker to select for gl4a-Mu or -ref alleles. However, because the su1 gene is ∼25 cM from the gl4a locus (our unpublished data), one-quarter of the plants selected after cross 3 (Figure S1) would be expected to not carry a gl4a-Mu allele but to instead carry gl4a-ref. This is consistent with the fact that Mu insertions could be identified only for 13/15 of the gl4a alleles isolated via direct tagging. Five mutant seedlings from each selfed family of gl4a/gl4a mutants and five nonmutant seedlings from each selfed family that did not segregate for gl4a mutants (cross 4, Figure S1) were pooled separately for DNA isolation and used for cosegregation analysis.
The DLA method (Figure 1):
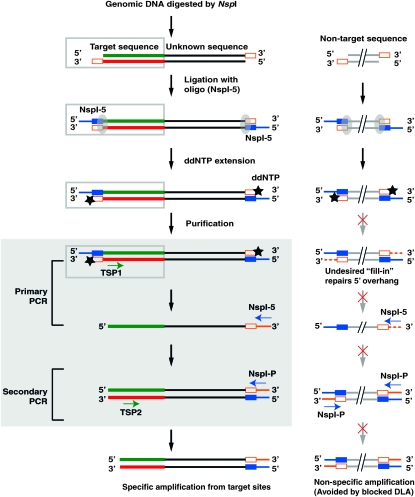
Figure 1.—
Digestion–ligation–amplification (DLA): a tool for genome walking. Genomic DNA is digested with the restriction enzyme, NspI, generating a 3′ overhang (orange open box). The single-stranded oligo, NspI-5 is used as an adaptor for ligation using T4 DNA ligase (shaded ellipses) followed by purification to remove unligated adaptors. Single-base extension with ddNTP (star) is performed to block further strand synthesis. Blocked and nonblocked DLA are defined as the procedure with and without this ddNTP extension, respectively. After purification to remove unincorporated ddNTP, nested PCR (shaded rectangle) is performed with target-specific primers (TSP1 and TSP2) in combination with the adaptor primers, NspI-5 and NspI-P, respectively. In the absence of ddNTP blocking, the 5′ overhang could be repaired, resulting in DNA fragments flanked by the adaptor sequence on both sides, which would serve as a template for nonspecific amplification with only adaptor primers (as illustrated on the right). Blocked DLA essentially prevents the PCR reaction shown on the right.
Approximately 1 μg of genomic DNA was digested with 10 units NspI [New England Biolabs (Beverly, MA), no. R0602L] at 37° for 1.5 hr. The single-stranded oligo, NspI-5, was added as the adaptor (5 μm) for ligation with T4 DNA ligase (New England Biolabs, no. M0202L). Ligation was performed at 16° for 3 hr in a 50-μl volume. The digestion–ligation product was then purified using the Qiaquick PCR purification kit [QIAGEN (Valencia, CA), no. 28106]. Unblocked and blocked (next section) ligation products were used as templates for the primary PCR reaction with the target-specific primer (TSP1, e.g., Mu-TIR, see Table S1) and the adaptor primer (NspI-5), using AmpliTaq Gold DNA Polymerase [Applied Biosystems (Foster City, CA), no. N808-0241]. The blocking step is described in the following section. This PCR program consisted of 94° for 10 min; 15 cycles of 94° for 30 sec, 60° for 45 sec, 72° for 2.5 min; and a final extension at 72° for 10 min in a 20-μl volume. The primary PCR product was diluted 10 times and 2 μl of diluted product was used for secondary PCR with nested primers TSP2 (e.g., aMu25, see Table S1) and NspI-P, using AmpliTaq Gold DNA Polymerase. This PCR program consisted of 94° for 10 min; suitable numbers of cycles (35 and 20 cycles were employed for MuClone and DLA-454, respectively) of 94° for 30 sec, 60° for 45 sec, and 72° for 2.5 min; and a final extension at 72° for 10 min.
Oligo sequences are as follows: NspI-5 20 mer, 5′-GAACGTCACAGCATGTCATG-3′; NspI-P 19 mer, 5′-AACGTCACAGCATGTCATG-3′.
ddNTP single-base extension (blocked DLA) and validation:
To prevent the single-stranded ends of ligation products from being filled during primary PCR and thus serving as undesired priming sites in the secondary PCR reaction, single-base extension with a ddNTP was performed prior to primary PCR to block further strand synthesis (Figure 1). Blocked and nonblocked DLA is defined as DLA with and without the ddNTP extension, respectively. In a 50-μl volume, 50–200 ng purified ligated DNAs were used for single-base ddNTP extension with 80 μm ddNTP and 4 units Polymerase I Large Fragment (Klenow) [Promega (Madison, WI), no. 9PIM220]. The blocking step was performed at 30° for 30 min, followed by 75° for 10 min to inactivate the enzyme. Prior to PCR, the Qiaquick PCR purification kit was used to remove extra ddNTP. To validate the application of DLA to amplifying MFSs and compare the specificity of blocked and nonblocked DLA, a pool of DNA from 12 sibling seedlings obtained via the self-pollinating of a single Mu-active plant was amplified via both blocked and nonblocked DLA. The primary and secondary PCR steps employed primer pairs Mu-TIR/NspI-5 and aMu25/NspI-P (Table S1), respectively. The nested PCR products from blocked and nonblocked DLA were TOPO cloned per the protocol of the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA) and plasmids from >90 clones from each set were isolated and sequenced with the M13f primer (5′-GTAAAACGACGGCCAG-3′).
Direct comparison between blocked DLA and splinkerette PCR:
Sau3AI digestion and adaptor ligation procedures of splinkerette PCR were performed according to Uren et al. (2009). The adaptor sequences and PCR primers (Splink 1 and 2) of Uren et al. (2009) were used. NspI digestion and ligation of blocked DLA were performed independently, using the same B73 genomic DNA as used for splinkerette PCR. Both ligated DNAs were subjected to cleanup, using the Qiaquick PCR purification kit prior to PCR amplification. The same sets of target-specific primers in combination with method-appropriate adaptor primers (Splink 1 and 2 for splinkerette PCR and NspI-5 and NspI-P for blocked DLA) and the same PCR program (94° for 10 min; 35 cycles of 94° for 30 sec, 60° for 45 sec, and 72° for 2.5 min; and 72° for 10 min) were used for both methods. The same volumes of the resulting PCR products were loaded during agarose gel electrophoresis.
MuClone:
DLA was adapted to facilitate cosegregation analysis that enables the cloning Mu tagged mutants. Instead of employing NspI-P primer (with CATG 3′ ends) used in DLA as the adaptor primer for the PCR, a set of nested primers with the addition of unique three-nucleotide tags at the 3′ ends was used in separate reactions to selectively amplify sequences flanking distinct subsets of Mu insertions. Because the 3′-end sequence generated by NspI must be (A/G) with a CATG overhang, a total of 32 tagged primers are sufficient to anneal to all NspI sites during DLA (see Table S2). The presence of these tags provides additional specificity and eliminates the need to use blocked DLA in the MuClone method. Resulting PCR reactions were analyzed via agarose gel (2%) electrophoresis. Mutant-specific bands were cut from the gel and purified with the QIAGEN gel extraction kit (QIAGEN no. 28704) and direct sequencing.
Adapting DLA primers for 454 sequencing:
To enable subsequent 454 sequencing the nested Mu-specific and adaptor primers were concatenated with the 454 sequencing primer to generate various composite primers (Table S1). A given bar-coded Mu composite primer contains sequences from 454 primer A (GCCTCCCTCGCGCCATCAG), the barcode, and the Mu TIR nested primer. The resulting composite adaptor primer (BnspI-P) contains 454 primer B (GCCTTGCCAGCCCGCTCAG) and adaptor sequence. The primary PCR product of DLA was diluted 10 times and 2 μl of diluted product was used for secondary PCR, using one of several bar-coded Mu composite primers (AeMu, AfMu, AgMu, and AhMu, Table S1) and BnspI-P primer (5′-GCCTTGCCAGCCCGCTCAGAACGTCACAGCATGTCATG-3′).
Processing DLA-454 reads:
1. Categorizing DLA-454 reads on the basis of their barcodes and sequencing trimming:
Raw 454 reads were categorized by their barcode sequences. SeqClean (http://compbio.dfci.harvard.edu/tgi/software/) was used to trim barcodes, primers, and partial Mu TIR sequences. A two-step trimming strategy was used. First, the Mu primer and the adaptor primer (default overlapping requirement, ≥80% identity with primers) were removed. Trimmed sequences with lengths ≥60 bp were subjected to a second round of trimming to remove Mu TIRs (≥30 bp overlapping, ≥80% identity with 34 bp known TIRs and a set of novel TIRs collected from 454 reads, our unpublished data).
2. Mapping DLA-454 MFS reads to the B73 reference genome:
Trimmed MFSs were aligned to the maize B73 reference genome (http://www.maizesequence.org/, released on 3/20/09), using BLASTN. Only MFSs that had five or fewer best BLAST alignments (lowest E-value hits) of MFS queries with ≥95% identity and ≥90% coverage were retained for further analysis.
3. Removing redundant Mu insertion sites:
To allow for trimming errors, MFSs with the same orientation that had apparent insertion sites within 3 bp of each other were regarded as having been derived from the same Mu insertion site. This is conservative because Mu insertions are known to cluster (Dietrich et al. 2002). Because the MFSs on both sides of a Mu insertion can be amplified and sequenced, two sequences having opposite alignment orientations but with apparent insertion sites exactly 8 bp apart (due to the 9-bp target site duplication) were counted as a single Mu insertion.
RNA isolation and quantitative RT–PCR:
RNA from 8-day-old leaves was isolated using the RNeasy plant mini kit (QIAGEN). RNase-free DNase (QIAGEN) was used during RNA isolation to remove genomic DNA from RNA. The RNA samples were confirmed to be gDNA free when only cDNA corresponding amplicons but no gDNA amplicons were detected after PCR, using actin (GenBank accession no. AY273142) primers that can amplify the gDNA and cDNA templates simultaneously, yielding amplicons of different sizes. qRT–PCR was conducted using an Mx4000 multiplex quantitative PCR system (Stratagene, La Jolla, CA). Quantitative (q)RT–PCR data were initially analyzed by using Mx4000 analysis software. Ct values were calculated using baseline-corrected, ROX-normalized parameters. Two technical replicates were included in each plate, and the average Ct value of these two replicates was used for further data analysis. The housekeeping gene, cytosolic GAPDH (GapC, GenBank accession no. X07156) was used as the internal control. The relative expression (RE) of gl4 genes, including gl4a and its paralog (gl4b), was calculated using the formula RE = 100 × 2Ct,gapC−Ct,gl4. The primers used are as follows: actin-408f 20mer, 5′-CCAGGCTGTTCTTTCGTTGT-3′; actin-520r 20mer, 5′-GCAGTCTCCAGCTCCTGTTC-3′; gap1106f 20mer, 5′-GCTTCTCATGGATGGTTGCT-3′; gap1224r 20mer, 5′-CAGGAAGGGAAGCAAAAGTG-3′; pgl4-17L 18mer, 5′-GACCGGGTGAAGCCCTAC-3′; gl4para17a 18mer, 5′-AACCGGATCAGGCCCTAC-3′; and pgl4-16R 20mer, 5′-TAGGCGAGCTCGTACCAGAG-3′.
RESULTS
DLA, a highly specific method for amplifying unknown flanking sequences:
We developed DLA to overcome technical limitations associated with existing genome-walking technologies. To test the utility of DLA for isolating unknown sequences adjacent to known sequences (i.e., genome walking), it was used as shown in Figure 1 to amplify sequences from the maize a1 (GenBank accession no. X05068) and rf2a genes (GenBank accession no. AF215823). First, genomic DNA was digested with NspI, which generates 3′ 4-bp overhangs. A single-stranded oligo was then used as the adaptor for ligation to the digested fragments. Note that each end of digested genomic DNA can ligate to only a single adaptor. To prevent the single-stranded ends of ligation products from being filled during primary PCR (5′-overhang fill-in), which would later serve as undesired primer annealing sites that are not target specific, single-base extension with a ddNTP can be performed prior to primary PCR to block further strand synthesis. Blocked and nonblocked DLA are defined as DLA with and without this single ddNTP extension step, respectively. Primary PCR was performed using TSP1 (target-specific primer 1, Table S1) and the first adaptor primer. Finally, secondary PCR was performed to amplify the flanking genomic sequences with TSP2 (target-specific primer 2, Table S1) and the second adaptor primer.
Two primers designed for each a1 gene and rf2a gene (Table S1, Figure S2) were used to perform DLA in combination with the adaptor primers, NspI-5 and NspI-P, respectively. Blocked and nonblocked DLAs were performed separately. The nonblocked DLAs yielded more nonspecific product (Figure S2) as shown after electrophoresis. The amplified fragments of expected sizes were excised from the gel and subjected to Sanger sequencing. Although both blocked and nonblocked DNA yielded the expected sequences, blocked DLA was, as expected, more specific. The results of these tests demonstrate that DLA can be applied to amplify unknown flanking sequences (i.e., genome walking).
To test the ability of DLA to amplify MFSs, PCR products from both blocked and nonblocked DLA were cloned and sequenced (materials and methods). The desired amplicons (termed Mu-Nsp amplicons) are expected to be flanked by the Mu-specific primer and the adaptor primer. Undesirable nonspecific amplicons having the adaptor primer at both ends were termed Nsp-Nsp amplicons. The sequencing data showed that 41/93 (44%) of the sequences from nonblocked DLA reactions were found to be the desired Mu-Nsp amplicons, with the remainder being undesired Nsp-Nsp amplicons. This suggests that about half of the nonblocked molecules resulted from the 5′-overhang fill-in (Figure 1). In contrast, all (94/94, 100%) sequences from the blocked DLA were from the desired Mu-Nsp amplicons. Therefore, blocking with ddNTP during DLA dramatically increases the specificity of amplification.
Comparison of DLA to splinkerette PCR:
Most ligation-based approaches for genome walking employ adaptors and most adaptors are partially double-stranded DNAs that have been subjected to specific modifications. In the absence of such modifications, partially double-stranded adaptors can participate in interadaptor ligation, reducing their effective concentration. In addition, unmodified adaptors can generate DNA molecules with adaptors at both ends (adaptor–adaptor molecules) that can serve as templates for nonspecific amplification. DLA was designed to solve these problems by using the single-stranded oligo instead of partially double-stranded DNAs used in most adaptor-mediated PCR methods as the adaptor. This slight modification makes DLA simple and efficient. Direct experimental comparisons were performed between blocked DLA and splinkerette PCR, a widely used method of genome walking that employs a partially double-stranded DNA as an adaptor (Uren et al. 2009). These comparisons were performed on three known genic targets (rf2a, a1, and rad51a). Results obtained using rf2a are shown in Figure S3. Both blocked DLA and splinkerette PCR exhibited high specificity. But the bands obtained via blocked DLA were much stronger, demonstrating that when using these templates and conditions, blocked DLA is more sensitive than splinkerette PCR. Similar results were obtained for a1 and rad51a (data not shown). It is, however, difficult to make general conclusions about sensitivity given that the performance of genome-walking methods is dependent upon choices of substrates, restriction enzymes, adaptor primers, and PCR conditions.
Cloning a Mu-tagged allele of gl4 via MuClone:
DLA was adapted to clone a gene using a Mu-tagged allele. In MuClone a set of nested adaptor primers with unique three-nucleotide tags at their 3′ ends are used in separate reactions (N = 32) of the final PCR step to selectively amplify sequences flanking distinct subsets of Mu insertions as is done in AFLP reactions. Since these selective adaptor primers dramatically decrease the nonspecific amplifications even under the presence of Nsp-Nsp templates, the ddNTP blocking step can be skipped during the MuClone procedure. MuClone reactions are performed on DNA samples isolated from a family that is segregating for a Mu-insertion mutant. PCR bands that cosegregate with the Mu-tagged mutant allele in this family (the candidate gene) are excised, purified, and sequenced.
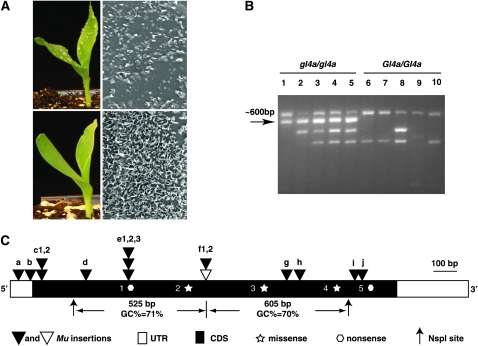
To test the MuClone protocol we made use of Mu-tagged alleles of gl4. Seedlings homozygous for gl4 mutant alleles express a “glossy” phenotype (Figure 2A; Emerson 1935) associated with alterations in the composition and/or amount of epicuticular waxes, which are derived from very-long-chain fatty acids. Mu-tagged alleles of gl4 were isolated as described (materials and methods). A set of sibling plants, each of which had the genotype gl4a-mu-94B560/gl4a-mu-94B560 or Gl4/Gl4, were self-pollinated. From each of five families derived from the self-pollination of gl4a-mu/gl4a-mu plants, DNA was extracted from pooled tissues of five glossy seedlings, resulting in five pooled DNA samples. The same process was applied to five nonglossy plants from each of five families derived from the self-pollination of Gl4/Gl4 plants. These 10 pooled DNA samples (5 mutants and 5 wild types) were used for cosegregation analysis. MuClone analyses of these 10 samples were performed using a set of primers containing the unique three-nucleotide tags (Table S2). Among the 32 three-nucleotide tags tested, only one (Nsp15ctc) could amplify a specific PCR band from all 5 mutant pools and not from any of the wild-type pools (Figure 2B). This band was purified and sequenced. The sequence of this PCR product aligned to a region of a maize BAC that contains a gene (the gl4 candidate) that has, on the basis of FGENESH prediction and EST alignments, a single exon. A collection of primers was designed on the basis of the sequence of the gl4 candidate gene. These primers, in combination with the Mu-TIR primer, were able to amplify Mu-flanking sequences from 13 of the 15 additional independently isolated Mu insertion alleles of gl4 (Table 1). All of the resulting amplified sequences aligned to the gl4 candidate gene (Table 1 and Figure 2C). The candidate gene was PCR amplified from a stock homozygous for the spontaneous reference allele of gl4 (gl4a-ref). This amplification product was shown to contain a G/C-to-A/T nonsense mutation at base position 1361 (codon 454, Table 1) of the coding region of the candidate gl4 gene (Figure 2C). In addition, four previously identified EMS-induced alleles of gl4 were shown to contain lesions of G/C-to-A/T transition (Table 1) that are typical EMS-induced point mutations (Greene et al. 2003). Among these four mutants, three are nonsynonymous missense point mutations and one generated a premature stop codon (Table 1). In combination, these data establish that the DNA sequence amplified from the gl4a-mu allele via MuClone is a portion of the gl4 gene and thereby establish the utility of MuClone for cloning insertion alleles.
Figure 2.—
Cloning of gl4a via MuClone. (A) gl4a mutants express a glossy phenotype due to altered accumulation of epicuticular waxes. Water is sprayed on seedlings to distinguish mutant (top left) from wild type (bottom left). Following scanning electron microscopy (10,000×), a mutant leaf shows fewer wax crystals (top right) than a wild-type leaf (bottom right). (B) Agarose gel electrophoresis results of modified DLA (MuClone) using the adaptor primer (Nsp-15ctc). The bands indicated by the horizontal arrow were produced from all five pools of five gl4a/gl4a (gl4a-mu-94B560) plants but not in the five pools of five Gl4a/Gl4a plants. (C) Structure of the gl4a gene and lesions associated with the mutants. a, gl4a-mu-94B366; b, gl4a-mu-93B170; c1, gl4a-mu-94B548; c2, gl4a-mu-94B327; d, gl4a-mu-93B321; e1, gl4a-mu-94B545; e2, gl4a-mu-94B324; e3, gl4a-92-1178-64; f1, gl4a-mu-93B103; f2, gl4a-mu-94B560; g, gl4a-mu-93B105; h, gl4a-mu-93-4700-2; i, gl4a-mu-93B102; j, gl4a-mu-93B107; the open triangle designates the allele used for MuClone; solid triangles designate Mu insertions amplified after cloning the gl4a locus. Nos. 1–5 are single-nucleotide missense mutants (star) or nonsense mutants (pentagon). Nos. 1–4 are EMS alleles. 1, gl4a-EMS-Ac1182-368, Gln137 to stop codon; 2, gl4a-EMS-94-1001-2705, Gly214Asp; 3, gl4a-EMS-94-1001-170, Gly314Asp; 4, gl4a-EMS-94-1001-1850, Gly412Ser; 5, gl4a-ref, Trp454 to stop codon.
TABLE 1.
gl4a mutant alleles
| Codea | Allele | Lesion | Nucleotide change or TSDb (positionc) | Represented in DLA-454d | Mutagen (sourcee) |
|---|---|---|---|---|---|
| 1 | gl4a-EMS-Ac1182-368 | Gln137 to stop codon | G/C to A/T (409) | NA | EMS |
| 2 | gl4a-EMS-94-1001-2705 | Gly214Asp | G/C to A/T (641) | NA | EMS |
| 3 | gl4a-EMS-94-1001-170 | Gly314Asp | G/C to A/T (941) | NA | EMS |
| 4 | gl4a-EMS-94-1001-1850 | Gly412Ser | G/C to A/T (1234) | NA | EMS |
| 5 | gl4a-ref | Trp454 to stop codon | NA (1361) | NA | NA (Emerson et al. 1935) |
| a | gl4a-mu-94B366 | Mu insertion at 5′-UTR | CGCCAGACC (−54–(−46)) | Yes | Mu |
| b | gl4a-mu-93B170 | Mu insertion at 5′UTR | CCTTGTCCA (−8–1) | Yes | Mu |
| c1 | gl4a-mu-94B548 | Mu insertion at CDS | CTGTACCAG (37–45) | No | Mu |
| c2 | gl4a-mu-94B327 | Mu insertion at CDS | CTGTACCAG (37–45) | No | Mu |
| d | gl4a-mu-93B321 | Mu insertion at CDS | CTGTACCTG (208–216) | Yes | Mu |
| e1 | gl4a-mu-94B545 | Mu insertion at CDS | GGCTGGGGG (377–385) | Yes | Mu |
| e2 | gl4a-mu-94B324 | Mu insertion at CDS | GGCTGGGGG (377–385) | Yes | Mu |
| e3 | gl4a-92-1178-64 | Mu insertion at CDS | GGCTGGGGG (377–385) | Yes | Mu |
| f1 | gl4a-mu-93B103 | Mu insertion at CDS | TGCTGCAGG (683–691) | Yes | Mu |
| f2 | gl4a-mu-94B560 | Mu insertion at CDS | GCTGCAGGT (684–692) | Yes | Mu |
| g | gl4a-mu-93B105 | Mu insertion at CDS | GGCCCCTGG (1010–1018) | Yes | Mu |
| h | gl4a-mu-93-4700-2 | Mu insertion at CDS | TCGCGCGCA (1061–1069) | Yes | Mu |
| i | gl4a-mu-93B102 | Mu insertion at CDS | GCCAAGGGC (1282–1290) | Yes | Mu |
| j |
gl4a-mu-93B107 |
Mu insertion at CDS |
CGTCTGGAT (1311–1319) |
Yes |
Mu |
See Figure 2.
TSD, 9-bp target site duplication.
Position relative to the translation start site (ATG). ATG is located at positions 1–3; the position just before the ATG is designated −1.
Indicates whether the Mu insertion allele is present among DLA-454 reads (see Table 2 and Figure 4).
This study if not otherwise specified.
Combining DLA with 454 sequencing technology:
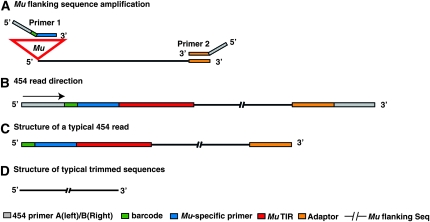
DLA was adapted for sequencing MFSs using 454 technology as illustrated in Figure 3. DNA barcodes (Qiu et al. 2003) were inserted between the 454 primer A and the Mu-specific primer to allow different input samples that were pooled in the same 454 run to be distinguished after sequencing. The resulting library amplified with primer 1 and primer 2 (Figure 3A) was subjected to 454 sequencing, using 454 primer A. Consequently, reads start from the barcode followed by portions of the Mu TIR and the MFS (Figure 3, B and C). The numbers of reads that carry the different barcodes were consistent with the relative amounts of DNA in each sample prior to pooling (data not shown). A total of 99.7% of reads carry the Mu-specific primer and 94% carry TIR sequences, demonstrating that “DLA-454” specifically and efficiently amplifies MFSs.
Figure 3.—
DLA-454 strategy to obtain sequences flanking Mu elements (A) DLA was used to amplify sequences flanking Mu elements. DLA primers were modified to facilitate 454 sequencing. 454 primer A and 454 primer B were appended to the Mu-specific primer and the Nsp-P primer, respectively. Additionally, barcodes were inserted between 454 primer A and the Mu-specific primer to allow different input samples to be analyzed in the same 454 run. (B) Typical structure of amplicons subjected to 454 sequencing. Only 454 primer A was used for pyrosequencing. (C) Structure of a typical 454 read. (D) Mu flanking sequences after removal of barcode, TIR primer, amplified TIR, and adaptor sequences.
Isolating Mu-tagged alleles of gl4 via DLA-454:
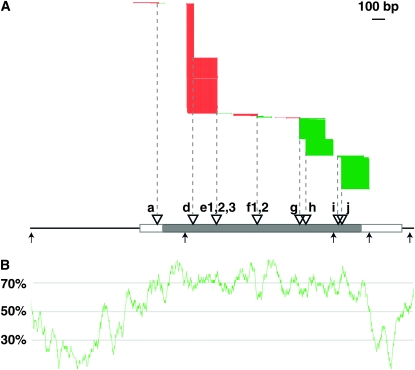
The 16 independent Mu-tagged gl4 alleles used in the MuClone experiment were subjected to DLA-454 to test the utility of this strategy for isolating multiple alleles (or mutants) derived from insertional mutagenesis screens (Table 2). Two approaches were used to prepare pooled DNAs for 454 sequencing. In the first approach 16 DNA samples each of which carried a single different gl4-Mu allele were separately subjected to the modified DLA protocol illustrated in Figure 3 prior to pooling amplification products across alleles. In the second approach DNA samples from plants carrying the 16 alleles were pooled prior to conducting the modified DLA. Subsequently, DLA products from both approaches were sequenced using 454 technology. After trimming the barcodes, the Mu-specific primer, the amplified portion of the Mu-TIR, and the Nsp-P adaptor primer, remaining MFSs were mapped to the maize genome (materials and methods). Only reads that mapped to fewer than five genomic locations were retained for further analysis. The number of Mu insertion sites in each 5-kb window across the genome (using 2.5-kb steps) was determined (Table 2). Previous genetic mapping data indicated that gl4 is located on chromosome 4 (Schnable et al. 1994). Only a single annotated gene on chromosome 4 that contains multiple independent Mu insertions was identified (Figure 4, Table 2) and this gene is the same one identified via MuClone. Similar results were obtained from both pooling strategies. The 5′ end of each alignment, which indicates the position of the Mu insertion site, is located within the gl4 transcriptional unit for each allele. The numbers of reads obtained from the different Mu insertions vary dramatically possibly due to GC content and the sizes of the allele-specific DLA products (Figure 4). Even so, of the 14 alleles that can be amplified using gene-specific primers, 11 were recovered from pooling strategy 1 and 12 from pooling strategy 2 (Table 2).
TABLE 2.
Summary of DLA-454 data
| No. 454 raw reads | No. 454 trimmed readsa | No. Mu alleles identifiedb | No. 5-kb nonoverlapping windows with the indicated number of independent Mu insertionsc |
|||
|---|---|---|---|---|---|---|
| Strategy | ≥7 | ≥8 | ≥9 | |||
| Amplify–poold | 152,483 | 100,390 | 11/14 | 1/7 | 1/5 | 1/2 |
| Pool–amplifye | 124,468 | 75,582 | 12/14 | 1/3 | 1/2 | 1/1 |
| Union of two strategies |
276,951 |
175,972 |
12/14 |
1/8 |
1/5 |
1/2 |
Trimming as per materials and methods.
No. of alleles identified via DLA-454/no. of Mu insertion alleles (Table 1).
Windows on chromosome 4/windows across the entire genome.
Conduct DLA amplification on independent alleles prior to pooling for sequencing.
Pool genomic DNAs from independent alleles prior to DLA amplification and sequencing.
Figure 4.—
DLA-454 reads from the gl4 candidate gene. (A) Alignments of DLA-454 reads (generated via the first pooling approach by DLA amplification of different alleles separately prior to pooling for 454 sequencing) that were mapped to the gl4 candidate locus. Green and red lines indicate reads with plus and minus orientations relative to the reference sequence, respectively. Inferred Mu insertion sites based on alignments of reads were projected onto the gl4 candidate locus (dashed line). Triangles represent the putative Mu insertions. NspI recognition sites are indicated by vertical arrows. The Mu insertion alleles (a, gl4a-mu-94B366; d, gl4a-mu-93B321; e1, gl4a-mu-94B545; e2, gl4a-mu-94B324; e3, gl4a-92-1178-64; f1, gl4a-mu-93B103; f2, gl4a-mu-94B560; g, gl4a-mu-93B105; h, gl4a-mu-93-4700-2; i, gl4a-mu-93B102; j, gl4a-mu-93B107) are designated in DLA-454 reads. (B) GC profile of the gl4 candidate locus (window size, 50 bp; step size, 1 bp).
gl4 is a homolog of CUT1 that encodes a very-long-chain fatty acid condensing enzyme required for epicuticular wax biosynthesis:
The amino acid sequence of GL4 shares 61% identity with the CUT1 (CER6) protein of Arabidopsis. CUT1/CER6 is a very-long-chain fatty acid (VLCFA) condensing enzyme required for epicuticular wax biosynthesis (Millar et al. 1999; Fiebig et al. 2000). Using BLASTN, the gl4 genomic sequence aligns to two loci in the maize genome (≥90% identity; ≥90% coverage). The copy (gl4a) cloned from the Mu-tagged allele is located on chromosome 4 and is predicted to encode a 485-amino-acid protein. The other copy identified by sequence homology (gl4b) is located on chromosome 5 and is predicted to encode a 480-amino-acid protein. The coding region of gl4b that has full-length cDNA (GenBank accession no. BT063439) support exhibits 96% identity with gl4a. Therefore, the maize genome contains two presumably functional copies of CUT1. In contrast, the Arabidopsis and rice genomes each contain only a single copy of CUT1. Consistent with the known subcellular localization of the VLCFA elongase (Bessoule et al. 1989; Millar et al. 1999; Xu et al. 2002), the Arabidopsis CUT1 and maize GL4A proteins are predicted to be integral membrane proteins containing two N-terminal transmembrane helices (Cserzo et al. 1997) (http://www.sbc.su.se/∼miklos/DAS/). Subcellular localization predictions indicate the GL4A protein is targeted to the plasma membrane or the endoplasmic reticulum (Horton et al. 2007) (http://wolfpsort.org/). These predictions are consistent with the fact that the membrane-bound elongase is localized in the microsomal fraction that includes the plasma membrane or the endoplasmic reticulum (Bessoule et al. 1989; Millar et al. 1999; Xu et al. 2002).
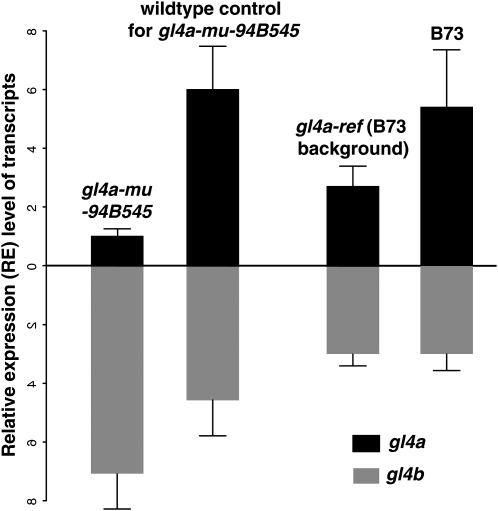
qRT–PCR experiments indicate gl4a transcripts accumulate to only low levels in gl4a-mu-94B545 homozygous seedlings, but to higher levels in gl4a-ref homozygous seedlings and the highest levels in wild type (B73) seedlings (Figure 5). These results are consistent with the fact that the gl4a-mu-94B545 mutant contains exonic Mu insertion, while the gl4a-ref mutant results from a nonsense mutant at the 3′ end of the gl4a gene (Figure 2C). These transcript accumulation levels are also consistent with the phenotypes of these mutants; gl4a-mu-94B545 mutant seedlings exhibit a strong glossy phenotype, whereas gl4a-ref mutant seedlings exhibit a weaker glossy phenotype (data not shown). Interestingly, the gl4a paralog, gl4b is upregulated in gl4a-mu-94B545 mutant seedlings, indicating the expression of gl4b is regulated at some level by the accumulation of gl4a transcripts. However, no significant change in the expression of gl4b was detected in the gl4a-ref mutants. We hypothesize this is due to the fact that gl4a-ref is a leaky mutant.
Figure 5.—
Relative accumulation of gl4a and gl4b transcripts in gl4a mutant and wild-type seedlings. DNase-treated RNAs from gl4a mutants and wild types were used for real-time RT–PCR. The relative expressions of gl4a and its paralog, gl4b, were adjusted using the expression level of the housekeeping gene, cytosolic GAPDH (GapC, GenBank accession no. X07156). RT–PCR was conducted on five to eight biological replications per genotype. Two technical replicates were included in each RT–PCR plate, and average Ct values were used for further analyses.
DISCUSSION
Comparisons to other methods:
Several ligation-based genome-walking strategies have been developed (Table S3). Some (Table S3, E) rely on self-annealing, which can result in low efficiency in large genomes. In comparison, adaptor ligation-mediated PCR exhibits improved genome-walking efficiency in large genomes. Multiple versions of adaptor ligation-mediated PCR have been developed (e.g., Riley et al. 1990; Jones and Winistorfer 1992; Devon et al. 1995; Hengen 1995; Frey et al. 1998; Kilstrup and Kristiansen 2000; Edwards et al. 2002; O'Malley et al. 2007; Uren et al. 2009). Typically, partially double-stranded DNA is used as the adaptor to ligate with digested genomic DNA. Two ligation-related problems are recognized. First, the double-stranded adapters can ligate to each other (interadaptor ligation), reducing the frequency of desired products (Table S3, C and D). Second, adaptors can ligate to both strands of both ends of genomic DNA fragments (adaptor–adaptor molecules). Such adaptor–adaptor molecules can serve as templates for nonspecific amplification (Table S3, B and D). Several modifications (Riley et al. 1990; Jones and Winistorfer 1992; Devon et al. 1995; Hengen 1995; Frey et al. 1998; O'Malley et al. 2007; Uren et al. 2009) have been developed to overcome these problems, but none solves both problems.
In the DLA ligation system, an oligo is used as the adaptor. The adaptor oligo anneals to the 3′-digested overhang. Nicks between the adaptor oligo and the digested 5′ end can be repaired by T4 DNA ligase. The new overhang, containing a portion of the adaptor sequence, is formed after ligation. Unfortunately, subsequent PCR steps could fill this 5′ overhang to form adaptor priming sites at both ends of a DNA molecule (Figure 1). If so, such a DNA molecule could be amplified by the adaptor primer even in the absence of target-specific primers, resulting in nonspecific amplification. To avoid 5′-overhang fill-in that introduces such adaptor–adaptor DNAs, ddNTPs were used to block the 3′ ends. The ddNTP blocking was shown to improve amplification specificity, which can be a problem with several other adaptor-mediated PCR methods (Table S3, B and D). The single-stranded oligo used in DLA is not expected to participate in interadaptor ligation. This results in higher concentrations of oligos, which is expected to increase ligation efficiency as compared to methods listed in Table S3, C and D. In addition, during DLA amplification, an adaptor primer works only on the newly synthesized strand primed by the target-specific primer, further increasing specificity. Taken together, these features make DLA efficient and highly specific.
DLA is a general method for amplifying DNA flanking known sequences:
It should be possible to apply the DLA method generally for obtaining unknown sequences flanking known target sequences, such as transposons, T-DNA, and transgenes. For unsequenced genomes, this method will be useful for determining sequences upstream and/or downstream of a known target locus (e.g., the isolation of promoter sequences associated with a full-length cDNA). It should be possible to broaden the application of DLA by utilizing different restriction enzymes for genomic digestion. At least three criteria should be considered when doing so: (1) restriction enzymes that produce 3′ overhangs are preferred (this is because the oligo adaptor can be ligated to the resulting restriction fragments without phosphorylation), (2) the average size of the restriction fragments should be suitable for PCR amplification, and (3) no recognition sites for the selected restriction enzyme should exist in the portion of the known sequence to be amplified.
MuClone is a simple and efficient approach for cloning Mu-tagged genes:
Mu transposons are widely used for forward genetics in maize. Active Mu transposon lines contain ∼50–200 copies of Mu and even commercial inbred lines have several dozen Mu-related sequences (Walbot and Warren 1988; Settles et al. 2004). These high copy numbers greatly complicate the visualization of individual Mu transposons following PCR amplification. The DLA method addresses this problem using an approach similar to that used during AFLP to amplify subsets of Mu transposons in separate reactions. Selective amplification of Mu flanking sequences is achieved by appending a series of unique three-nucleotide tags to the adaptor primer during the nested PCR (NspI-P). This achieves sufficient complexity reduction to permit Mu-derived amplification products to be individually visualized via agarose gel electrophoresis. Taking advantage of this feature, DLA can be used to identify sequences that flank a Mu transposon and that cosegregate with mutant phenotypes. The amplification of Nsp-Nsp templates requires that the selective adaptor primer matches both ends of Nsp-Nsp DNAs, thus dramatically decreasing the nonspecific amplifications even under the presence of Nsp-Nsp templates. Therefore, the blocked step need not be performed during the MuClone procedure.
DLA-454, a specific and practicable strategy for high-throughput sequencing of Mu flanking sequences:
The DLA-454 strategy combines DLA with 454 pyrosequencing to analyze MFSs. Approximately 94% of DLA-454 sequencing reads contain the expected Mu-TIR sequences, demonstrating the specificity of DLA. We were among the first to use barcodes to deconvolute pooled cDNA libraries after sequencing ESTs (Qiu et al. 2003). We have now applied this technology to pool samples prior to DLA-454 sequencing. To avoid the potential of confusion among samples due to sequencing errors an edit distance of at least two is required between distinct barcodes. Considering the number of input DNA samples, for this project 6-base barcodes were sufficient, but because 454 reads are long relative to the lengths of barcodes, large numbers of samples could potentially be pooled in a single 454 run.
Because we were able to clone the gl4 gene using both DLA-454 and MuClone, we conclude that DLA-454 is an efficient method for cloning genes for which multiple Mu-tagged alleles are available. A significant advantage of DLA-454 over MuClone is that it is not necessary to develop a segregating population that can save calendar time. On the other hand, if multiple independent alleles are not available for a given gene, DLA-454 could still be used by comparing Mu insertion sites in a pool of mutants vs. a pool of nonmutant siblings from a segregating population. The use of DNA barcodes allows multiple mutants to be analyzed in a single 454 run, reducing the per gene cost of cloning via DLA-454. Given its efficiency, it would be possible to use DLA-454 in combination with barcodes to create a sequence-indexed collection of Mu-insertion alleles of maize for reverse genetics (Fernandes et al. 2004; Settles et al. 2007; Vandenbussche et al. 2008). With minor modifications DLA-454 could also be used to construct a sequence-indexed collection of other maize insertions or insertion alleles in other species.
gl4a is a maize homolog of CUT1:
Epicuticular waxes cover all aerial surfaces of land plants, where they prevent nonstomatal water loss and protect plants from frost-induced injury, UV radiation, and pathogens (Post-Beittenmiller 1996). In addition, they play important roles in pollen–stigma and plant–insect interactions (Post-Beittenmiller 1996). Several maize genes related to the accumulation of epicuticular waxes have been cloned, including gl1 (Hansen et al. 1997; Sturaro et al. 2005), gl2 (Tacke et al. 1995), gl8 (Xu et al. 1997), and gl15 (Moose and Sisco 1996). Similarly, epicuticular wax genes have been cloned from Arabidopsis, including CER1 (Aarts et al. 1995), CER2 (Negruk et al. 1996; Xia et al. 1996), CER3 (Hannoufa et al. 1996), CER4 (Rowland et al. 2006), CER5 (Pighin et al. 2004), CER6/CUT1 (Millar et al. 1999; Fiebig et al. 2000), CER7 (Hooker et al. 2007), CER10 (Zheng et al. 2005), and PAS2 (Bach et al. 2008).
Epicuticular waxes are composed of derivatives of VLCFAs that are synthesized via four enzymatic reactions: condensation, reduction, dehydration, and a second reduction. In maize, the reduction step is encoded by gl8 (Xu et al. 1997) and in Arabidopsis, the dehydration step is encoded by PAS2 (Bach et al. 2008). The CUT1 protein functions in the condensation reaction, which is thought to be rate limiting (Millar and Kunst 1997; Millar et al. 1999). The phenotype of gl4 mutants demonstrates that this gene is required for epicuticular wax accumulation in maize. Our finding that the gl4a gene encodes a CUT1 homolog is consistent with the function of this protein in the biosynthesis of epicuticular waxes in Arabidopsis, where defects in CUT1 result in waxless stems (Fiebig et al. 2000; Hooker et al. 2002). Like several Arabidopsis mutants that interrupt the accumulation of epicuticular waxes, CUT1 mutants confer conditional male sterility (Fiebig et al. 2000; Hooker et al. 2002). This is thought to be a consequence of an interruption in the tapetal cells of the biosynthesis of lipids destined for inclusion in the exine (Ariizumi et al. 2004). In contrast, only two of the maize glossy mutants (not including gl4) affect male fertility (C. R. Dietrich and P. S. Schnable, unpublished data). This could be a consequence of differing roles of epicuticular wax genes in tapetal cells and pollen development between Arabidopsis and maize or due to genetic redundancy in maize. Although the observed expression interactions between gl4a and its paralog, gl4b, are at least consistent with the latter argument, we favor the former hypothesis because no gl4a or gl4b transcripts were observed among 333,174 454-ESTs isolated from tapetal cells of maize anthers (J. Cao and P. S. Schnable, unpublished data).
Acknowledgments
We thank Philip Stinard for technical assistance with the gl4 tagging experiment; Li Fan for generating families segregating for the gl4 mutants; Jun Cao for sharing unpublished data; Michael Miller, An-Ping Hsia, and Yan Fu for useful discussions; Lisa Coffey, Hailing Jin, and undergraduate students Tint-fen Low and Jia-Ling Pik for technical assistance; and the maize genome sequencing project (DBI-0527192) for sharing the sequences of BACs prior to publication. This research was supported in part by grants from the National Science Foundation (award no. DBI-6344852) to P. S. Schnable and B. J. Nikolau and from the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service, grant no. 2005-35301-15715 to P. S. Schnable.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.108936/DC1.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. FJ829343–FJ829363.
Sequence read archive accession no: SRX007377.
References
- Aarts, M. G., C. J. Keijzer, W. J. Stiekema and A. Pereira, 1995. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen et al., 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., K. Hatakeyama, K. Hinata, R. Inatsugi, I. Nishida et al., 2004. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39 170–181. [DOI] [PubMed] [Google Scholar]
- Bach, L., L. V. Michaelson, R. Haslam, Y. Bellec, L. Gissot et al., 2008. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 105 14727–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, M. I., and V. Walbot, 1997. Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoule, J. J., R. Lessire and C. Cassagne, 1989. Partial purification of the acyl-CoA elongase of Allium porrum leaves. Arch. Biochem. Biophys. 268 475–484. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., 2002. Transposon tagging in maize. Funct. Integr. Genomics 2 4–12. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and L. J. Conrad, 2003. Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236 157–176. [DOI] [PubMed] [Google Scholar]
- Cserzo, M., E. Wallin, I. Simon, G. von Heijne and A. Elofsson, 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10 673–676. [DOI] [PubMed] [Google Scholar]
- Devon, R. S., D. J. Porteous and A. J. Brookes, 1995. Splinkerettes—improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23 1644–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. R., F. Cui, M. L. Packila, J. Li, D. A. Ashlock et al., 2002. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, D., J. Coghill, J. Batley, M. Holdsworth and K. J. Edwards, 2002. Amplification and detection of transposon insertion flanking sequences using fluorescent muAFLP. Biotechniques 32 1090–1092, 1094, 1096–1097. [DOI] [PubMed] [Google Scholar]
- Emerson, R. A., G. W. Beadle and A. C. Fraser, 1935. A summary of linkage studies in maize. Cornell University Agric. Exp. Stn. Memoir 180 1–83. [Google Scholar]
- Fernandes, J., Q. Dong, B. Schneider, D. J. Morrow, G. L. Nan et al., 2004. Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 5 R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig, A., J. A. Mayfield, N. L. Miley, S. Chau, R. L. Fischer et al., 2000. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., C. Stettner and A. Gierl, 1998. A general method for gene isolation in tagging approaches: amplification of insertion mutagenised sites (AIMS). Plant J. 13 717–721. [Google Scholar]
- Greene, E. A., C. A. Codomo, N. E. Taylor, J. G. Henikoff, B. J. Till et al., 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannoufa, A., V. Negruk, G. Eisner and B. Lemieux, 1996. The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J. 10 459–467. [DOI] [PubMed] [Google Scholar]
- Hansen, J. D., J. Pyee, Y. Xia, T. J. Wen, D. S. Robertson et al., 1997. The glossy1 locus of maize and an epidermis-specific cDNA from Kleinia odora define a class of receptor-like proteins required for the normal accumulation of cuticular waxes. Plant Physiol. 113 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen, P. N., 1995. Vectorette, splinkerette and boomerang DNA amplification. Trends Biochem. Sci. 20 372–373. [DOI] [PubMed] [Google Scholar]
- Hooker, T. S., A. A. Millar and L. Kunst, 2002. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker, T. S., P. Lam, H. Zheng and L. Kunst, 2007. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, P., K. J. Park, T. Obayashi, N. Fujita, H. Harada et al., 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M. G., D. S. Robertson and A. M. Myers, 1995. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. H., and S. C. Winistorfer, 1992. Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res. 20 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilstrup, M., and K. N. Kristiansen, 2000. Rapid genome walking: a simplified oligo-cassette mediated polymerase chain reaction using a single genome-specific primer. Nucleic Acids Res. 28 E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik, T., I. Szeverenyi, D. Bachmann, C. S. Kumar, S. Jiang et al., 2004. Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J. 37 301–314. [DOI] [PubMed] [Google Scholar]
- Kumar, C. S., R. A. Wing and V. Sundaresan, 2005. Efficient insertional mutagenesis in rice using the maize En/Spm elements. Plant J. 44 879–892. [DOI] [PubMed] [Google Scholar]
- May, B. P., H. Liu, E. Vollbrecht, L. Senior, P. D. Rabinowicz et al., 2003. Maize-targeted mutagenesis: a knockout resource for maize. Proc. Natl. Acad. Sci. USA 100 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D. R., A. M. Settles, M. Suzuki, B. C. Tan, S. Latshaw et al., 2005. Steady-state transposon mutagenesis in inbred maize. Plant J. 44 52–61. [DOI] [PubMed] [Google Scholar]
- Millar, A. A., and L. Kunst, 1997. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12 121–131. [DOI] [PubMed] [Google Scholar]
- Millar, A. A., S. Clemens, S. Zachgo, E. M. Giblin, D. C. Taylor et al., 1999. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao, A., K. Tanaka, K. Murata, H. Sawaki, S. Takeda et al., 2003. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose, S. P., and P. H. Sisco, 1996. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10 3018–3027. [DOI] [PubMed] [Google Scholar]
- Negruk, V., P. Yang, M. Subramanian, J. P. McNevin and B. Lemieux, 1996. Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J. 9 137–145. [DOI] [PubMed] [Google Scholar]
- O'Malley, R. C., J. M. Alonso, C. J. Kim, T. J. Leisse and J. R. Ecker, 2007. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2 2910–2917. [DOI] [PubMed] [Google Scholar]
- Pighin, J. A., H. Zheng, L. J. Balakshin, I. P. Goodman, T. L. Western et al., 2004. Plant cuticular lipid export requires an ABC transporter. Science 306 702–704. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller, D., 1996. Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 405–430. [DOI] [PubMed] [Google Scholar]
- Qiu, F., L. Guo, T. J. Wen, F. Liu, D. A. Ashlock et al., 2003. DNA sequence-based “bar codes” for tracking the origins of expressed sequence tags from a maize cDNA library constructed using multiple mRNA sources. Plant Physiol. 133 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, J., R. Butler, D. Ogilvie, R. Finniear, D. Jenner et al., 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18 2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, O., H. Zheng, S. R. Hepworth, P. Lam, R. Jetter et al., 2006. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud, C., C. Gay, P. Larmande, M. Bes, P. Piffanelli et al., 2004. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 39 450–464. [DOI] [PubMed] [Google Scholar]
- Schnable, P. S., P. S. Stinard, T. J. Wen, S. Heinen, D. Weber et al., 1994. The genetics of cuticular wax biosynthesis. Maydica 39 279–287. [Google Scholar]
- Settles, A. M., S. Latshaw and D. R. McCarty, 2004. Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 32 e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles, A. M., D. R. Holding, B. C. Tan, S. P. Latshaw, J. Liu et al., 2007. Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala, V., and G. F. Ames, 1989. Genome walking by single-specific-primer polymerase chain reaction: SSP-PCR. Gene 84 1–8. [DOI] [PubMed] [Google Scholar]
- Speulman, E., P. L. Metz, G. van Arkel, B. te Lintel Hekkert, W. J. Stiekema et al., 1999. A two-component enhancer-inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturaro, M., H. Hartings, E. Schmelzer, R. Velasco, F. Salamini et al., 2005. Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke, E., C. Korfhage, D. Michel, M. Maddaloni, M. Motto et al., 1995. Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 8 907–917. [DOI] [PubMed] [Google Scholar]
- Uren, A. G., H. Mikkers, J. Kool, L. van der Weyden, A. H. Lund et al., 2009. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 4 789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, M., A. Janssen, J. Zethof, N. van Orsouw, J. Peters et al., 2008. Generation of a 3D indexed Petunia insertion database for reverse genetics. Plant J. 54 1105–1114. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and C. Warren, 1988. Regulation of Mu element copy number in maize lines with an active or inactive Mutator transposable element system. Mol. Gen. Genet. 211 27–34. [DOI] [PubMed] [Google Scholar]
- Xia, Y., B. J. Nikolau and P. S. Schnable, 1996. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 8 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., C. R. Dietrich, M. Delledonne, Y. Xia, T. J. Wen et al., 1997. Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 115 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., C. R. Dietrich, R. Lessire, B. J. Nikolau and P. S. Schnable, 2002. The endoplasmic reticulum-associated maize GL8 protein is a component of the acyl-coenzyme A elongase involved in the production of cuticular waxes. Plant Physiol. 128 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis, I., M. Salazar and E. J. Yunis, 1991. HLA-DR generic typing by AFLP. Tissue Antigens 38 78–88. [DOI] [PubMed] [Google Scholar]
- Zheng, H., O. Rowland and L. Kunst, 2005. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]