Abstract
Centromeres, because of their repeat structure and lack of sequence conservation, are difficult to assemble and compare across organisms. It was recently discovered that rice centromeres often contain genes. This suggested a method for studying centromere homologies between wheat and rice chromosomes by mapping rice centromeric genes onto wheat aneuploid stocks. Three of the seven cDNA clones of centromeric genes from rice centromere 8 (Cen8), 6729.t09, 6729.t10, and 6730.t11 which lie in the Cen8 kinetochore region, and three wheat ESTs, BJ301191, BJ305475, and BJ280500, with similarity to sequences of rice centromeric genes, were mapped to the centromeric regions of the wheat group-7 (W7) chromosomes. A possible pericentric inversion in chromosome 7D was detected. Genomewide comparison of wheat ESTs that mapped to centromeric regions against rice genome sequences revealed high conservation and a one-to-one correspondence of centromeric regions between wheat and rice chromosome pairs W1-R5, W2-R7, W3-R1, W5-R12, W6-R2, and W7-R8. The W4 centromere may share homology with R3 only or with R3 + R11. Wheat ESTs that mapped to the pericentromeric region of the group-5 long arm anchored to the rice BACs located in the recently duplicated region at the distal ends of the short arms of rice chromosomes 11 and 12. A pericentric inversion specific to the rice lineage was detected. The depicted framework provides a working model for further studies on the structure and evolution of cereal chromosome centromeres.
CENTROMERES and their associated kinetochores are protein–DNA complexes that mediate spindle microtubule attachment during mitosis and meiosis and are necessary for the accurate segregation of the chromosomes into daughter nuclei. Despite the conservation of centromere function, centromere sequence composition consisting of highly repetitive satellite DNA and retrotransposons varies widely among different organisms (Henikoff et al. 2001; Sullivan et al. 2001; Jiang et al. 2003; Lee et al. 2005; Ma et al. 2007; Kanizay and Dawe 2009). The most abundant sequences in plant centromeres are the 180-bp satellite repeat pAL1 in Arabidopsis, CentO satellite repeats in rice, CentC repeats in maize, and the B-specific repeats in the centromere of maize B chromosome (Round et al. 1997; Ananiev et al. 1998; Dong et al. 1998; Copenhaver et al. 1999; Cheng et al. 2002; Jin et al. 2004, 2005; Birchler et al. 2009). Centromeric satellites serve as the core of the centromere, which is flanked by pericentric heterochromatin rich in middle repetitive elements, including retroelements and transposons. Because of the abundance of various repeats, centromeres of most eukaryotic chromosomes are upward of 1 Mb in size, mostly devoid of genes, and their sequencing and assembly pose a big challenge (Su et al. 1997; Hosouchi et al. 2002). Among the sequenced genomes of many multicellular eukaryotes, including Drosophila melanogaster, human, mouse, Arabidopsis thaliana, and rice, only the centromeres of rice chromosomes 3, 4, 5, and 8 have been fully assembled (Nagaki et al. 2004; Wu et al. 2004; Y. Zhang et al. 2004; International Rice Genome Sequencing Project 2005; Yan et al. 2006).
Unlike centromere DNA sequences, a group of proteins specific to the centromere/kinetochore complex is highly conserved among diverse organisms, including fungi, animals, human, and plants. The centromere-specific histone H3 variants (CENH3s) were found in fungi (Cse4), insect (Cid), nematodes (HCP-3), mammals (CENP-A), Arabidopsis (HTR-12), rice, and maize (Palmer et al. 1991; Dawe et al. 1999; Henikoff et al. 2001; Talbert et al. 2002; Zhong et al. 2002; Jiang et al. 2003). Previous studies have reported expressed genes and transcripts in the flanking regions of some centromeres (Copenhaver et al. 1999; Schueler et al. 2001) and a human neocentromere (Saffery et al. 2003). Nagaki et al. (2004) reported active genes in the sequenced centromere of rice chromosome 8 (Cen8); at least 16 active genes reside within a ∼750-kb core domain associated with CENH3 of Cen8 (Yan et al. 2005). In the centromere of rice chromosome 3, 19 transcribed genes have been localized to the ∼1881-kb CENH3 domain (Yan et al. 2006). The genes present in the conserved domains open possibilities for the comparative mapping of centromeric regions among groups of organisms where gene synteny is conserved.
The cereal crops wheat (1x = 7), maize (1x = 10), sorghum (1x = 10), and rice (1x = 12) share 65 million years of evolutionary history, differ in basic chromosome number and genome size (40-fold), and yet maintain large syntenic blocks and in some cases whole chromosome homologies (Ahn et al. 1993; Gale and Devos 1998; Huang et al. 2002; Sorrells et al. 2003; Paterson et al. 2004, 2009; Singh et al. 2007; Wei et al. 2007; Salse et al. 2008). However, the information about centromere synteny between rice, wheat, and other species is still limited because most of the rice centromere cores are in sequencing gaps. The observation that active genes were found in the centromeres of rice chromosomes, combined with the fact that wheat telosomic chromosomes can be used to precisely map genes to centromeric and pericentromeric regions (Sandhu et al. 2001; Francki et al. 2002; Qi et al. 2006) provides an excellent opportunity to study syntenic relationships between centromeres of wheat and rice. In the present study, we report the mapping of centromeric genes of rice chromosome 8 to wheat chromosome centromeric regions using wheat aneuploid stocks. These data, together with bioinformatics analysis of previous data (Sandhu et al. 2001; Francki et al. 2002; Qi et al. 2006), provide a framework for genomewide comparisons of homology among the centromeric regions of wheat, rice, and other cereal species and novel insights into their karyotypic evolution.
MATERIALS AND METHODS
Wheat aneuploid stocks:
Twenty-one wheat nullitetrasomic (NT) lines in Triticum aestivum L. cv Chinese Spring (CS) background were used to assign rice genes to specific wheat chromosomes. Sixteen CS ditelosomic (Dt) lines and 20 wheat-alien ditelosomic addition (DtA) lines involving homologous group 3, 5, and 7 chromosomes were used to locate genes to a specific arm or centromeric or pericentromeric region of wheat and alien chromosomes (supporting information, Table S1). In addition, 64 chromosome deletion (del) lines with the distal segment deleted for a specific chromosome were used to assign genes to specific chromosome segments with respect to the centromere (pericentromeric or not) (Table S2, Endo and Gill 1996). The del5DL-7 line was reselected from a cross of the original del5DL-7 + monosmic 5D with N5DT5B to remove chromosome 5D from this line. In an alien DtA line, the first number designates the homologous group, followed by the genome symbol; the # sign is used to distinguish between chromosomes belonging to the same homologous group but derived from different accessions, and last, the arm location. The genetic stocks and Triticeae species are maintained at the Wheat Genetic and Genomic Resources Center at Kansas State University (http://www.k-state.edu/wgrc/).
RFLP analysis:
Procedures for genomic DNA isolation, restriction endonucleases digestion, gel electrophoresis, and DNA gel blot hybridization were as described in Qi et al. (2003). The genomic DNAs of the selected genetic stocks were digested with four enzymes of EcoRI, HindIII, DraI, and BamHI. The rice centromere clones and wheat ESTs were provided by J. Jiang, University of Wisconsin, Madison and Y. Ogihara, Kyoto Prefectural University, Shimogamo, Sakyo-ku, Japan.
Nucleic acid sequence alignments:
The sequences of three rice centromeric clones, 6729.t09, 6729.t10, and 6730.t11, and Cen8 BAC B1052H09, were subjected to BLASTN searches of the National Center for Biotechnology and Information (NCBI) dbEST database (http://www.ncbi.nlm.nih.gov/dbEST/) to identify corresponding wheat ESTs and/or tentative consensus (TC) sequences. The sequences of selected wheat bin-mapped ESTs and one RFLP clone were anchored to the 12 rice pseudomolecules composed of ordered BACs/PACs to compare microcolinearity in the centromeric regions between wheat and rice (http://rice.plantbiology.msu.edu/pseudomolecules/info.shtml).
BAC library screening:
The filters of BAC libraries of Aegilops tauschii Coss. and Ae. speltoides were provided by J. Dvorak, University of California, Davis. Each high-density colony filter contains 18,432 clones. Library screening was performed using four filters that contain 73,728 clones for each EST marker. The procedure for colony filter hybridization was similar to the one used for Southern blot hybridization.
BAC-FISH analysis:
In addition to centromere assignment based on aneuploid stocks, BAC-fluorescence in situ hybridization (FISH) was used to map genes to the centromeres. Positive BAC clones for specific genes digested with HindIII were first screened with three centromere-specific clones pAet6-09, Hi10, and pRCS1 to reveal their potential centromeric locations (Abbo et al. 1995; Dong et al. 1998; P. Zhang et al. 2004). BAC clones with strong hybridization signals with the centromere-specific repeats were selected as probes for FISH experiments. BAC-FISH was as described by P. Zhang et al. (2004). Slides were analyzed with an epifluorescence Zeiss Axioplan 2 microscope. Images were captured using a SPOT 2.1 CCD (charge-coupled device) camera (Diagnostic Instruments; http://www.diaginc.com) and processed with Photoshop v5.5 software (Adobe Systems; http://www.adobe.com).
RESULTS
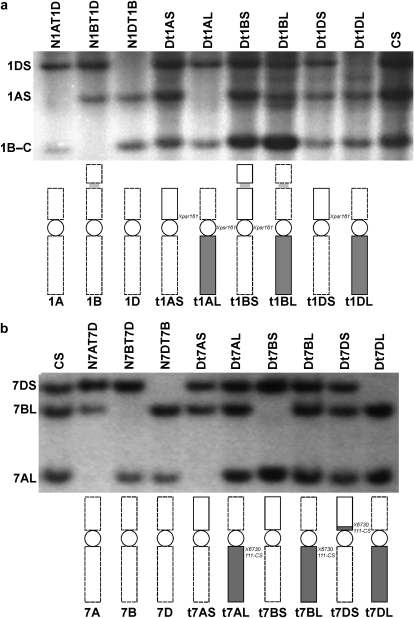
The methodology for centromere mapping of RFLP fragments in wheat and the Triticeae is illustrated in Figure 1 (see also Qi et al. 2006). The Dt stocks of wheat and those of other Triticeae species often arise from breaks in the centromere (Sears and Steinitz-Sears 1978; Zhang et al. 2001). Briefly, if an RFLP fragment is assigned to a specific chromosome but is not missing in either of the telosomic stocks for that chromosome then it is assigned to the centromeric region (Figure 1a). More often, a fragment maps to the short arm in some telosomic stocks and to the long arm in other telosomic stocks, and then it is assigned to the pericentromeric region (Figure 1b). All DtA lines, in which a pair of alien chromosome arms is added to the wheat complement, were developed in CS wheat background. Physical mapping of loci to an alien chromosome arm using DtA lines is based on intergenomic polymorphism. If a polymorphic fragment is observed in a specific DtA line when compared to the hybridization pattern of CS, this fragment can be mapped to a specific arm of an alien chromosome. In wheat, these stocks can be used to assign RFLP loci to centromeres or centromeric bins in the specific Triticeae species.
Figure 1.—
An example of the localization of RFLP loci to the centromere and pericentromeric regions of wheat homologous chromosomes. Chromosome ideograms in a and b indicate the specific chromosome constitution in the corresponding genetic stocks. The dashed line indicates a missing chromosome or chromosome arm. The open and shaded bars represent the short and long arms, respectively. (a) An autoradiograph of a Southern hybridization of genomic DNA of standard Chinese Spring (CS) and aneuploid stocks for group-1 chromosomes. Genomic DNA of nullisomic-tetrasomic (NT) and ditelosomic (Dt) lines was digested with EcoRI and hybridized with clone PSR161. The top fragment detected by this clone was missing in N1DT1B and Dt1DL, respectively, and was mapped to the short arm of chromosome 1D (1DS). Similarly, the second fragment was mapped to the short arm of chromosome 1A (1AS). However, the third fragment was missing in N1BT1D, but present in both telosomics of the short and long arms of chromosome 1B, indicating the centromere location of PSR161 on chromosome 1B. The position of wheat cDNA clone PSR161 is shown in italics on the right of each chromosome. (b) An autoradiograph of a Southern hybridization of genomic DNA of standard CS and aneuploid stocks for group-7 chromosomes. Genomic DNA of the nullisomic-tetrasomic and ditelosomic lines was digested with HindIII and hybridized with the PCR product 6730.t11-CS. The top fragment detected by this clone was missing in N7DT7B and Dt7DL, respectively, and was mapped to the short arm of chromosome 7D (7DS). However, the second fragment was missing in N7BT7D and Dt7BS, respectively, and was mapped to the long arm of chromosome 7B (7BL). Similarly, the third fragment was mapped to the long arm of chromosome 7A (7AL). It revealed a pericentric inversion in chromosome 7D in comparison to the standard arrangement in homologous chromosomes 7A and 7B. The position of clone 6730.t11-CS is shown in italics on the right of each chromosome.
Mapping of rice R8 centromeric genes in wheat and the Triticeae:
Rice chromosome 8 centromere (Cen8) genes were selected for a test of centromeric region homology between rice and wheat. The seven rice centromeric clones selected in the present study are expressed genes located in the centromere of rice chromosome 8, and three of them lie in the Cen8 kinetochore region (Nagaki et al. 2004, Figure S1). These clones were mapped by Southern hybridization to determine their chromosome, arm, and deletion bin location in wheat using a set of wheat aneuploid stocks (Table S1 and Table S2). Three clones, 3507.t05, 6731.t10, and 6731.t12, failed to yield clear hybridization signals and could not be mapped. Of the remaining four clones, 6733.t09 was mapped to the distal regions of the short arms of 3A, 3B, and 3D of the wheat group-3 chromosomes (Table 1, Figure 2). Three clones, 6729.t09, 6729.t10, and 6730.t11, which were previously located on the kinetochore region of the rice chromosome 8 centromere, mapped in the centromeric bins of wheat chromosomes 7A, 7B, and 7D. Within the centromeric bin, 6729.t09 and 6729.t10 mapped to the short arms of chromosomes 7A and 7B, clone 6730.t11 mapped to the centromeric bin of the long arms of chromosomes 7A and 7B, and all three mapped to the short arm centromeric bin in chromosome 7D. A paralogous locus for 6729.t09 was detected at a proximal position in the long arm of all three group-5 chromosomes, as well as in the long arm of the barley chromosome 5H, indicating a sequence duplication event in the Triticeae (Figure 2).
TABLE 1.
Clone/enzyme combination and chromosome location of clones
| Clone/enzyme | Chromosome location |
|---|---|
| 6733.t09/EcoRI | 3AS, 3BS, 3DS |
| 6733.t09/HindIII | 3SS, 3HS |
| 6729.t10/EcoRI | 7AS, 7BS, 7DS, 7SS, 7HS |
| 6729.t10/BamHI | 7RL |
| 6729.t10/HindIII | 5AL, 5BL, 5DL |
| 6729.t10/DraI | 5HL |
| 6729.t09/EcoRI | 7AS |
| 6729.t09/HindIII | 7BS, 7DS, 7SS |
| 6730.t11/HindIII | 7AL, 7BL, 7DS |
| 6730.t11/DraI | 7RS |
| 6730.t11-CS/HindIII | 7AL, 7BL, 7DS |
| 6730.t11-CS/DraI | 7RS |
| BJ280500/HindIII | 7AL, 7BL, 7DS, 7SL, 7RL, 7HL |
| BJ305475/BamHI | 7AS, 7BS, 7DS, 7SS, 7RL, 7HS |
|
BJ301191/EcoRI |
7AS, 7BS, 7DS, 7RL, 7HS |
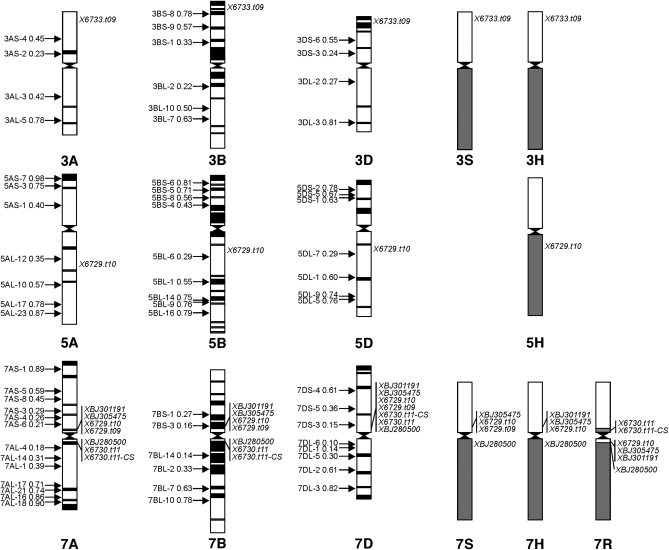
Figure 2.—
Physical mapping of rice centromeric genes and wheat ESTs in individual chromosome bins of wheat and the Triticeae chromosomes. The deletion names and breakpoints (indicated as fraction length from the centromere) are on the left of each chromosome. The rice cDNA clones and wheat ESTs mapped to the bins are in italics on the right of each chromosome. The ideogram of C-banded chromosomes of groups 3, 5, and 7 is after Gill et al. (1991).
A PCR-amplified fragment from CS wheat using RT–PCR primer of clone 6730.t11 (Nagaki et al. 2004), named 6730.t11-CS, gave an RFLP pattern similar to that of 6730.t11, but the signal intensity was much stronger (Figure 1b). 6730.t11-CS was mapped to the same centromeric bins as 6730.t11 (Figure 2).
A collection of ditelosomic addition stocks of Ae. speltoides, barley, and rye was used to determine the location of rice centromeric genes in these species. Digested genomic DNA of CS and six CS-alien DtA lines of DtA3S#3S, DtA3Sl#2L, DtA3RS, DtA3EL, DtA3HS, and DtA3HL were probed with clone 6733.t09. The polymorphic fragments were only detected in DtA3S#3S and Dt3HS, mapping them to the short arms of chromosome 3 of Ae. speltoides and barley, respectively (Table 1, Figure 2). It confirmed that the rice Cen8 clone 6733.t09 is located on the short arm of homologous group-3 chromosomes in the Triticeae.
Southern hybridization with four clones, 6729.t10, 6729.t09, 6730.t11, and 6730.t11-CS, to DtA lines of homologous group-7 revealed that clone 6729.t10 mapped to the short arms of chromosome 7S of Ae. speltoides and chromosome 7H of barley but to the long arm of rye chromosome 7R. Clone 6729.t09 was only mapped to the short arm of chromosome 7S of Ae. speltoides. Two other clones, 6730.t11 and 6730.t11-CS, were mapped to the short arm of rye chromosome 7 (Table 1, Figure 2). The mapping of these clones to the group-7 chromosomes in Ae. speltoides, barley, and rye again indicated that similar to wheat, these clones are located in the pericentromeric region of all the Triticeae chromosomes.
Wheat ESTs homologous to the rice Cen8 clones and their physical mapping:
To find homologous centromeric sequences of three rice centromeric clones, 6729.t09, 6729.t10, and 6730.t11, in wheat, we aligned these rice clone sequences against all the sequences present in the wheat EST database using BLASTN searches at a higher stringency level (E > e−20) (Altschul et al. 1997). The clones 6729.t09 and 6729.t10 had similarity to the same wheat contig TC249611, coding a poly (A)-binding protein. The clone 6730.t11 had significant similarity to the contig TC255802, which is predicted to encode a CBS domain-containing protein (Table 2). In addition, a Cen8 BAC B1052H09 (http://rice.plantbiology.msu.edu/pseudomolecules/centromere.shtml) was also used to search the wheat EST database and gave hits to wheat EST contigs TC265287, TC265289, TC265290, and TC255432. These contigs were again subjected to BLAST searches of the rice pseudomolecule database. Only two contigs, TC265290 and TC255432, matched against the centromeric BAC B1052H09. TC265290 encodes a TGF-β receptor-interacting protein-like protein and TC255432 encodes a putative Rer1 protein (Table 2).
TABLE 2.
Blast search results of rice centromeric clones and BAC against wheat EST database
| Rice clone or BAC | Wheat EST | E-value | Description | Selected EST from TCa for mapping |
|---|---|---|---|---|
| B1052H09 | TC265290 | 2.40E-96 | TGF-beta receptor-interacting protein-like protein | BJ244076, BJ222044 |
| TC255432 | 3.30E-59 | Rer1 protein, putative, expressed | BJ280500 | |
| 6730.t11 | TC255802 | 2.70E-43 | CBS domain-containing protein | BJ219066 |
| 6729.t09, 6729.t10 | TC249611 | 2.60E-253 | Polyadenylate-binding protein, putative, expressed | BJ301191 |
|
BJ305475 |
Wheat EST tentative consensus sequence.
Searching all ESTs in the four contigs that matched rice sequences to the mapped wheat EST database revealed that none of them has been mapped (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). Six wheat ESTs from the four contigs were selected and hybridized to a set of wheat NT, Dt, DtA, and del lines to compare their mapping positions with corresponding rice clones in wheat (Table 2). Of the six ESTs, BJ244076, BJ222044, and BJ219066 did not provide useful information because of high copy number. Two ESTs, BJ301191 and BJ305475, which matched the rice clones 6729.t09 and 6729.t10, mapped to the short arm of group-7 chromosomes, similar to the previous mapping of these two rice clones in wheat. In addition, these two wheat ESTs mapped to the short arms of Ae. speltoides 7S and barley 7H, but to the long arm of rye 7R (Table 1, Figure 2). The EST BJ280500, which aligned to the rice Cen8 BAC, mapped to the long arms of chromosomes 7A and 7B but to the short arm of 7D, similar to rice clone 6730.t11. However, this EST mapped to only the long arms of Ae. speltoides 7S, barley 7H, and rye 7R, indicating a long arm origin.
Genomewide comparison of wheat ESTs mapped to centromeric regions with rice genomic sequences:
In previous studies, a total of 24 ESTs and one RFLP clone were physically mapped in the centromeric regions of wheat chromosomes. These ESTs are diagnostic markers detecting pericentric inversions in homologous groups 2, 3, 4, 5, and 6 in the Triticeae and are markers for the pericentromeric regions of wheat chromosomes (Conley et al. 2004; Linkiewicz et al. 2004; Miftahudin et al. 2004; Qi et al. 2004, 2006). Two cDNA clones, PSR161 and BCD1072, were previously mapped to the centromere of chromosome 1B (Sandhu et al. 2001; Francki et al. 2002). Searches against the rice genomic DNA database revealed that 22 of the 24 mapped wheat ESTs and PSR161 matched rice expressed genes (http://rice.plantbiology.msu.edu/pseudomolecules/info.shtml, Table S3). The positions of the anchored rice BACs were compared with that of the rice centromeric BACs in each rice chromosome to discover the conservation of colinearity of the centromere regions between wheat and rice (Figure 3). The rice centromeric BACs were selected from “Information about the Centromeres in the Rice Genome Annotation Project Pseudomolecules Release 6” (http://rice.plantbiology.msu.edu/pseudomolecules/centromere.shtml).
Figure 3.—
Blast search results of wheat EST sequences against rice pseudomolecules. The wheat ESTs previously mapped to the centromeric regions showed pericentromeric inversions in chromosomes of homologous groups 2–7. The ESTs mapped in the D-genome chromosomes were selected as ancestral except for chromosomes 1B and 7A. The rice BAC positions in the maps are based on megabase distances in the pseudomolecule and were taken from the Rice Genome Annotation Project–MSU Rice Genome Annotation (Osa1) Release 6 (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). *Rice centromeric BAC.
Wheat cDNA PSR161 is the only centromeric clone in chromosome 1B identified so far (Sandhu et al. 2001; Francki et al. 2002), and it had similarity to a sequence on rice BAC OJ1234_D05. This BAC clone was located at position 13.5 Mb in the pseudomolecule of rice chromosome 5 (R5) and was only 1 Mb away from the centromeric BAC P0697B04 (Figure 3). Of the two group-2 pericentromeric ESTs, BE404630 anchored to a BAC on the interstitial region of the R7 short arm. The EST BE500625 did not match any rice sequence (Figure 3). All four wheat group-3 (W3) pericentromeric ESTs gave hits on the short or long arm of rice chromosome R1 encompassing the centromeric BAC B1061G08 (Figure 3). The closest rice BAC OsJNBa0086A10 was at a distance of 2.7 Mb from the Cen1 BAC.
The pericentromeric region of the group-4 chromosomes of wheat and the Triticeae is highly dynamic involving multiple and independent inversion events, and eight EST clones mark the region (Qi et al. 2006). Seven of the eight clones (seven ESTs and one RFLP clone) gave hits on rice chromosomes R3, R4, and R11. Four gave hits on rice chromosome R3 and EST BE497635 aligned to the Cen3 BAC OsJNBb0047D08 (Figure 3). Two gave hits on rice chromosome R11, and the rice BAC OSJNBb0018P20 anchored by EST BE637507 was adjacent to the Cen11 BAC OSJNBa0046A04 within 0.2 Mb distance (Figure 3). However, the second hit on R11 BAC OSJNBa0042J05 by EST BF202706, which was physically mapped to the centromeric region of wheat chromosomes by BAC-FISH (see below), was located toward the terminal end in the long arm. EST BE494281, a multicopy clone detecting 7–13 restriction fragments with different probe/enzyme combinations, gave a hit in the long arm of R4.
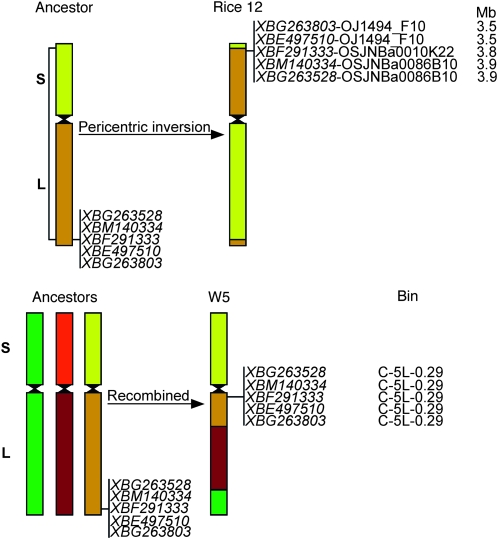
All W5 ESTs except one first hit on R12 and second on R11, which shares a duplicated segment with R12 (Table S3). The EST BE403618 of the W5 short arm gave a hit on the R12 BAC at 4.4 Mb from the Cen12 BAC and a second hit on the Cen11 BAC OsJNBa0046A04 (Figure 3). Further sequence search of the W5 short arm ESTs mapped to the centromeric bin against rice genome sequences (see below) discovered two rice BACs, OJ1060_G11 anchored by EST BG314119 and OSJNBb115B15 anchored by EST BE604729, were located at distances of 0.7 Mb and 1.6 Mb from the Cen12 BAC OsJNBa0088J04, respectively (Table S4, Figure 3). The other five pericentromeric ESTs of the W5 long arm gave hits on the distal ends of the short arms of both R12 and R11, representing a 3.5- to 3.9-Mb region in R12 pseudomolecules and a 3.4- to 4.1-Mb region in R11, a part of the duplicated regions on R11S and R12S (Figure 3, Wu et el. 1998; Rice Chromosomes 11 and 12 Sequencing Consortia 2005; Yu et al. 2005).
Four pericentromeric ESTs of group-6 gave hits on rice chromosome R2, two in each arm. EST BE405809 aligned to rice BAC P0705A04 1.2 Mb from the Cen2 BAC B1120G10d (Figure 3). Our study provided direct evidence that the rice Cen8 is related to the centromeres of the W7 chromosomes. Three rice Cen8 clones were mapped to the pericentromeric region of W7. Three wheat ESTs, with matches to rice Cen8 active genes, along with the Cen8 BAC, also mapped to the pericentromeric regions of the W7 chromosomes (Figure 3).
Syntenic block between W5 and the distal region of rice chromosome R12:
As noted above, W5 pericentromeric ESTs were aligned to the distal end of rice chromosome 12S. To investigate this discrepancy further in the context of chromosome homology in the proximal region of the centromere, we searched 179 wheat bin-mapped ESTs against rice genome sequences. Of those, 22 were mapped to the region proximal to the centromere in the short arm and 157 in the long arm of W5. W5 short-arm ESTs had the most hits in rice chromosome 12 (36%), three in the long arm and five in the short arm (Table S4). Of 157 W5 long-arm ESTs, 32 had no hit to any rice pseudomolecules. The highest percentage (50%) of W5 long-arm ESTs were mapped to rice chromosome 9 and about 23% (29 ESTs) were mapped to rice chromosome 12 (Table S4). Most W5 long-arm ESTs aligned to rice chromosome 12 are located in the region close to the W5 centromere, including 9 ESTs, which detected the pericentromeric inversion in chromosome 5A. Twenty-nine W5 long-arm ESTs mapped to the short arm of rice chromosome 12, covering a region from 0.1 Mb to 5.7 Mb with an opposite gene order compared to the position of these ESTs in wheat (Table S4). Most of them mapped to the distal end of the short arm of rice chromosome 12 known as a recently duplicated region in R11S and R12S.
Wheat ESTs assigned to the centromere by BAC-FISH:
In wheat, with its complex and large genome, single-copy ESTs cannot be mapped by FISH (P. Zhang et al. 2004). We used BAC-FISH to further confirm the possible centromeric location of the ESTs mentioned above. A total of 10 wheat ESTs previously mapping to the pericentromeric regions of wheat group-1–group-6 chromosomes and three rice centromeric clones were used to screen high-density BAC filters from Ae. tauschii or Ae. speltoides (Table 3). Positive BAC clones were detected for all except rice clones 6729.t09 and 6730.t11 (Table 3). HindIII-digested positive BAC clones were probed with the centromere-specific clones pAet6-09, Hi10, and pRCS1 to identify BAC clones containing centromere-specific repeats. More than half of the positive BAC clones did not hybridize with centromere-specific repeats. However, most BAC clones of Ae. tauschii or Ae. speltoides that were anchored with W4 ESTs strongly hybridized with centromeric clones and showed a tandem repeat pattern with a range of 6–17 fragments (Table 3).
TABLE 3.
Hybridization results of BACs with centromeric-specific clones and BAC-FISH results with the selected BACs
| No. fragments of BAC hybridizing to: |
||||||||
|---|---|---|---|---|---|---|---|---|
| Group | Marker | Associated BAC | Contigb | BAC library | pAet6-09 | Hi10 | PRCS1 | BAC-FISH signal |
| 1 | PSR161 | 13K16a | NA | Ae. speltoides | 0 | 0 | 0 | P/+++ |
| 27O24 | NA | Ae. speltoides | 0 | 0 | 0 | |||
| 49E02 | NA | Ae. speltoides | 0 | 0 | 0 | |||
| 83M17 | NA | Ae. speltoides | 0 | 0 | 0 | |||
| 144A17 | NA | Ae. speltoides | 0 | 0 | 0 | |||
| 2 | BE404630 | RI017L168a | ctg3127 | Ae. tauschii | 1 | 1 | 1 | − |
| RI018G22 | ctg3127 | Ae. tauschii | 1 | 1 | 1 | |||
| RI032A4 | Singleton | Ae. tauschii | 1 | 1 | 1 | |||
| 3 | BE485348 | HD90D18 | ctg734 | Ae. tauschii | 0 | 0 | 0 | |
| RI031J15a | ctg734 | Ae. tauschii | 1 | 1 | 1 | P/+++ | ||
| HD082G15 | Singleton | Ae. tauschii | 1 | 1 | 1 | |||
| HI068F4 | Singleton | Ae. tauschii | 0 | 0 | 0 | |||
| BE404580 | HD012G15 | ctg1754 | Ae. tauschii | 0 | 0 | 0 | ||
| 4 | BE497309 | HD008H1a | Singleton | Ae. tauschii | 17 | 13 | 6 | C/++ |
| BE497635 | RI003E6a | ctg2862 | Ae. tauschii | 2 | 1 | 0 | P/+++ | |
| HD073F23a | Singleton | Ae. tauschii | 2 | 1 | 0 | C/+++; P/++ | ||
| BF202706 | RI004I21 | ctg8723 | Ae. tauschii | 0 | 0 | 0 | ||
| HD024H2a | Singleton | Ae. tauschii | 3 | 2 | 1 | C/++; P/++ | ||
| HD015P19a | Singleton | Ae. tauschii | 3 | 1 | 1 | C/++; P/++ | ||
| HD003D13 | Singleton | Ae. tauschii | 3 | 2 | 1 | |||
| 21E12a | NA | Ae. speltoides | 11 | 5 | 3 | C/+++ | ||
| 256K19a | NA | Ae. speltoides | 11 | 4 | 6 | C/+++ | ||
| 5 | BE403618 | HD67C12 | ctg5061 | Ae. tauschii | 0 | 0 | 0 | |
| HI004K2a | ctg3651 | Ae. tauschii | 1 | 1 | 1 | − | ||
| HI074N14a | Singleton | Ae. tauschii | 1 | 1 | 1 | − | ||
| HD90B04 | Singleton | Ae. tauschii | 0 | 0 | 0 | |||
| 6 | BF428533 | HD062L14 | ctg3048 | Ae. tauschii | 0 | 0 | 0 | |
| HD80B22 | ctg3048 | Ae. tauschii | 0 | 0 | 0 | |||
| HI80I03a | ctg3048 | Ae. tauschii | 1 | 1 | 1 | P/+++ | ||
| HD14P08 | Singleton | Ae. tauschii | 0 | 0 | 0 | |||
| BE405809 | RI31A22 | ctg3817 | Ae. tauschii | 1 | 1 | 1 | ||
| 7 | 6729.t10 | HD28P20a | ctg6291 | Ae. tauschii | 0 | 0 | 1 | P/+++ |
| HD57F11a | ctg6291 | Ae. tauschii | 0 | 0 | 0 | P/++ | ||
| 6730.t11-CS |
HD32N11a |
Singleton |
Ae. tauschii |
1 |
1 |
1 |
P/+++ |
|
Localization of FISH signals: C, centromere; P, paint along entire chromosomes. − and + represent, respectively, the presence and absence of hybridization signals: +++, strong signal; ++, intermediate signal. NA, contig information is not available for Ae. speltoides BACs.
Selected BACs for BAC-FISH.
Contig information is taken from http://wheatdb.ucdavis.edu:8080/wheatdb. FPC assembly is 1.1 version.
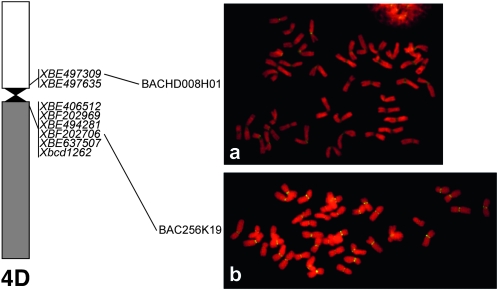
To further confirm their physical location, the BAC clones anchored by W4 ESTs were mapped by BAC-FISH to mitotic metaphase chromosomes of CS wheat. Wheat EST BE497635 mapped to the R3 centromere. Of the Ae. tauschii BACs harboring this locus, HD073F23 preferentially hybridized to the centromeres of CS chromosomes, but also hybridized weakly over their entire length. The second BAC RI003E6 gave a dispersed signal (Table 3). The Ae. tauschii BAC clone HD008H01 harboring BE497309 (mapped to the short arm of R3 at a distance of 3.4 Mb from the centromere) was exclusively localized at the primary constriction of CS chromosomes (Figure 4). Of the Ae. tauschii and Ae. speltoides BAC clones harboring BE202706, the Ae. speltoides BACs gave a strong signal exclusively at the centromeres of CS chromosomes, whereas Ae. tauschii BACs gave a signal at the centromeres as well as along their entire length (Table 3, Figure 4). The apparently centromeric BE202706 clone in wheat was mapped in the terminal end of the long arm of R11 at 11.5 Mb from the centromere in rice.
Figure 4.—
FISH patterns of (a) Ae. tauschii BAC HD008H01 and (b) Ae. speltoides BAC 256K19. Both BACs anchored by group-4 ESTs are exclusively located in the wheat centromeres visualized by yellow-green FITC florescence.
Nine BAC clones that showed only one fragment with the centromere-specific repeats harboring pericentromeric ESTs from chromosomes W1, W2, W3, W5, W6, and W7 were also analyzed by BAC-FISH. Six gave a FISH signal over the entire length of all CS chromosomes due to their high content of noncentromere-specific repetitive DNA, and three did not hybridize to any of the wheat chromosomes (Table 3).
DISCUSSION
The impetus for the present study came from the discovery of transcribed genes in the CENH3 core domains of the centromeres of rice chromosomes 3 and 8 (Nagaki et al. 2004; Yan et al. 2005, 2006). In wheat, a collection of telosomic chromosomes with breaks at the centromere are available for all 42 arms of the 21 chromosomes and for many of the related Triticeae species added to wheat as telosomic additions (Sears and Steinitz-Sears 1978; Islam et al. 1981; Mukai et al. 1992; Friebe et al. 1993, 2000). These stocks allow mapping of any probe to the short arm, the long arm, or the centromere (see Figure 1). Because gene synteny is conserved between wheat and rice, comparative mapping of centromeric genes of rice on wheat aneuploid stocks provides an elegant system for testing centromere homology between wheat and rice. For Cen8, contiguous rice genes 6729.t09 and 6729.t10 located in the CENH3 core mapped to the centromeric bin in the short arm of wheat chromosomes 7A, 7B, and 7D. Another CENH3-domain rice gene 6730.t11, located at 200 kb from 6729.t09/10, mapped to the long arm of wheat chromosomes 7A and 7B but to the short arm of 7D. Thus, these two genes that are 200 kb apart in the rice CENH3 domain span the large centromere in wheat group-7 chromosomes and are subjected to frequent inversions due to the dynamic nature of pericentromeric regions. Gene 6733.t09 located outside the CENH3 domain in rice mapped to the distal ends of group-3 chromosomes and provided evidence for the breakdown of synteny. The data presented here provide a method for determining centromere homology between wheat and rice. These and other aspects in the structure and evolution of the wheat and rice centromeric regions are discussed below.
Recurrent origin of pericentromeric inversions:
Previously Qi et al. (2006) reported that pericentric regions in the Triticeae, especially those of group-4 chromosomes, have undergone rapid and recurrent rearrangements. Pericentromeric inversions close to the centromere regions were detected in wheat chromosomes 2B, 3B, 4A, 4B, 5A, and 6B, as well as in other Triticeae species (Conley et al. 2004; Linkiewicz et al. 2004, Miftahudin et al. 2004; Qi et al. 2004, 2006). Analyzing W7-R8 centromere homology, we observed two independent pericentromeric inversions involving chromosome 7R of rye and 7D of wheat (Figures 1b and 2). This is the first report on a pericentromeric inversion in a D-genome chromosome of wheat, a relatively conserved genome compared to other genomes of hexaploid wheat (Qi et al. 2004, 2006).
Pericentromeric inversions were also reported between chimpanzee and human chromosomes on the basis of comparative karyotyping and on chromosome 4 of A. thaliana specific to several ecotypes (Yunis and Prakash 1982; Nickerson and Nelson 1998; Fransz et al. 2000; Goidts et al. 2005; Kehrer-Sawatzki et al. 2005). It is not clear why pericentromeric regions are prone to inversions. One possibility could be the occurrence of more frequent ectopic recombination, because the centromeres are enriched in tandem satellite repeat units. Another reason could be the stress imposed on the centromeres especially during movement to the spindle poles. Kinetochores trap the microtubules and act as “arm joints” that bear the chromosome load during their movement to the poles and may be damaged. The repair of damaged centromeres in the subsequent interphase may lead to structural changes including pericentromeric inversions. Nevertheless, as a result of pericentric inversions, the CENH3 domains and associated genes are moving targets changing position, moving in, out, and around the centromeric regions, and there may be an adaptive value to these perturbations for specific chromosome services rendered. The practical value and useful outcome of the identification of the genes and clones involved in pericentromeric inversions is that they must be close to the centromere and thus provide useful markers in analyzing centromere homology in taxa where gene synteny is conserved, as is the case between wheat and rice.
Centromere synteny between wheat and rice:
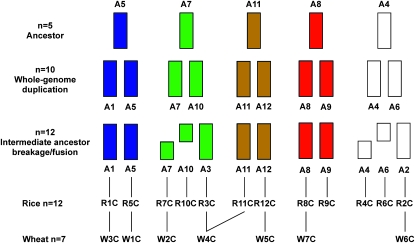
Wheat and the Triticeae with 1x = 7 have 7 centromeres and rice with 1x = 12 has 12 centromeres. If 1x = 12 was the ancestral chromosome number of the common progenitor of wheat and rice (Salse et al. 2008), then what was the fate of the 5 centromeres during the evolutionary reduction of the basic chromosome number from 1x = 12 to 1x = 7 in wheat and the Triticeae? During this reduction in basic chromosome number, some centromeres may have been conserved, whereas others were inactivated or eliminated, and 2 may have fused to form 1, or others may have arisen de novo. One of the important findings of the present study is that most wheat centromeres showed one-to-one correspondence to rice centromeres. We detected homology, with one possible exception, between 7 wheat and 7 rice centromeres.
Large-scale EST sequence comparisons using bin-mapped wheat ESTs and rice pseudomolecules had previously indicated colinearity between W3 and R1 and between W6 and R2 chromosomes (Sorrells et al. 2003; La Rota and Sorrells 2004; Munkvold et al. 2004; Randhawa et al. 2004). Their centromere homology was confirmed in this study. W3-R1 and W6-R2 chromosomes and their centromeres have maintained perfect synteny (Figure 3). The remaining wheat and Triticeae chromosomes are associated with linkage blocks corresponding to two or three rice chromosomes (Ahn et al. 1993; Gale and Devos 1998; Sorrells et al. 2003; Conley et al. 2004; Hossain et al. 2004; La Rota and Sorrells 2004; Linkiewicz et al. 2004; Miftahudin et al. 2004; Peng et al. 2004; Randhawa et al. 2004; Salse et al. 2008). Chromosome W1 arose from the fusion of chromosomes with homology to R5 and R10 but its centromere was derived from R5 (Figure 3). Chromosome W2 was derived from the fusion of ancestral chromosomes sharing homology with R4 and R7, and the W2 centromere is homologous to R7. Chromosome W5 arose from the fusion of ancestral chromosomes sharing homology with R3, R9, and R12, but its centromere was derived from R12. Chromosome W7 arose from the fusion of a chromosome sharing homology with R6 and R8 and, as deduced previously, we confirmed that its centromere was derived from R8.
The centromere region of W4 appears to be more complex and may be an exception to the single-centromere origin in wheat (Figure 3). Chromosome W4 arose mainly from the fusion of chromosomes sharing homology with R3 and R11 and its centromere may be derived from R3, but a hybrid origin containing parts of R3 and R11 centromeres cannot be ruled out (Figure 6). The EST BF202706 was mapped to the pericentromeric region of W4 by both deletion mapping and BAC-FISH analysis (Figure 4). However, rice BAC OsNBa0042J05 aligned by this EST is located in the distal region of rice chromosome 11 (Figure 3). A possible paracentric inversion could explain the difference in location of this EST between wheat and rice.
Figure 6.—
Model for centromere evolution of rice and wheat from a common ancestor with n = 5 chromosomes, modified from Salse et al. (2008). C, centromere.
We did not detect centromere homology of any of the wheat chromosomes to the centromeres of R4, R6, R9, and R10. On the basis of these results, the most likely hypothesis is that centromeres sharing homologies with R4, R6, R9, and R10 were either eliminated or inactivated (Birchler et al. 2009). However, a rigorous test of this hypothesis must await the complete assembly of the centromeres of these chromosomes (similar to R3 and R8; Nagaki et al. 2004; Wu et al. 2004; Yan et al. 2006) followed by comparative mapping in wheat aneuploid stocks using the approach outlined in this article.
Surprisingly, our present study indicated that most of the pericentromeric ESTs of the W5 long arm matched the short arm BACs located on the duplicated block of rice R11S and R12S (Table S4, Figure 3). Sequence alignment of the same W5 ESTs against the Brachypodium 8× release database and BAC-FISH using Brachypodium BAC clones anchored by W5 ESTs as probes indicated that the Brachypodium homologous sequences to W5 ESTs are located in the distal end of the long arm of Brachypodium distachyon chromosome 4 with similar gene order to that in wheat (L. L. Qi, B. Friebe, Y. Q. Gu, Q. Chen, B. S. Gill, unpublished data, http://www.brachybase.org/blast/) A hypothesis of evolutionarily independent inversion events is illustrated in Figure 5 to explain the difference in location of these homologous sequences in wheat and rice. In an ancestral chromosome, homologous sequences represented by selected wheat ESTs reside in the distal end of the long arm. A pericentric inversion occurred to form rice chromosome 12. In wheat, an insertion or translocation moved the sequences from the distal end of the ancestral chromosome to the region proximal to the centromere of the long arm of W5 chromosomes with the same gene order as that in the ancestor. It is evident that the long arm of wheat group-5 chromosomes was known as a recombined chromosome arm, with sequence similarity to rice chromosomes R12, R9, and R3 (Figure 5, La Rota and Sorrells 2004; Linkiewicz et al. 2004; Rice Chromosomes 11 and 12 Sequencing Consortia 2005). The pericentric inversion is specific to the rice lineage. It is supported by data that more than 90% of wheat ESTs that mapped to the short arms of group-5 chromosomes aligned to the long arm of rice chromosome 12, and most of wheat ESTs that mapped to the long arms of group-5 chromosomes aligned to the short arm of rice chromosome 12 (Sorrells et al. 2003; Linkiewicz et al. 2004; Rice Chromosomes 11 and 12 Sequencing Consortia 2005). In barley, most ESTs in the distal part of the short arm of chromosome 5H mapped to the distal region of the long arm of rice chromosome 12 (Stein et al. 2007).
Figure 5.—
Chromosome evolution of rice chromosome 12 and wheat group-5 chromosome. Different ancestral chromosomes are color coded. Five wheat ESTs that mapped to the pericentromeric region of the long arm of group-5 chromosomes were selected and represent homologous sequences in the ancestor, wheat, and rice. S, short arm; L, long arm.
In the model of cereal karyotype evolution proposed by Salse et al. (2008), a common ancestor with five chromosomes, A5, A7, A11, A8, and A4, underwent whole-genome duplication to produce an intermediate ancestor with n = 10 chromosomes that, following breakage and fusion events, produced the n = 12 karyotype of rice (Figure 6). The centromeres of wheat chromosomes can be traced to the ancestral chromosomes as follows: W1 and W3 trace their centromere to A5 through duplicated chromosomes A1 and A5; W2 and W4 trace their lineages to ancestral chromosome A7. In addition, W4 may contain a part of a centromere tracing to the A11 lineage (Figure 6). W5, W6, and W7 trace their lineages directly to ancestral chromosomes A11, A8, and A4, respectively. The depicted framework provides a working model for further studies on the structure and evolution of cereal chromosome centromeres.
Acknowledgments
We thank Jiming Jiang for sharing information on the rice R8 centromeric clones, W. John Raupp for editorial assistance of the manuscript, and Duane L. Wilson for excellent technical help. This research was supported by a special U.S. Department of Agriculture–Cooperative State Research, Education, and Extension Service grant to the Wheat Genetic and Genomic Resources Center. This article is contribution number 09-334-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan, KS.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107409/DC1.
References
- Abbo, S., R. P. Dunford, T. Foote, S. M. Reader, R. B. Flavell et al., 1995. Organization of retroelement and stem-loop repeat families in the genomes and nuclei of cereals. Chromosome Res. 3 5–15. [DOI] [PubMed] [Google Scholar]
- Ahn, S., J. A. Anderson, M. E. Sorrells and S. D. Tanksley, 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241 483–490. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. Chromosome specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., G. Zhi and F. Han, 2009. A tale of two centromeres—diversity of structure but conservation of function in plants and animals. Funct. Integr. Genomics 9 7–13. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., F. Dong, T. Langdon, S. Ouyang, C. R. Buell et al., 2002. Functional rice centromees are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, E. J., V. Nduati, J. L. Gonzalez-Hernandez, A. Mesfin, M. Trudeau-Spanjers et al., 2004. A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics 168 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., K. Nickel, T. Kuromori, M. I. Benito, S. Kaul et al., 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468–2474. [DOI] [PubMed] [Google Scholar]
- Dawe, R. K., L. M. Reed, H. G. Yu, M. G. Muszynski and E. N. Hiatt, 1999. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., T. Miller, S. A. Jacson, G. L. Wang, P. C. Ronald et al., 1998. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87 295–307. [Google Scholar]
- Francki, M. G., W. A. Berzonsky, H. M. Ohm and J. M. Anderson, 2002. Physical location of a HSP70 gene homologue on the centromere of chromosome 1B of wheat (Triticum aestivum L.). Theor. Appl. Genet. 104 184–191. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., S. Armstrong, J. H. de Jong, L. D. Parnell, C. van Drunen et al., 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana. Cell 100 367–376. [DOI] [PubMed] [Google Scholar]
- Friebe, B., Y. Mukai and B. S. Gill, 1993. Standard karyotype of Triticum longissimum and its cytogenetic relationship with T. aestivum. Genome 36 731–742. [DOI] [PubMed] [Google Scholar]
- Friebe, B., L. L. Qi, S. Nasuda, P. Zhang, N. A. Tuleen et al., 2000. Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theor. Appl. Genet. 101 51–58. [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Plant comparative genetics after 10 years. Science 282 656–659. [DOI] [PubMed] [Google Scholar]
- Gill, B. S., B. Friebe and T. R. Endo, 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34 830–839. [Google Scholar]
- Goidts, V., J. M. Szamalek, P. J. de Jong, D. N. Cooper, N. Chuzhanova et al., 2005. Independent intrachromosomal recombination events underlie the pericentric inversions of chimpanzee and gorilla chromosomes homologous to human chromosome 16. Genome Res. 15 1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Hosouchi, T., N. Kumekawa, H. Tsuruoka and H. Kotani, 2002. Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 9 117–121. [DOI] [PubMed] [Google Scholar]
- Hossain, K. G., V. Kalavacharla, G. R. Lazo, J. Hegstad, M. J. Wentz et al., 2004. A chromosome bin map of 2148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. X., A. Sirikhachornkit, X. J. Su, J. D. Faris, B. S. Gill et al., 2002. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Islam, A. K. M. R., K. W. Shepherd and D. B. H. Sparrow, 1981. Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 46 161–174. [Google Scholar]
- Jiang, J., J. A. Birchler, W. A. Parrott and R. K. Dawe, 2003. A molecular view of plant centromeres. Trends Plant Sci. 8 570–575. [DOI] [PubMed] [Google Scholar]
- Jin, W. W., J. R. Melo, K. Nagaki, P. B. Talbert, S. Henikoff et al., 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W.W., J. C. Lamb, J. M. Vega, R. K. Dawe, J. A. Birchler et al., 2005. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay, L., and R. K. Dawe, 2009. Centromeres: long intergenic spaces with adaptive features. Funct. Integr. Genomics 9 287–292. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., C. Sandig, N. Chuzhanova, V. Goidts, J. M. Szamalek, et al., 2005. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes). Hum. Mutat. 25 45–55. [DOI] [PubMed] [Google Scholar]
- La Rota, M., and M. E. Sorrells, 2004. Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct. Integr. Genomics 4 34–46. [DOI] [PubMed] [Google Scholar]
- Lee, H. R., W. Zhang, T. Langdon, W. W. Jin, H. Yan et al., 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkiewicz, A. M., L. L. Qi, B. S. Gill, A. Ratnasiri, B. Echalier et al., 2004. A 2500-locus bin map of wheat homoeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics 168 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., R. A. Wing, J. L. Bennetzen and S. A. Jackson, 2007. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 23 134–139. [DOI] [PubMed] [Google Scholar]
- Miftahudin, K. Ross, X.-F. Ma, A. A. Mahmoud, J. Layton, et al., 2004. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai, Y., B. Friebe and B. S. Gill, 1992. Comparison of C- banding patterns and in situ hybridization sites using highly repetitive and total genomic rye DNA probes of 'Imperial' rye chromosomes added to 'Chinese Spring' wheat. Jpn. J. Genet. 67 71–83. [Google Scholar]
- Munkvold, J. D., R. A. Greene, C. E. Bermudez-Kandianis, C. M. La Rota, H. Edwards et al., 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Nickerson, E., and D. L. Nelson, 1998. Molecular definition of pericentric invasion breakpoints occurring during the evolution of humans and chimpanzees. Genomics 50 368–372. [DOI] [PubMed] [Google Scholar]
- Palmer, D. K., K. O'Day, H. L. Trong, H. Charbonneau and R. L. Margolis, 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histon. Proc. Natl. Acad. Sci. USA 88 3734–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bowers and B. A. Chapman, 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA 101 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bower, R. Bruggmann, I. Dubchak, J. Grimwood et al., 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556. [DOI] [PubMed] [Google Scholar]
- Peng, J. H., H. Zadeh, G. R. Lazo, J. P. Gustafson, S. Chao et al., 2004. Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homology with rice and Arabidopsis. Genetics 168 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for using in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3 39–55. [DOI] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, S. Chao, G. R. Lazo, G. E. Butler et al., 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploidy wheat. Genetics 168 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L. L., B. Friebe and B. S. Gill, 2006. Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome 49 1628–1639. [DOI] [PubMed] [Google Scholar]
- Randhawa, H. S., M. Dilbirligi, D. Sidhu, M. Erayman, D. Sandhu et al., 2004. Deletion mapping of homologous group 6-specific wheat expressed sequence Tags. Genetics 168 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosomes 11 and 12 Sequencing Consortia 2005. The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 3 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, E. K., S. K. Flowers and E. J. Richards, 1997. Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res. 7 1045–1053. [DOI] [PubMed] [Google Scholar]
- Saffery, R., H. Sumer, S. Hassan, L. H. Wong, J. M. Craig et al., 2003. Transcription within a functional human centromere. Mol. Cell 12 509–516. [DOI] [PubMed] [Google Scholar]
- Salse, J., S. Bolot, M. Throude, V. Jouffe, B. Piegu et al., 2008. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, D., J. A. Champoux, S. N. Bondareva and K. S. Gill, 2001. Identification and physical localization of useful genes and markers to a major gene-rich region on wheat 1S chromosomes. Genetics 157 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler, M. G., A. W. Higgins, M. K. Rudd, K. Gustashaw and H. F. Willard, 2001. Genomic and genetic definition of a functional human centromere. Science 294 109–115. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., and L. M. Steinitz-Sears, 1978. The telocentric chromosomes of common wheat, pp. 389–407 in: Proceedings of the Fifth International Wheat Genetics Symposium, edited by S. Ramanujam. Ind. Soc. Gen. Plt. Breed. New Delhi, India.
- Singh, N. K., V. Dalal, K. Batra, B. K. Singh, G. Chitra et al., 2007. Single-copy genes define a conserved order between rice and wheat for understanding differences caused by duplication, deletion, and transposition of genes. Funct. Integr. Genomics 7 17–35. [DOI] [PubMed] [Google Scholar]
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, N., M. Prasad, U. Scholz, T. Thiel, H. G. Zhang et al., 2007. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor. Appl. Genet. 114 823–839. [DOI] [PubMed] [Google Scholar]
- Su, X., J. Wahlstrom and G. Karpen, 1997. Molecular structure of a functional Drosophila centromere. Cell 91 1007–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B. A., M. D. Blower and G. H. Karpen, 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2 584–596. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., R. Masuelli, A. P. Tyagi, L. Comai and S. Henikoff, 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F., E. Coe, W. Nelson, A. K. Bharti, F. Engler et al., 2007. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3 1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., N. Kurata, H. Tanoue, T. Shimokawa, Y. Umehara et al., 1998. Physical mapping of duplicated genomic regions of two chromosome ends in rice. Genetics 150 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., H. Yamagata, M. Hayashi-Tsugane, S. Hijishita, M. Fujisawa et al., 2004. Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H., W. Jin, K. Nagaki, S. Tian, S. Ouyang et al., 2005. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17 3227–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H., H. Ito, K. Nobuta, S. Ouyang, W. Jin et al., 2006. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18 2123–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., J. Wang, W. Lin, S. Li, H. Li et al., 2005. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3 266–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis, J. J., and O. Prakash, 1982. The origin of man: a chromosomal pictorial legacy. Science 215 1525–1530. [DOI] [PubMed] [Google Scholar]
- Zhang, P., B. Friebe, A. J. Lukaszewski and B. S. Gill, 2001. The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110 335–344. [DOI] [PubMed] [Google Scholar]
- Zhang, P., W. L. Li, J. Fellers, B. Friebe and B. S. Gill, 2004. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112 288–299. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Y. Huang, L. Zhang, Y. Li, T. Lu et al., 2004. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32: 2023–2030. [DOI] [PMC free article] [PubMed]
- Zhong, C. X., J. B. Marshall, C. Topp, R. Mroczek, A. Kato et al., 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]