Abstract
Yeast replication checkpoint mutants lose viability following transient exposure to hydroxyurea, a replication-impeding drug. In an effort to understand the basis for this lethality, we discovered that different events are responsible for inviability in checkpoint-deficient cells harboring mutations in the mec1 and rad53 genes. By monitoring genomewide replication dynamics of cells exposed to hydroxyurea, we show that cells with a checkpoint deficient allele of RAD53, rad53K227A, fail to duplicate centromeres. Following removal of the drug, however, rad53K227A cells recover substantial DNA replication, including replication through centromeres. Despite this recovery, the rad53K227A mutant fails to achieve biorientation of sister centromeres during recovery from hydroxyurea, leading to secondary activation of the spindle assembly checkpoint (SAC), aneuploidy, and lethal chromosome segregation errors. We demonstrate that cell lethality from this segregation defect could be partially remedied by reinforcing bipolar attachment. In contrast, cells with the mec1-1 sml1-1 mutations suffer from severely impaired replication resumption upon removal of hydroxyurea. mec1-1 sml1-1 cells can, however, duplicate at least some of their centromeres and achieve bipolar attachment, leading to abortive segregation and fragmentation of incompletely replicated chromosomes. Our results highlight the importance of replicating yeast centromeres early and reveal different mechanisms of cell death due to differences in replication fork progression.
CENTROMERES have long been known to be one of the earliest regions of the budding yeast genome to replicate during S phase (McCarroll and Fangman 1988). However, the biological significance of early replication of centromeres remains speculative, partly owing to the lack of mutants showing altered or delayed timing of centromere replication. During our investigation of chromosome replication dynamics during nucleotide shortage brought upon by the treatment with hydroxyurea (HU), we discovered that problems with centromere replication can lead to fundamentally different forms of genome instability.
The two mutations that exhibit interesting centromere replication phenotypes are in the genes encoding two essential protein kinases, Mec1 and Rad53, which play pivotal roles in the cellular response to DNA damaging agents as well as in cell cycle arrest in response to HU (Branzei and Foiani 2006; Tourriere and Pasero 2007). Mutations in the kinase domains of Mec1 and Rad53 render the proteins checkpoint deficient and cause the cells carrying such mutations to be hypersensitive to HU. Because Mec1 is an upstream effector of Rad53 in the replication checkpoint pathway, checkpoint-deficient alleles of the two genes are thought to lead to similar phenotypes in response to replication impediments. When rad53 cells encounter HU during S phase, they fail to slow the temporal program of origin firing, expose large regions of single-stranded DNA (ssDNA) at effectively all origins, and elongate their spindles, a phenotype that indicates that the cells are attempting premature chromosome partitioning (Allen et al. 1994; Weinert et al. 1994; Desany et al. 1998; Santocanale and Diffley 1998; Sogo et al. 2002; Feng et al. 2006). Similarly, mec1 cells have also been shown to initiate precocious segregation of unreplicated chromosomes upon exposure to HU (Weinert et al. 1994; Sanchez et al. 1996). Even after the removal of HU, both mec1 and rad53 cells show considerable reduction in their ability to produce progeny. However, the reason for inviability after HU exposure is ill defined. We reasoned that understanding the molecular basis of cell death would help elucidate the role of checkpoint control in DNA replication and cell cycle regulation and in the maintenance of genome integrity.
Previous reports indicated that following transient exposure to HU, rad53 checkpoint-deficient cells are unable to complete DNA replication, which in turn was thought to constitute the primary reason for the loss of viability (Desany et al. 1998; Lopes et al. 2001). This conclusion was corroborated by the observation that delaying premature chromosome segregation by inhibiting microtubule assembly (via nocodazole treatment) following HU exposure was unable to improve the viability of rad53 cells (Desany et al. 1998). Together, these studies suggested that incomplete replication rather than premature chromosome segregation per se was the major reason for loss of viability in both rad53 and mec1 cells, following treatment with HU. By flow cytometric measurements it appeared that rad53 cells were able to replicate slowly a significant amount of genomic DNA following transient (30–60 min) exposure to HU, but that these chromosomes did not enter pulse field gels (Desany et al. 1998). The authors proposed that the DNA synthesized by rad53 cells must contain gaps, branches, or other structures that retard the mobility of chromosomal DNA in the pulse field gels. The presence of putative abnormal replication intermediates was later revealed by two-dimensional gel electrophoresis and by electron microscopy (Lopes et al. 2001; Sogo et al. 2002).
Intrigued by the results of Desany et al. (1998), we decided to ask where in the genome DNA replication occurred during recovery from HU—in particular, whether forks established during HU treatment were able to resume or whether replication was occurring from unfired origins. By addressing this question, we hoped to identify what precisely was the defect, if any, in DNA replication during the recovery phase from HU in cells lacking the checkpoint function.
In this study, we examine the cellular responses of two checkpoint-deficient mutants, mec1-1 sml1-1 and rad53K227A (Weinert et al. 1994; Sogo et al. 2002), upon exposure to HU during the initiation of S phase and during recovery after HU is removed. We chose the mec1-1 sml1-1 strain for our studies because it is in an isogenic background (A364a) as the rad53K227A mutant that we have previously studied. The mec1-1 allele is phenotypically identical to a mec1 null allele and it requires the sml1-1 mutation for viability (Zhao et al. 1998; Basrai et al. 1999). In contrast, the rad53K227A mutation (Feng et al. 2006) is not lethal and the strain harboring this mutation does not contain the sml1-1 allele (data not shown). Through the usage of these specific mutations in the checkpoint pathway, we observed that the mec1-1 sml1-1 and rad53K227A cells, upon exposure to HU, lose viability through distinct mechanisms that arise from differences in centromere replication in the two mutants. We further demonstrate that the extent to which the two mutants recover DNA synthesis upon the removal of HU also differs. Our study reveals the importance of early replication of centromeres and underscores the involvement of the replication checkpoint pathway in the establishment of chromosome biorientation.
MATERIALS AND METHODS
Yeast strains and media:
Yeast strains used in this study are listed in supporting information, Table S2. Cells were grown at 30° in synthetic complete medium unless otherwise indicated. α-Factor was used at 200 nm for bar1 strains and 3 μm for BAR1 strains. Pronase was used at 25 μg/ml and 300 μg/ml for bar1 and BAR1 strains, respectively, to remove α-factor from the culture medium. HU was added at 200 mm and nocodazole was used at 15 μg/ml.
Measurement of cell viability by colony formation assay:
Cells synchronized in G1 by α-factor treatment were released into medium containing 200 mm HU or in combination with 15 μg/ml nocodazole as detailed in the main text. Aliquots of 100 μl were removed for serial dilutions in ice-cold minimal medium lacking a nitrogen source. The cell suspension of the appropriate concentration was sonicated briefly before plating aliquots in triplicate on solid medium. The plates were incubated at 30° for 2–3 days before colonies were counted and analyzed.
Contour-clamped homogeneous electric field gel electrophoresis and Southern blotting:
Contour-clamped homogeneous electric field (CHEF) gel analysis was performed as described previously (van Brabant et al. 2001). Electrophoresis was conducted at 14° for 25 hr with a switch time ramped from 60 to 120 sec at 200 V. The Chr IX probe was amplified from genomic DNA with primers of the following sequences: forward, 5′-CTATGACGAGGGCGAAGAAG-3′; reverse, 5′-ATT TCACAGGGCCAGACACG-3′. Southern blotting was performed according to standard procedures.
Flow cytometry:
Cells were collected and mixed with 0.1% NaN3, followed by fixing with 70% ethanol. Flow cytometry was performed using standard procedures after staining the cells with Sytox Green (Molecular Probes) and the data were analyzed with CellQuest software (Becton-Dickinson).
Chromosome biorientation assay:
Cells carrying the CEN4-GFP tag and the cdc23-1 mutation were blocked in G1 with α-factor and released into YPD medium at 35° (to block cells at the metaphase/anaphase transition by inactivating the Cdc23 protein) in the presence or absence of 200 mm HU. After 1 hr, cells were allowed to recover in fresh medium lacking HU at 35°. At the indicated times, samples were evaluated for the percentage of cells with two separated CEN4-GFP foci, indicative of successful biorientation at the cdc23 block.
Indirect end labeling:
Yeast chromosomes embedded in agarose gels were prepared as previously described (van Brabant et al. 2001). A detailed protocol for in-gel restriction digestion can be found at http://fangman-brewer.genetics.washington.edu/fork-D.html. The digested agarose plugs were then placed in wells of a 0.4% agarose gel (without ethidium bromide) and electrophoresed at 1 V/cm for 26 hr at room temperature. Standard Southern blotting techniques were used. The primer sequences for the DSF2 probe are: DSF2-F, 5′-TTTCATTACCTCCAACGCCA-3′; DSF2-R, 5′-TTTCGGACCTTGTTTCATGT-3′. The TRP1 probe was isolated as a HindIII fragment from plasmid pTA-DIR (M. K. Raghuraman, unpublished results.).
Genomic ssDNA mapping:
ssDNA analysis on rad53 and mec1 sml1 cells was performed as previously described (Feng et al. 2006, 2007).
Density transfer:
Dense-isotope substitution experiments were performed essentially as described earlier (McCarroll and Fangman 1988; Raghuraman et al. 2001) with modifications. A detailed protocol can be found at http://fangman-brewer.genetics.washington.edu/density_transfer.html. Thirty minutes prior to the release from α-factor the culture was transferred to isotopically light medium to equilibrate nucleotide pools. HU was added at 200 mm just prior to pronase addition. After 1 (for rad53 and mec1 sml1 cells) or 2 [for wild-type (WT) cells] hr, cells were filtered to remove HU and allowed to recover in fresh medium without HU. Samples were collected at the times indicated in the figure legends and genomic DNA was extracted followed by fractionation after ultracentrifugation in a CsCl density gradient. An aliquot of each gradient fraction was slot blotted, followed by hybridization with centromere DNA probes (CEN2 and CEN4) or a whole-genomic DNA probe to identify the unreplicated (HH) DNA and replicated (HL) DNA. The CEN2 fragment was amplified from genomic DNA with the following primers: forward, 5′-TAGTCTATCAGCCTCCGAAG-3′; reverse, 5′-GTA GGTGCCAGTTGAATAGC-3′. The CEN4 fragment was excised as a XhoI-SpeI restriction fragment from the plasmid YCpG.EDamtd (M. K. Raghuraman, unpublished results). For microarray analysis, those fractions containing HH and HL DNA identified by slot blotting and hybridization with a genomic DNA probe were pooled, respectively. The HH and HL DNA for each timed sample were differentially labeled with cyanine (Cy3 or Cy5)-conjugated dUTP (Perkin Elmer) as described by the Brown laboratory (http://cmgm.stanford.edu/pbrown/protocols/4_genomic.html) followed by purification through a Sephadex G-50 column and ethanol precipitation for microarray hybridization.
Microarray hybridization and analysis:
See File S2.
RESULTS
Extent of replication fork progression in HU inferred from ssDNA profiles:
Treatment of checkpoint-deficient rad53K227A cells (henceforth referred to as rad53 cells) with HU during a synchronized S phase generates regions of ssDNA restricted to the immediate vicinity of virtually all replication origins in the yeast genome (Feng et al. 2006, 2007). The colocalization of ssDNA with origins suggests that replication forks are unable to progress very far into adjacent regions before they become arrested. To assess whether the accumulation of ssDNA at origins is unique to the rad53 mutation, we analyzed a mec1-1 sml1-1 (henceforth referred to as mec1 sml1) checkpoint deficient mutant under similar conditions.
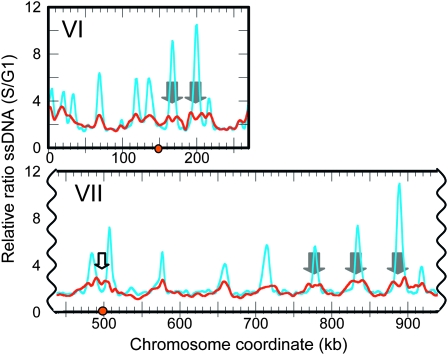
When we released mec1 sml1 cells from a G1 arrest into HU for 1 hr, ssDNA also appeared at virtually all replication origins in the genome. However the peaks of ssDNA in mec1 sml1 cells, though significantly above background, were lower, broader, and often “split” in comparison to those in rad53K227A cells (data from Feng et al. 2006; Figure 1 and Figure S1), suggesting that replication forks in mec1 sml1 cells were able to proceed further than were forks in rad53 cells. While the majority of centromeres in rad53 cells are located in troughs between ssDNA peaks, in mec1 sml1 cells, ssDNA extended into some centromeric regions (such as Chr VII, Figure 1, open block arrow). This observation prompted us to investigate whether centromere duplication during HU treatment differs between these two mutants.
Figure 1.—
Genomic ssDNA mapping reveals more extensive migration of replication forks in mec1 sml1 (yMP10913) cells than in rad53 (WFY34) cells. Cells were released from G1 arrest to enter S phase in the presence of 200 mm HU. Samples were collected at 0 (α-factor arrested) and 1 hr (S phase) and DNA from the two samples was isolated, differentially labeled with Cy-conjugated dUTPs without the denaturation of template DNA (to limit synthesis to the single-stranded portions of the genome; details in Feng et al. 2007), and cohybridized to a microarray to obtain the relative ratio of ssDNA in the S phase sample to that in the G1 control sample (S/G1) at any region of the genome. The ratio of ssDNA for rad53 (cyan) and mec1 sml1 (red) for chromosome VI and a portion of VII are shown. Gray block arrows mark “split peaks” and the open block arrow marks ssDNA near CEN7. Orange dots indicate centromere locations. See Figure S1 for all 16 chromosomes.
Centromere replication in HU:
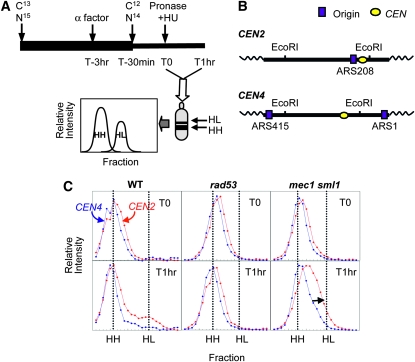
We examined the extent of centromere replication in rad53 and mec1 sml1 cells during exposure to HU using a density transfer method (Figure 2A). Following EcoRI digestion, unreplicated (HH) and replicated (HL) genomic DNA were hybridized with probes to detect the EcoRI fragments that contained CEN2 (4.078 kb, 65.1% A+T) and CEN4 (5.778 kb, 63.5% A+T) (see materials and methods; Figure 2A). These centromeres differ in the distance to the nearest potential origin of replication or autonomously replicating sequence (ARS): CEN2 is ∼500 bp from ARS208 and CEN4 is 13.7 kb from ARS1 (Figure 2B). Surprisingly, neither of the fragments in the rad53 or the mec1 sml1 sample showed any DNA of fully hybrid density (HL) at T1hr in HU (Figure 2C). However, the profile of CEN2 in mec1 sml1 cells at T1hr is slightly broader and shifted more to the HL position than that at T0 (Figure 2C), consistent with partial replication. No such shift is observed for either centromere in the rad53 sample. Given the presence of ssDNA in the sample, the actual amount of replication that had occurred at CEN2 may be underrepresented, as indicated by the observation that filling in the ssDNA gaps by in vitro synthesis increased the shift to the HL position in the mec1 CEN2 but not CEN4 fragment (Figure S2). From the extent of the shift we estimate that replication forks from ARS208 on the CEN2 fragments synthesized an average of <1 kb. On the basis of the calculated distances between each of the 16 centromeres and their nearest ARSs (using a compiled origin list by Nieduszynski, http://www.oridb.org) only two centromeres (CEN3 and CEN12) are closer to an ARS than CEN2 is (Table S1). While we have not done an exhaustive test to ask whether any of the other centromeres was replicated in rad53 cells in HU, we note that we have never been able to detect any shift toward hybrid DNA for the genome as a whole or even for efficient, early firing origins such as ARS305 and ARS306 (data not shown). We conclude that a very small percentage of rad53 cells are capable of replicating even a single centromere whereas, under the same conditions, mec1 sml1 cells can potentially duplicate at least three centromeres (CEN2, -3, and -12).
Figure 2.—
Differences in centromere replication in mec1 sml1 (WFY73) and rad53 (WFY34) cells in HU. HM14-3a cells were used as a WT control. (A) Experimental scheme for density transfer and subsequent analyses. Cells were propagated in isotopically dense (13C and 15N) medium for at least eight generations prior to G1 arrest. At 30 min prior to release into S phase (time = T-30 min), cells were switched to light medium (12C and 14N) to allow nucleotide pools to equilibrate. At time 0 (T0), cells were released into S phase in the presence of 200 mm HU. The culture was harvested an hour later (T1hr). DNA was extracted, digested with EcoRI and subjected to equilibrium centrifugation in CsCl gradients to separate HH (unreplicated) and HL (replicated) DNA. The resulting gradients were fractionated and each fraction slot blotted onto nylon membranes to allow hybridization with specific DNA probes. (B) CEN2 and CEN4 locations in relation to their neighboring origins on their respective EcoRI fragments (not drawn to scale). (C) Relative amounts of replication of the EcoRI fragments containing CEN4 (blue) and CEN2 (red), for rad53 and mec1 sml1. The spreading of the HH peak toward the position of hybrid DNA (HL) is indicated by a black arrow.
Two concurrent studies showed that chromosomes remained monopolarly tethered to one of the spindle pole bodies (SPBs) during premature spindle extension in HU-treated checkpoint mutants (Krishnan et al. 2004; Bachant et al. 2005). It was proposed that, for rad53 cells, it was due to the lack of centromere replication in HU that the chromosomes fail to biorient and the cells undergo precocious spindle extension (Bachant et al. 2005). This hypothesis is consistent with our finding that rad53 cells failed to replicate centromeres during exposure to HU.
Replication resumption after HU exposure:
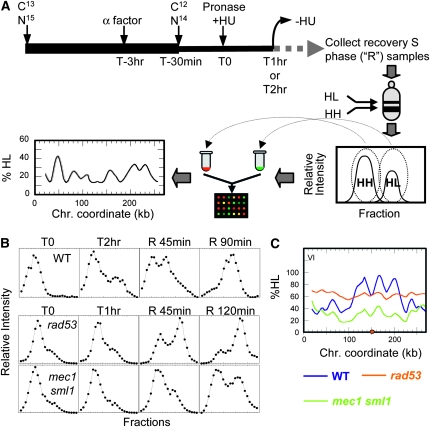
To ask whether the difference in the extent of replication by rad53 and mec1 sml1 cells in the presence of HU has an impact on how cells recover once HU is removed, we measured the progress of replication after removal of HU. Density transfers (Figure 3A) were identical to those described earlier with the exception that after cells were exposed to HU for 1 hr, they were filtered and allowed to recover in fresh light medium without HU for up to 2 hr. The extent of genomic DNA replication was measured by hybridization with total genomic DNA (Figure 3B). We found that rad53 cells were able to recover from HU and replicate the bulk of the genome (Figure 3B), including the centromeres (Figure 3C; Figure S3), by 120 min. In contrast, mec1 sml1 cells accumulated far less HL DNA (<50%) during the same recovery period (Figure 3, B and C). These observations suggest that mec1 sml1 cells were less capable of resuming synthesis after exposure to HU than rad53 cells.
Figure 3.—
Resumption of DNA synthesis after transient HU exposure in mec1 sml1 (WFY73) vs. rad53 (WFY34) cells. HM14-3a cells were used as a WT control. (A) Experimental scheme for density transfer coupled with microarray analysis. Density transfer and DNA isolation procedures were identical to those described in Figure 2 except that after cells had been transferred from dense to light medium and exposed to HU for 1 hr (for rad53 and mec1 sml1 cells) or for 2 hr (for WT cells), HU was removed from the culture by filtration and the cells were resuspended in fresh medium containing light isotopes and allowed to recover. Samples were removed at intervals during this “recovery” S phase (labeled as “R” samples). The “HH” and “HL” pools were differentially labeled with Cy-conjugated dUTP (Amersham) and cohybridized to a microarray. The smoothed %HL values were plotted against chromosome coordinates to generate replication profiles. (B) Slot blot results of CsCl gradients of WT (top), rad53 (middle), and mec1 sml1 (bottom) samples using genomic DNA as probe. (C) Replication dynamics of rad53 R45 sample (orange) and mec1 sml1 R45 sample (green), along with a WT control sample recovering for 45 min from a 2-hr HU exposure (blue). The orange dot indicates the centromere. For whole genome plots, see Figure S3.
We hybridized HH and HL DNA from these recovery samples to microarrays to investigate which parts of the genome were able to resume replication. By comparing samples that had recovered for 45 min after a 1-hr HU exposure to a sample of WT control cells that were also given a 45-min recovery period following a 2-hr exposure to HU, we observed that the temporal order of replication in the rad53 and mec1 sml1 mutants was altered: WT cells simply resumed a normal pattern of chromosome replication; however, for both mec1 sml1 and rad53 cells accumulation of HL DNA in normally early replicating regions was delayed relative to later replicating regions, resulting in rather flat replication profiles (Figure 3C, Figure S3). These results are consistent with the notion that in the mutants, replication forks become “damaged” and are less likely to resume replication after HU exposure. Therefore, the cells would need to rely on initiation of new forks from any unused late/dormant origins to complete genome replication.
High level of chromosome breakage after HU exposure in mec1 sml1 but not rad53 cells:
As budding yeast cells assemble intranuclear mitotic spindles during S phase, it has been suggested that early replication of centromeres might be crucial for ensuring bipolar attachment of the chromosomes during S phase and thus their proper segregation later in mitosis (Tanaka et al. 2005). If mec1 sml1 but not rad53 cells were able to duplicate at least some of their centromeres during HU exposure, then mec1 sml1 cells would have a much higher probability of at least a few chromosomes achieving bipolar attachment during HU treatment and thereby reducing overall spindle extension. However, because mec1 sml1 cells have difficulty in completing replication after HU is removed, we would predict that they might be prone to chromosome breakage during recovery from HU.
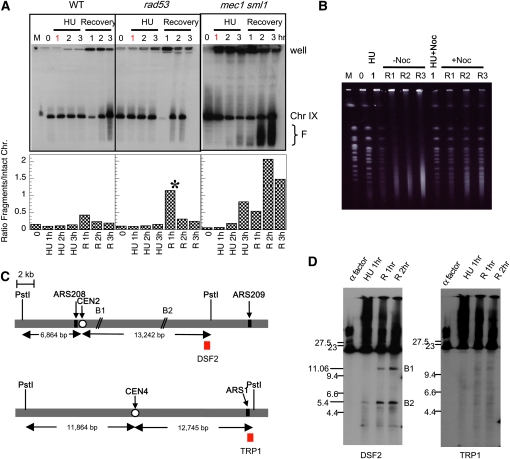
We used CHEF gel electrophoresis to examine the chromosomes in cells following transient exposure to HU. Cells that had been exposed to HU for 1 hr were washed and resuspended in fresh medium without HU and samples were collected every hour for up to 3 hr. Chromosome breakage was assayed by Southern blotting using probes located near the ends of specific chromosomes. For all three strains (WT, rad53, and mec1 sml1) chromosomes remained mostly intact for up to 3 hr in HU, with mec1 sml1 cells experiencing a slightly elevated level of chromosome IX breakage (Figure 4A). However, upon the removal of HU at 1 hr, mec1 sml1 cells showed substantial chromosome breakage, while rad53 cells were comparable to WT cells (Figure 4A). Similar results were obtained with a chromosome V probe (data not shown).
Figure 4.—
mec1 sml1 but not rad53 cells show chromosome breakage during recovery from transient (1 hr) exposure to HU in S phase. (A) Top: CHEF gel and Southern blot (probed with a telomeric-proximal Chr IX probe) of WT (HM14-3a), mec1 sml1 (WFY73), and rad53 (WFY34) cells exposed to HU continuously for up to 3 hr (HU: 1, 2, and 3 hr) or, after 1-hr exposure to HU, allowed to recover for another 3 hr (recovery: 1, 2, and 3 hr). M, yeast chromosome markers; 0, α factor-arrested cells. The positions of sample wells, intact chromosomes, and fragmentation products (F) are indicated. Bottom: Chromosome breakage expressed as relative ratios of fragmented to intact chromosomes, quantified from relative pixel counts from the Southern blots on a Packard InstantImager Electronic Autoradiography system. Note that the ratio for the R1h sample of rad53 (marked by an asterisk) is unreliable because most of the chromosomes remained trapped in the well. (B) Comparative analyses of chromosome breakage in mec1 sml1 cells exposed to HU only (HU) or HU and nocodazole (HU+Noc) for 1 hr and released into media without nocodazole (−Noc, R1-3; i.e., recovery: 1, 2, and 3 hr) or with nocodazole (+Noc, R1-3; i.e., recovery: 1, 2, and 3 hr). CHEF gel electrophoresis followed by ethidium bromide staining is shown. (C and D) Chromosome breakage detected near CEN2 but not CEN4. (C) The structures of the PstI fragments containing CEN2 and CEN4, respectively. (D) Chromosomal DNA from mec1 sml1 cells recovering from 1-hr exposure to HU was embedded in agarose and in-gel digested with PstI before electrophoretic separation. Southern blotting was performed with probes indicated in C as red blocks. The two breakage sites (B1 and B2) near CEN2 are indicated.
If the chromosome breakage in mec1 sml1 cells resulted from tension exerted by the spindle on the partially replicated chromosomes, we would expect that blocking spindle extension via nocodazole treatment would prevent or reduce chromosome breakage during recovery from HU. To test this hypothesis, we released mec1 sml1 cells from the G1 block into medium containing HU (“HU”) or HU and nocodazole (“HU+Noc”) and, after a 1-hr exposure, allowed them to recover in just fresh medium (“−Noc”) or in the continued presence of nocodazole (“+Noc”) (Figure 4B). As expected, for the samples recovering in the absence of nocodazole (“−Noc”) most of the genomic DNA was reduced to fragments of a median size of ∼200 kb (Figure 4B). In contrast, fragmentation was greatly reduced in cells that recovered in the presence of nocodazole (“+Noc”) leaving chromosomes mostly intact. This result supports our hypothesis that the incompletely replicated chromosomes in mec1 sml1 cells break directly or indirectly as a consequence of tension exerted by the spindle. The residual breakage occurring in the presence of nocodazole could be attributed to either incomplete blockage of spindle extension by nocodazole or spontaneous breakage that occurred independently of tension.
Chromosome breakage near the centromere in mec1 cells:
If spindle tension on the bioriented centromeres of partially duplicated chromosomes is responsible for some of the breaks in the mec1 sml1 cells recovering from HU treatment, then breaks should be detected near centromeres such as CEN2 but not CEN4 because CEN2 has a higher probability of being duplicated during HU treatment than CEN4. We employed an indirect end-labeling method to ask whether in vivo breakage occurs near CEN2 and CEN4. mec1 sml1 cells at different stages of HU treatment and recovery was embedded in agarose plugs and genomic DNA was digested in-gel with PstI, which generates 20-kb and 24.6-kb fragments containing CEN2 and CEN4, respectively (Figure 4C). Hybridizing the Southern blot with probes located near the end of each fragment (DSF2 for Chr II and TRP1 for Chr IV, Figure 4C) allowed us to detect and map the potential sites of chromosome breakage on each fragment.
We detected two breakage sites in the pericentric region of CEN2, labeled B1 and B2, located ∼2 kb and 10 kb to the right of CEN2, respectively (Figure 4, C and D). Breakage at B1 is observed only during recovery; breakage at B2 is detected during HU treatment but increases in frequency during recovery (Figure 4D). The relative percentages of breakage at these sites at 2 hr during recovery are 3.8 and 4.9% for B1 and B2, respectively. These values are consistent with the notion that chromosome breakage occurs at a low rate at any given locus but collectively contribute to the overall level of chromosome breaks. Neither breakage is detected in α-factor arrested cells, suggesting that replication is required to generate both breaks (Figure 4D). In contrast, no significant sites of breakage near CEN4 were observed using a TRP1 probe on the right end of the PstI fragment on Chr IV (Figure 4D). These observations are consistent with our hypothesis that incompletely replicated chromosomes in HU-treated mec1 sml1 cells are subject to breakage, and that at least some of this breakage occurs as a direct consequence of bipolar spindle force.
Partial improvement of rad53 cell viability by eliminating reorientation of kinetochore-to-SPB connections:
If rad53 cells can resume replication after HU removal rapidly enough to escape chromosome fragmentation, why do they die after exposure to HU? We propose that at least one type of lethal event in rad53 cells in HU is the precocious segregation of chromosomes with an unreplicated centromere that can only experience monopolar attachments. Furthermore, if the spindle assembly checkpoint (SAC) is active during the HU-challenged S phase, then attachments to the old spindle will be repeatedly broken as the error correction mechanisms attempt to biorient chromosomes by distributing attachments between the two spindle poles. With randomly oriented, unreplicated centromeres, the genome would undergo a reductional segregation toward the two spindle poles.
This proposed scenario is reminiscent of the phenotype of a cdc6 mutant that is defective in the initiation of DNA synthesis—cdc6 cells undergo reductional mitosis to randomly segregate their intact unreplicated chromosomes (Hartwell 1976; Bueno and Russell 1992; Liang et al. 1995; Piatti et al. 1995). However, in a cdc6 ipl1 double mutant (IPL1 encodes the yeast homolog of the mammalian Aurora B kinase) (Chan and Botstein 1993; Biggins et al. 1999), the chromosomes predominantly remain attached to the old spindle pole (Tanaka et al. 2002). Consequently, all the chromosomes segregate to the daughter cell (Tanaka et al. 2002). Therefore, we hypothesized that inactivating the Ipl1 kinase to avert the reductional mitosis might improve rad53 cell viability in HU by increasing the chance that one of the cells would inherit the entire genome and proceed with replication once HU is removed.
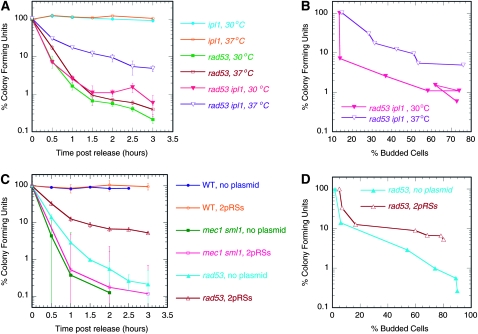
A rad53 ipl1 culture was split upon release from α-factor arrest, with one half incubated at 30° (the permissive temperature for the ipl1-321 mutation), and the other half at 37° (the nonpermissive temperature). Cells incubated at 37° showed significantly higher levels of viability than did the cells incubated at 30° (Figure 5A). This observation held true even after we accounted for the difference in cell cycle entry kinetics at the two temperatures (Figure 5B). The viability of either single mutant was unaffected by temperature (Figure 5A). These results support our predictions that (1) the inactivation of the Ipl1 kinase would allow at least some of the rad53 cells to inherit a full genome in the presence of HU, and (2) during recovery from HU, rad53 cells would be able to finish replicating the bulk of their chromosomes and retain viability. Among the rad53 ipl1 cells that had survived HU exposure, we would expect to find some cells with greater than one genome equivalent of DNA content. We screened 14 colonies of rad53 ipl1 HU survivors by flow cytometry. Most of these colonies gave rise to cultures that were heterogeneous in DNA content, ranging from one to two genome equivalents, indicative of aneuploidy (Figure S4).
Figure 5.—
Lethality in rad53 cells in HU can be partially rescued by inactivating the Ipl1 kinase (A and B) or by the introduction of CEN-ARS plasmids (C and D). The percentage of colony forming units was calculated for each sample by normalizing to the control T0 sample. Error bars represent standard deviations. (A) Effect of the ipl1-321 temperature sensitive mutation on viability of rad53 cells released into HU at 30° or 37°. ipl1 (SBY630), rad53 (WFY88), and ipl1 rad53 (WFY80) cells are shown. (B) The cell viability of ipl1 rad53 cells at 30° and 37° described in A was plotted against percentage of budded cells at the respective temperature. (C) Viability of cells with or without CEN-ARS plasmids (pRS315 and pRS316) in HU. (D) Viability of rad53 cells transformed with the two plasmids or no plasmid described in C was plotted against the percentage of budded cells for comparison of cell viability at equivalent cell cycle stages.
Because the ipl1 mutation is defective in the tension-sensing branch of the SAC, it is formally possible that it is the inactivation of the SAC rather than the elimination of the kinetochore-to-pole reorientation that rescues rad53 lethality in HU. If this postulate were true, then one would expect that the inactivation of the SAC via a mutation in the MAD2 gene would also rescue rad53 lethality in HU. We found that the double mutant rad53 mad2Δ did not have elevated viability in HU compared to the rad53 single mutant (Figure S5). Therefore, we conclude that ipl1 mutation rescues rad53 HU sensitivity by forcing all chromosomes to cosegregate with the same pole, thereby preventing reductional segregation of the genome.
Partial restoration of rad53 cell viability in HU by CEN-ARS plasmids:
We have estimated from the mec1 sml1 cells that a minimum of three duplicated centromeres is sufficient to block precocious spindle extension and random segregation of the genome. Previously, it was demonstrated that the introduction of multiple CEN-ARS plasmids in which a centromere was placed immediately adjacent to an ARS to increase the number of replicated centromeres was able to reduce the level of spindle extension in rad53 cells in HU (Bachant et al. 2005). We wanted to ask whether reinforcing bipolar attachment would actually ameliorate the loss of viability in rad53 cells in HU. We tested both mec1 sml1 and rad53 cells bearing two CEN-ARS plasmids (pRS314 and pRS316) for their ability to form colonies after HU treatment.
Cells grown under selection for the plasmid markers were released from a G1 arrest into selective medium containing HU. Samples were collected during S phase and plated on nonselective medium lacking HU. At the time of transfer the culture was quite heterogeneous with only 30% of the population containing both plasmids. Therefore we chose not to select for the presence of the plasmids after cells were plated because we wanted to examine specifically whether the subset of cells that actually contained plasmids were able to improve cell viability during HU exposure, not whether the ensuing plasmid replication and segregation were successful. After 3 days of incubation, colonies were counted to calculate the relative viability of cells over time in HU.
The culture of rad53 cells transformed with the two CEN-ARS plasmids had improved viability (∼10% at 3 hr in HU) when compared to the control cells without the plasmids (<1% at 3 hr in HU) (Figure 5C). To ensure that the improved survival was not simply due to slowed S phase entry of the transformed cells, we also compared cell viability at times when the two cell cultures had reached similar levels of budding or cell cycle entry. To make this comparison, we plotted the cell viability as a function of the budding indices (Figure 5D). This plot confirms that the viability of cells that contained plasmids was higher than that of control cells at the same cell cycle stage. Moreover, rescue was specific to the rad53 mutant, as we did not observe any increase in viability of mec1 sml1 cells with the plasmids (Figure 5C). Thus, we conclude that increasing the number of centromeres in rad53 cells that could potentially be replicated in the presence of HU, and thus become bioriented, stabilizes the mitotic spindle and prevents the precocious random segregation of chromosomes with only monopolar attachment.
rad53 mutants are unable to biorient or separate chromosomes after HU treatment:
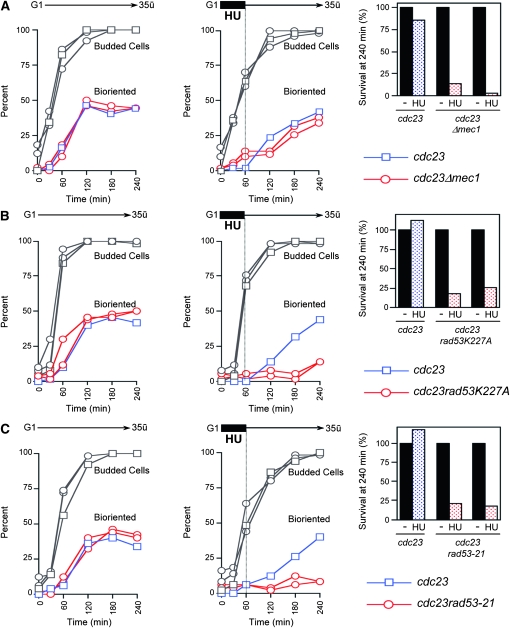
Our observations thus far are consistent with the hypothesis that rad53 cells suffer from reductional chromosome segregation during HU treatment, but can substantially replicate centromeres following removal of HU. To determine if these replicated centromeres can establish bipolar connections to the spindle, we compared the ability of rad53K227A, rad53-21, and mec1Δ GAP-RNR3 cells to recover chromosome biorientation following a 1-hr HU treatment; rad53-21 is a well-characterized S phase checkpoint defective allele (Allen et al. 1994) and mec1Δ GAP-RNR3 (where the essential function of MEC1 is compensated by the overproduction of RNR3 under the glyceraldehyde 3-phosphate dehydrogenase promoter, similar to the sml1 mutation; Desany et al. 1998) is phenotypically identical to mec1-1 sml1. A cdc23-1 mutation was introduced to block anaphase entry and allow ample time for chromosome biorientation. The centromere on Chr IV was tagged with GFP (CEN4-GFP) to assay biorientation (Goshima and Yanagida 2000); bipolar attachment normally causes sister CENs to split into two distinct GFP foci. Following a 1-hr exposure to HU, we observed that WT and the mec1Δ GAP-RNR3 mutant exhibited fairly similar kinetics of biorientation. Both rad53 mutants, however, largely failed to achieve biorientation (Figure 6).
Figure 6.—
Lack of biorientation of chromosomes in rad53K227A cells after removal of HU. (A) cdc23 (JBY686) and cdc23 mec1Δ GAP-RNR3 (JBY1720; two experiments) cells carrying CEN4-GFP were released from α-factor arrest into S phase in YPD media at 35° in the presence or absence of 200 mm HU for 1 hr, followed by recovery in fresh media lacking HU at 35°. At the indicated times, samples were evaluated for the percentage of cells with two separated CEN4-GFP foci, indicative of successful biorientation at the cdc23 block. At the 240-min end point, cell viability was measured as the percentage of colony forming units of both HU-treated and -untreated cells; viability in the HU-untreated culture was normalized to 100%. (B) cdc23 (JBY1726) and cdc23 rad53K227A (JBY1728, JBY1729; two independent isolates) cells and (C) cdc23 (JBY1732) and cdc23 rad53-21 (JBY1735, JBY1736; two independent isolates) cells with CEN4-GFP were processed and scored as described in A.
Defective biorientation in rad53 cells would be expected to activate the SAC, leading to a Mad2-dependent delay in sister chromatid separation and progression through the metaphase-to-anaphase transition. Indeed, we found the anaphase inhibitor Pds1/securin (Yamamoto et al. 1996) was stabilized in rad53 cells for up to 5 hr after a 1-hr HU exposure. This stabilization was largely alleviated in a rad53 mad2Δ double mutant (Figure S6, A and B). We also examined sister chromatid separation during HU recovery using TRP1-GFP near CEN4. Although our density transfer experiments revealed efficient replication of both TRP1 and CEN4 in rad53 cells during HU recovery, very little sister chromatid separation was observed in rad53 and rad53 mad2Δ strains, either when cells were washed out of HU into mating pheromone to restore a second G1 arrest or washed out into nocodazole (Figure S6, C and D). In contrast, mad2Δ control cells were proficient for TRP1-GFP separation under both conditions (Figure S6, C and D). Since the chromosome segregation defects in rad53 cells appears to activate the SAC, these observations suggest a physical impediment to chromatid disjunction is associated with HU recovery defects in rad53 cells.
DISCUSSION
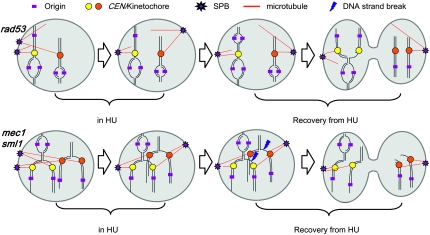
Why replication-checkpoint-deficient yeast cells die when challenged with a replication impediment is a long-standing question. Do cells die because they cannot complete some aspect of genome replication after the impediment is removed, or is it because, in the absence of the checkpoint, they have executed some critical cell cycle events out of order? Characterizing replication and segregation phenotypes of mutants in two checkpoint kinases, we have been able to distinguish how failure of each of these two processes contributes to distinct types of genome instability (Figure 7).
Figure 7.—
A model for the differential chromosomal behaviors in rad53 and mec1 cells upon exposure to HU. Two representative chromosomes are depicted. The centromeres and their associated kinetochores, origins of replication, spindle pole bodies, and spindles are labeled as indicated in the legend. In rad53 cells, upon HU exposure, replication forks initiated from nearby origins do not reach the centromeres, failing to duplicate them. Meanwhile, SPB duplicates and chromosomes are randomly attached to one of the two SPBs in a monopolar fashion, followed by SPBs separation in the absence of tension on the spindle. After HU is removed, despite substantial replication through resumption of replication forks (on the chromosome with orange centromere) or new initiation events (on the chromosome with yellow centromere), the genome is randomly segregated to opposite poles. In mec1 sml1 cells, replication forks are able to reach and duplicate some centromeres and establish bipolar attachment of the chromosomes. But the failure to finish replicating the chromosomes causes them to break under tension on the spindle after HU is removed.
We found that when rad53 cells encounter HU in S phase they are unable to duplicate their centromeres and therefore do not achieve the bipolar attachment that normally occurs early in S phase to generate tension within the spindle. As a consequence of premature spindle elongation, unreplicated chromosomes become randomly partitioned to the two spindle poles. Though rad53 cells are capable of nearly completing genomic DNA replication after the removal of HU, we propose that by the time that replication restarts, the window of opportunity to establish bioriented chromosomes has passed and/or that rad53 cells suffer from specific defects that preclude chromosome biorientation (such as structural defects of the centromeres and/or kinetochores) or sister chromatid disjunction during recovery from HU. Thus, rad53 cells are destined for random chromosome segregation by both precocious spindle extension during HU treatment and lethal defects in chromosome attachment/separation after HU removal. In contrast, mec1 sml1 cells are largely proficient for bipolar attachment but experience lethal chromosome breaks due to nondisjunction of incompletely replicated chromosomes. Although the force exerted by a spindle microtubule (Nicklas 1983; Bloom 2008) is thought to be insufficient to make a dsDNA break (Bensimon et al. 1995), the presence of ssDNA brings spindle force-induced breakage into the realm of possibility. Alternatively, spindle force may induce chromatin unraveling, making regions more accessible to nucleases—an idea consistent with our observation of discrete breakage sites (Figure 4D; see File S1 for a fuller discussion).
While we refer to these differences in replication and cell cycle execution as resulting from the loss of Rad53 or Mec1 checkpoint function, we do not mean to imply that we have necessarily detected distinct functions for Rad53 and Mec1 kinases in the DNA replication checkpoint pathway. It may be the case that Rad53 and Mec1 kinases do have distinct phosphorylation targets, and that the two mutations reflect these differences. However, there are at least two other explanations for the phenotypic differences that we have observed in the two mutants. (1) The different point mutations in the rad53K227A and mec1-1 sml1 alleles may retain different residual levels of checkpoint activation and the specific phenotypes may reflect this difference. (2) The mec1 sml1 strain may have higher level of dNTPs than rad53 due to the sml1 mutation. However, we were informed that by chromatin immuoprecipitation of bromodeoxyuridine-labeled DNA coupled with microarray analysis, it was observed that mec1 mutants (mec1-1 and mec1-100) consistently showed longer tracks or more extensive fork progression than a rad53 mutant (rad53-11) in the presence of HU (L. Crabbe, P. Pasero and A. Lengronne, personal communication). Because neither the mec1-100 nor the rad53-11 allele requires the sml1 mutation for survival, this observation suggests that the difference between mec1 sml1 and rad53 mutants may not be simply attributed to different levels of dNTPs in the cell. Regardless of which explanation is correct, the specific mutations we have used have allowed us to identify an important step in the chromosome duplication cycle that is regulated by the DNA replication checkpoint. Interestingly, another checkpoint mutant mrc1, which similarly exhibits precocious spindle elongation upon HU treatment, is able to maintain substantial viability (Alcasabas et al. 2001). However, because a detailed study of replication dynamics and chromosome segregation of mrc1 cells under replication stress has not been performed, it is possible that mrc1 cells can replicate some centromeres in HU as mec1 sml1 cells do and recover replication as rad53 cells do, thus averting both precocious chromosome segregation and chromosome breakage. Our study underscores the importance for yeast cells to execute linked steps of the cell cycle in their correct order and specifically the importance of replicating centromeres early in S phase to ensure proper spindle assembly and chromosome segregation.
A corollary of the hypothesis stated above is that restraining the spindle from extension via nocodazole treatment should ameliorate the loss of viability in rad53 cells in HU. The addition of nocodazole following transient exposure to HU failed to rescue the loss of viability of rad53 cells (Desany et al. 1998). We found that including nocodazole during the exposure to HU also did not result in improved viability (data not shown). However, we do not believe that these results should be taken to suggest that precocious spindle extension is not a principal reason for lethality in rad53 cells after HU treatment. Rather, we speculate that nocodazole affects not only microtubule dynamics but also the behavior of spindle poles. Indeed, we note that even treating RAD53 cells simultaneously with HU and nocodazole during S phase for just half an hour led to a reduction of viability by ∼50% (data not shown). Consistent with this finding is the observation that nocodazole not only prevents spindle pole body (SPB) separation (Yoder et al. 2003) but also abolishes centromere clustering near the SPBs during G1, causing them to drift away from the SPBs (Tanaka et al. 2002). Recent studies that followed kinetochore recapture after nocodazole treatment demonstrated that yeast mitotic chromosomes have difficulty attaching and biorienting on the spindle when the SPBs are in the unseparated (side by side) configuration, and require the functional Sgo1 protein for correction (Indjeian et al. 2005; Indjeian and Murray 2007). If the unduplicated kinetochores in rad53 cells in HU were released from the SPBs to which they were first attached, subsequent duplication and recapture by both SPBs might be an inefficient and error-prone event. Therefore, nocodazole's potential ability to rescue rad53 cell lethality in HU may be masked by its negative effect on SPB segregation and subsequent capture and biorientation of chromosomes.
We note that in the population of cells transformed with the CEN-ARS plasmids we achieved only modest increase in viability (from 3% without CEN-ARS plasmids to 12% with two CEN-ARS plasmids at 60 min in HU). While we do not know for certain how many replicated centromeres are required to restrain the spindle in WT cells, we can make estimates on the basis of our replication studies of rad53 cells in HU. As we were unable to detect any replication of CEN2, which is ∼500 bp from its closest origin of replication, it would leave only two centromeres as candidates, and one of these is adjacent to an ARS that is rarely used under normal circumstances (ARS308). If we were to assume a success rate for replication of either of these two centromeres of 20% (an estimate based on the sensitivity of our density transfer assay to detect even partial replication of fragments containing the efficient origins ARS305 and ARS306), and if two replicated centromeres were sufficient to restrain the spindle to ensure viability upon recovery from HU, then we could expect a survival rate of 4%. Introducing plasmids with similarly spaced centromeres and ARSs would theoretically improve viability to ∼20%. The fact that only 30% of the cells in the culture actually had both plasmids reduces the expectation of survival to be <20%. We predict that introducing origins of replication closer to additional chromosomal centromeres might demonstrate even better rescue of HU sensitivity in rad53 cells than the CEN-ARS plasmids. Likewise, the rescue of rad53 HU sensitivity by ipl1 mutation was only partial presumably because many of the cells that survived are aneuploid (Figure S4) and thus suffer a growth disadvantage (Torres et al. 2007).
Although we have shown that rad53K227A cells suffer from reductional mitosis, it is difficult to ascertain how much cell death of rad53 cells can be attributed to segregation defects as opposed to other causes. In fact, we believe that rad53 cells also suffer substantially from replication defects during recovery from HU, despite near completion of DNA synthesis. It is noteworthy that the observation of the substantial ability of rad53K227A cells to replicate DNA, following transient (1 hr) exposure to HU, is somewhat at odds with a previous study where it was reported that rad53K227A cells are virtually incapable of DNA synthesis after exposure to HU (Lopes et al. 2001). We believe the difference between the two studies lies in the fact that Lopes et al. employed a much more extensive (3 hr) exposure of rad53 cells to HU. We have observed that the ability of rad53 cells to recover from HU decreased as the duration of exposure to HU increased and that the replication occurring during recovery relied even more on new initiation events in cells exposed to HU for longer period than shorter ones, suggesting that replication forks are destabilized in a time-dependent fashion in HU (data not shown). The genomic replication profiles during recovery from HU also showed that the normal replication timing pattern was somewhat reversed in both mutants, suggesting that many replication forks established during HU exposure were defective in resuming synthesis and that some portion of the observed replication during recovery may have originated from new initiation events from previously unfired origins. We are currently investigating the extent to which replication resumption in rad53 and mec1 sml1 cells relies on new initiations vs. resumption from preexisting forks and whether ssDNA gaps are filled or become sites of chromosomal breakage.
Although there are no studies that specifically examine whole-genome replication and spindle assembly in the same WT culture, we can extrapolate our results with checkpoint mutants to propose a possible role of early centromere replication during normal and HU-challenged cell cycles in WT cells. Once the cyclin-dependent kinase (CDK1) becomes active in G1 a cascade of events is initiated that sets in motion two independent pathways leading to spindle pole duplication and chromosome replication. The intra-S-phase checkpoint is responsible for making sure that the two pathways remain coordinated. We imagine that one crucial event is the early replication of at least a few centromeres. Once duplicated, the assembly of kinetochores on these centromeres and their attachment to opposite spindle poles puts a physical restraint on the spindle. In the presence of a replication-inhibiting agent such as HU, the checkpoint cascade must intervene with both chromosome replication and spindle assembly. Engaging the checkpoint accomplishes two important replication functions: (1) stabilizing ongoing forks so that they can continue at a slow rate to incorporate nucleotides during HU treatment and can efficiently resume once HU is removed and (2) delaying activation of unfired origins so that the cell can concentrate the few available nucleotides to sites of the earliest activated replication forks—among which are those near a few centromeres.
Acknowledgments
We thank L. Breeden, B. Garvik, and S. Biggins for providing yeast strains and U. Surana for personal communications at early stages of this work. We are grateful to T. Davis, S. Biggins, B. Byers, J. Sidorova, and the Brewer/Raghuraman lab members for insightful discussions and to the anonymous reviewers for helpful suggestions. We also thank P. Pasero for personal communication of unpublished data. We extend our gratitude to the staff at the Center for Array Technologies in Seattle for microarray hybridization and scanning. This work was supported by the National Institute of General Medical Sciences (NIGMS) grant 18926 (to B.J.B. and M.K.R.). W.F. was supported by a Pathway to Independence award (1K99GM081378-01) from the National Institutes of Health.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107508/DC1.
References
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler et al., 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell. Biol. 3 958–965. [DOI] [PubMed] [Google Scholar]
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg and S. J. Elledge, 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8 2401–2415. [DOI] [PubMed] [Google Scholar]
- Bachant, J., S. R. Jessen, S. E. Kavanaugh and C. S. Fielding, 2005. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 168 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai, M. A., V. E. Velculescu, K. W. Kinzler and P. Hieter, 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7041–7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon, D., A. J. Simon, V. V. Croquette and A. Bensimon, 1995. Stretching DNA with a receding meniscus: experiments and models. Phys. Rev. Lett. 74 4754–4757. [DOI] [PubMed] [Google Scholar]
- Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman et al., 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K. S., 2008. Beyond the code: the mechanical properties of DNA as they relate to mitosis. Chromosoma 117 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei, D., and M. Foiani, 2006. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 312 2654–2659. [DOI] [PubMed] [Google Scholar]
- Bueno, A., and P. Russell, 1992. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C. S., and D. Botstein, 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany, B. A., A. A. Alcasabas, J. B. Bachant and S. J. Elledge, 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W., D. Collingwood, M. E. Boeck, L. A. Fox, G. M. Alvino et al., 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W., M. K. Raghuraman and B. J. Brewer, 2007. Mapping yeast origins of replication via single-stranded DNA detection. Methods 41 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida, 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100 619–633. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H., 1976. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol. 104 803–817. [DOI] [PubMed] [Google Scholar]
- Indjeian, V. B., and A. W. Murray, 2007. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 17 1837–1846. [DOI] [PubMed] [Google Scholar]
- Indjeian, V. B., B. M. Stern and A. W. Murray, 2005. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307 130–133. [DOI] [PubMed] [Google Scholar]
- Krishnan, V., S. Nirantar, K. Crasta, A. Y. Cheng and U. Surana, 2004. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell 16 687–700. [DOI] [PubMed] [Google Scholar]
- Liang, C., M. Weinreich and B. Stillman, 1995. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81 667–676. [DOI] [PubMed] [Google Scholar]
- Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani et al., 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412 557–561. [DOI] [PubMed] [Google Scholar]
- McCarroll, R. M., and W. L. Fangman, 1988. Time of replication of yeast centromeres and telomeres. Cell 54 505–513. [DOI] [PubMed] [Google Scholar]
- Nicklas, R. B., 1983. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 97 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, S., C. Lengauer and K. Nasmyth, 1995. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka et al., 2001. Replication dynamics of the yeast genome. Science 294 115–121. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang et al., 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271 357–360. [DOI] [PubMed] [Google Scholar]
- Santocanale, C., and J. F. Diffley, 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395 615–618. [DOI] [PubMed] [Google Scholar]
- Sogo, J. M., M. Lopes and M. Foiani, 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297 599–602. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., N. Mukae, H. Dewar, M. van Breugel, E. K. James et al., 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434 987–994. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. U., N. Rachidi, C. Janke, G. Pereira, M. Galova et al., 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108 317–329. [DOI] [PubMed] [Google Scholar]
- Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli et al., 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 916–924. [DOI] [PubMed] [Google Scholar]
- Tourriere, H., and P. Pasero, 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6 900–913. [DOI] [PubMed] [Google Scholar]
- van Brabant, A. J., C. D. Buchanan, E. Charboneau, W. L. Fangman and B. J. Brewer, 2001. An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7 705–713. [DOI] [PubMed] [Google Scholar]
- Weinert, T. A., G. L. Kiser and L. H. Hartwell, 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8 652–665. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., V. Guacci and D. Koshland, 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol. 133 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, T. J., C. G. Pearson, K. Bloom and T. N. Davis, 2003. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol. Biol. Cell 14 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., E. G. Muller and R. Rothstein, 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2 329–340. [DOI] [PubMed] [Google Scholar]