Abstract
Small regulatory RNAs are key regulators of gene expression. One class of small regulatory RNAs, termed the endogenous small interfering RNAs (endo siRNAs), is thought to negatively regulate cellular transcripts via an RNA interference (RNAi)-like mechanism termed endogenous RNAi (endo RNAi). A complex of proteins composed of ERI-1/3/5, RRF-3, and DICER (the ERI/DICER complex) mediates endo RNAi processes in Caenorhabditis elegans. We conducted a genetic screen to identify additional components of the endo RNAi machinery. Our screen recovered alleles of eri-9, which encodes a novel DICER-interacting protein, and a missense mutation within the helicase domain of DICER [DCR-1(G492R)]. ERI-9(−) and DCR-1(G492) animals exhibit defects in endo siRNA expression and a concomitant failure to regulate mRNAs that exhibit sequence homology to these endo siRNAs, indicating that ERI-9 and the DCR-1 helicase domain function in the C. elegans endo RNAi pathway. We define a subset of Eri mutant animals (including eri-1, rrf-3, eri-3, and dcr-1, but not eri-9 or ergo-1) that exhibit temperature-sensitive, sperm-specific sterility and defects in X chromosome segregation. Among these mutants we find multiple aberrations in sperm development beginning with cytokinesis and extending through terminal differentiation. These results identify novel components of the endo RNAi machinery, demonstrate differential requirements for the Eri factors in the sperm-producing germline, and begin to delineate the functional requirement for the ERI/DICER complex in sperm development.
EUKARYOTIC cells express a wide variety of 20–30 nucleotide small regulatory RNAs that function in a wide range of biological processes including, but not limited to, heterochromatin formation, developmental timing, defense against parasitic nucleic acids, and genome rearrangement (Lee et al. 1993; Wianny and Zernicka-Goetz 2000; Knight and Bass 2001; Hall et al. 2002; Mochizuki et al. 2002; Plasterk 2002; Mochizuki and Gorovsky 2004; Verdel et al. 2004; Cam et al. 2005). Small regulatory RNAs associate with ARGONAUTE (AGO) and PIWI proteins. Together, small regulatory RNAs and AGO/PIWI proteins seek out and regulate homologous nucleic acid sequences via a variety of mechanisms, including decreased mRNA stability, translational repression, transcriptional repression, meiotic silencing of unpaired DNA, and DNA elimination (Kim 2005).
One class of small regulatory RNA, termed the endogenous small interfering RNAs (endo siRNAs), was identified via biochemical purification in Caenorhabditis elegans (Ambros et al. 2003). Endo siRNAs have now been identified in a wide array of eukaryotic organisms, including mammals (Hamilton et al. 2002; Llave et al. 2002; Tang et al. 2003; Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Tam et al. 2008; Watanabe et al. 2008). In C. elegans, endo siRNAs are complementary to predicted coding and noncoding genomic sequences and map to a large number of clusters within the C. elegans genome (Ruby et al. 2006). Several proteins that are required for the biogenesis and/or stability of a subset of the cellular endo siRNAs in C. elegans have been identified, including the exonuclease ERI-1, the RNA-dependent RNA polymerase (RdRP) RRF-3, ERI-3, and the Tudor-domain protein ERI-5 (collectively, ERIs) (Simmer et al. 2002; Kennedy et al. 2004; Duchaine et al. 2006). ERI-1, RRF-3, ERI-3, and ERI-5 coprecipitate with DCR-1, suggesting that these factors assemble into a complex with Dicer (DCR-1 in C. elegans), an RNase III enzyme that converts double-stranded RNAs (dsRNAs) to small RNAs (Duchaine et al. 2006). This complex of proteins has been termed the ERI/DICER complex. Animals lacking components of the ERI/DICER complex fail to express some endo siRNAs and also overexpress cellular mRNAs that exhibit sequence homology to these endo siRNAs (Duchaine et al. 2006; Lee et al. 2006; Asikainen et al. 2007). Consequently, endo siRNAs are postulated to initiate or perpetuate the silencing of mRNAs via a process termed endogenous RNA interference (endo RNAi) Duchaine et al. 2006; Lee et al. 2006). Finally, C. elegans lacking ERI-1, RRF-3, or ERI-3 exhibit a temperature-sensitive (ts) sterile phenotype, hinting that the ERI/DICER complex and endo siRNAs may play important roles during germline development (Kennedy et al. 2004; Duchaine et al. 2006).
Here we report the molecular identification and characterization of genes required for endo siRNA expression in C. elegans. We identify a subset of endo RNAi genes that are required for endo RNAi in the male germline and document a role for these genes in sperm development.
MATERIALS AND METHODS
C. elegans strains:
Bristol strain N2 was used as the standard wild-type strain. See the supporting information, File S1, for a full list of strains used in these studies.
RNAi experiments:
RNAi experiments were conducted as described previously (Timmons et al. 2001). HT115 E. coli expressing dsRNA, including sqt-3, lir-1, unc-73, cel-1, tra-2, dpy-13, lin-1, unc-22, and lin-15a, were obtained from the Ahringer RNAi library (Kamath et al. 2003) and sequenced to verify their identities. lin-15b RNAi was performed as described previously (Guang et al. 2008).
RNA analysis:
Sequences for quantitative reverse transcriptase PCR (qRT–PCR) primers, Northern analysis probes and in situ probes can be found in File S1. Total RNA samples were prepared by dounce homogenization in TRIzol solution (Invitrogen) followed by isopropanol precipitation. Small RNAs were enriched utilizing a mirVana microRNA (miRNA) isolation kit (Ambion) according to the manufacturer's protocol. For Northern analysis, 10–20 μg of RNA (50 μg of RNA for ssp-16) was separated on 15% polyacrylamide (for small RNAs) or on 1.5% agarose (for mRNAs) denaturing gel, transferred to Hybond-N+ membrane (GE-Amersham) with a semi-dry apparatus (Hoefer), and blotted in Ultrahyb-Oligo hybridization buffer (Ambion). Strand-specific oligonucleotide probes were synthesized and labeled with [α-32P]dATP utilizing a StarFire kit (IDT). Membranes were washed in 2× SSC + 0.5% SDS, and signals were detected by PhosphorImager (Molecular Dynamics). For qRT–PCR, cDNA was generated from RNA with iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. qPCR was performed on an iCycler machine (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Whole-mount in situ hybridization was performed essentially as described previously (Motohashi et al. 2006). Digoxigenin (DIG)-labeled, strand-specific probes were synthesized from full-length ssp-16 cDNA by multiple cycles of primer extension in the presence of DIG-dNTP. Fixed worms were costained with DAPI and an α-digoxigenin antibody conjugated to alkaline phosphatase (Roche).
Sperm analysis:
Sperm and nuclear morphology were determined by gonad dissection into SM medium (50 mm HEPES, 45 mm NaCl, 25 mm KCl, 5 mm CaCl2, 1 mm MgSO4, pH 7.8), supplemented with 10 mg/ml polyvinyl pyrrolidone (average molecular weight = 40,000), containing DAPI (Shakes and Ward 1989). In vitro activation of spermatids was quantified after treatment with monensin at 100 nm concentration (Sigma Chemicals) on poly-l-lysine-coated slides (Shakes and Ward 1989). In vivo activation and sperm transfer were assessed by vital staining of adult males with the fluorescent dye MitoTracker Red CMXRos (Molecular Probes) followed by mating with unstained fem-1(hc17ts) adult hermaphrodites (Hill and L'Hernault 2001). Early germline development was visualized in intact young adult animals by fixation with cold methanol followed by DAPI staining. Microscopy was performed with Zeiss Axio Imager equipped for DIC Nomarski and fluorescence imaging.
RESULTS
A genetic screen identifies novel Eri genes:
We previously conducted a genetic screen for regulators of RNAi that identified the genes eri-1, rrf-3, eri-3, and eri-5 (Kennedy et al. 2004; Duchaine et al. 2006). Animals defective for these genes fail to accumulate endo siRNAs (Ambros et al. 2003; Duchaine et al. 2006). In addition to this endo siRNA defect, eri-1, rrf-3, eri-3, and eri-5 mutants respond more robustly than wild-type animals to exogenous sources of dsRNA, a phenotype referred to as enhanced RNAi (Eri) (Figure 1A and Simmer et al. 2002; Kennedy et al. 2004; Duchaine et al. 2006). To explain this phenomenon, a model was proposed in which different small regulatory RNA pathways compete for limiting amounts of shared components. We have taken advantage of the Eri phenotype to screen for additional factors required for the production and stability of endo siRNAs.
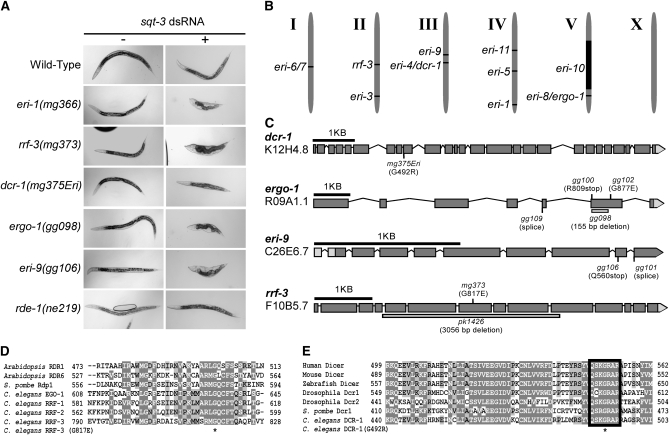
Figure 1.—
A genetic screen identifies Eri genes. (A) Strains of the indicated genotype were treated with double-stranded RNA derived from sqt-3 (Timmons et al. 2001). (B) Schematic of mapping position of Eri genes on the six C. elegans linkage groups. (C) Schematic of the eri-4/dcr-1, eri-8/ergo-1, eri-9, and rrf-3 genes. Positions and types of identified mutations are indicated, including mutations predicted to affect splicing (splice) patterns. Boxes indicate exons; lines indicate introns as predicted by WormBase release 190. (D) rrf-3(mg373) alters a conserved glycine residue. An asterisk indicates G817. (E) dcr-1(mg375Eri) alters a conserved residue within the DCR-1 helicase domain. Black outline indicates motif VI of DEAD-box helicases. An asterisk indicates G492.
We chemically mutagenized ∼150,000 haploid genomes and screened for mutant animals that exhibit enhanced sensitivity to dsRNAs (for details of screens, see Figure S1). We identified five new complementation groups that defined the genes eri-4, eri-8, eri-9, eri-10, and eri-11 (Figure 1B). Subsequent characterization (described below) identified eri-4(mg375) as an allele of dcr-1 and eri-8 as being allelic to ergo-1. For the sake of clarity, the designations dcr-1(mg375Eri) and ergo-1 are henceforth used where appropriate. Our screens identified a single allele of eri-4/dcr-1, mg375, and multiple alleles of eri-8/ergo-1 and eri-9. These screens also recovered alleles of the previously identified eri-1, rrf-3, and eri-3 genes (Figure S1). For example, we identified an allele of rrf-3, termed rrf-3(mg373), that encodes an amino acid substitution [RRF-3(G817E)] of a glycine residue that is highly conserved in related RdRPs (Figure 1, C and D). Here, we focus most of our analysis on eri-4/dcr-1, eri-8/ergo-1, and eri-9.
Animals mutant for eri-1, rrf-3, eri-3, or eri-5 exhibit enhanced sensitivity to dsRNAs targeting a wide-array of mRNAs (Simmer et al. 2002; Kennedy et al. 2004; Duchaine et al. 2006; Lee et al. 2006). We found that ergo-1(−), eri-9(−), and, to a lesser extent, dcr-1(mg375Eri) animals likewise exhibit a generalized enhanced sensitivity to RNAi (Figure 1A and Table 1). For example, hypomorphic alleles of sqt-3 trigger a dumpy phenotype (van der Keyl et al. 1994). Exposure of wild-type animals to dsRNA derived from the sqt-3 gene (sqt-3 RNAi) fails to induce a dumpy phenotype (Figure 1A). sqt-3 RNAi, however, is sufficient to induce a dumpy phenotype in eri-1(mg366), ergo-1(gg098), or eri-9(gg106) animals (Figure 1A). dcr-1(mg375Eri) animals exhibited an enhanced, but less pronounced, response to sqt-3 RNAi (Figure 1A). Genetic criteria indicate that these Eri genes are not components of the class B synthetic multi-vulva pathway (Table S1), mutations of which also exhibit enhanced sensitivity to RNAi (Wang et al. 2005). Rather, animals carrying mutations for eri-1;eri-9,eri-1; ergo-1, or eri-1;dcr-1(mg375Eri) exhibited Eri phenotypes similar to animals harboring the individual mutations, suggesting that eri-9, ergo-1, and dcr-1(mg375Eri) are components of the eri-1 genetic pathway (Table 1).
TABLE 1.
Enhanced sensitivity to exogenous dsRNA
| Feeding RNAi (phenotype scored) |
|||||||
|---|---|---|---|---|---|---|---|
| Strain | lir-1a (lethality) | unc-73a (uncoordinated) | cel-1a (larval arrest) | dpy-13a (dumpy) | tra-2b (male anatomy) | lin-1b (multi-vulva) | lin-15bb (multi-vulva) |
| Wild type (N2) | − | + | + | + | 1.3 ± 0 | 2.8 ± 3.7 | 0 ± 0 |
| eri-1(mg366) | ++++ | ++++ | ++++ | ++++ | 61.5 ± 20.6 | 68.6 ± 12.2 | 93.1 ± 4.9 |
| rrf-3(pk1426) | ++++ | ++++ | NS | ++++ | NS | 70.0 ± 6.9 | 86.7 ± 8.1 |
| rrf-3(mg373) | ++++ | ++++ | NS | ++++ | NS | 66.3 ± 10.2 | 78.8 ± 12.4 |
| eri-3(mg408) | ++++ | +++ | +++ | ++++ | 45.1 ± 21.9 | 60.5 ± 14.9 | 59.8 ± 20.2 |
| dcr-1(mg375Eri) | ++ | ++ | + | ++ | 2.4 ± 3.8 | 2.0 ± 2.8 | 2.1 ± 5.2 |
| eri-9(gg101) | ++++ | ++++ | ++++ | ++++ | 66.4 ± 12.9 | 70.8 ± 11.6 | 92.0 ± 4.3 |
| eri-9(gg106) | ++++ | ++++ | ++++ | ++++ | 55.1 ± 16.7 | 60.3 ± 9.8 | 90.7 ± 7.5 |
| eri-9(tm2346) | +++ | ++++ | +++ | ++++ | 57.9 ± 26.2 | 63.3 ± 12.8 | 83.7 ± 13.5 |
| ergo-1(gg098) | ++++ | ++++ | ++++ | ++++ | 67.3 ± 12.4 | 62.4 ± 14.0 | 85.6 ± 10.7 |
| rde-1(ne219) | − | − | − | − | 0.9 ± 2.0 | 0.3 ± 0.9 | 0 ± 0 |
| dcr-1(mg375Eri); eri-1(mg366) | ++ | ++ | + | + | 1.2 ± 2.5 | 0.9 ± 7.5 | 0.9 ± 1.4 |
| ergo-1(gg098);eri-1(mg366) | ++++ | ++++ | ++++ | ++++ | 63.3 ± 18.4 | 60.4 ± 10.2 | 84.5 ± 7.3 |
| eri-9(gg106);eri-1(mg366) | ++++ | ++++ | ++++ | ++++ | 60.9 ± 16.3 | 51.8 ± 11.2 | 80.2 ± 19.2 |
|
dcr-1(mg375Eri); HA∷DCR-1(+) |
+ |
+ |
NS |
NS |
NS |
NS |
NS |
Effectiveness of RNAi scored from unaffected (−) to maximally enhanced (++++) or not scored (NS).
Scored as percentage of F1 animals that display the indicated phenotype.
Molecular identification of eri-4, eri-8, and eri-9:
We mapped eri-8 to a <1.5-cM interval on chromosome V between Y50D4C (−19.99) and F52F10 (−18.54). The C. elegans genome encodes ∼27 Argonaute proteins. One of those Argonaute genes, R09A1.1, lies within this mapping interval. We sequenced R09A1.1 from DNA isolated from four eri-8 alleles; each contained a unique mutation within the R09A1.1 open reading frame (Figure 1C). During the course of our studies, animals harboring a deletion within the R09A1.1 locus were shown to exhibit an enhanced RNAi phenotype and defects in endo siRNA expression (Yigit et al. 2006). On that basis, R09A1.1 was named endogenous-RNAi deficient Argonaute-1 (ergo-1) (Yigit et al. 2006). eri-8(gg098) animals failed to complement ergo-1(tm1860) animals for enhancement of RNAi (data not shown). We conclude that eri-8 corresponds to ergo-1.
We mapped eri-9 between −7.4 and +1.5 on chromosome III. The ORF C26E6.7 lies within this interval. Four lines of evidence indicate that eri-9 corresponds to C26E6.7. First, like ERI-1, RRF-3, ERI-3, and ERI-5, the C26E6.7 gene product coprecipitates with DCR-1 (Duchaine et al. 2006). Second, we sequenced C26E6.7 from two eri-9 mutants and found that each had a unique mutation within the predicted C26E6.7 coding sequence (Figure 1C). Third, eri-9(gg106) failed to complement the RNAi enhancement phenotype shown by a C26E6.7 deletion mutant (data not shown). Fourth, a transgene that includes the wild-type C26E6.7 gene rescued the Eri phenotype associated with eri-9(gg106) animals (Table S2). We conclude that eri-9 corresponds to C26E6.7. Database searches reveal a single homolog of ERI-9 in C. briggsae and C. remanei (data not shown). We have not detected sequence homologs of ERI-9 in other, more divergent organisms (data not shown).
We mapped the single eri-4 allele, mg375, to a <1-cM interval on chromosome III, an interval that contains the dcr-1 gene. As DCR-1 is required for converting dsRNA to siRNAs (a necessary prerequisite for RNAi), we did not anticipate identifying alleles of dcr-1 in our screen for enhanced RNAi sensitivity. Surprisingly, sequencing of the dcr-1 locus from DNA isolated from mg375 animals identified a mutation that encodes a G492R substitution within the N-terminal helicase domain of DCR-1 (Figure 1C). Two additional lines of evidence indicate that mg375 is a mutant allele of dcr-1. First, mg375 fails to complement dcr-1(ok247), a deletion that likely represents a null allele of dcr-1 (Knight and Bass 2001), for sterility defects (Figure S2A, and see below). Second, a transgene expressing full-length dcr-1 partially rescues the sterility and enhanced RNAi phenotypes associated with mg375 hermaphrodite animals (Figure S2B and Table 1). We speculate that the partial rescue of dcr-1(mg375Eri) animals by transgenically expressed dcr-1 likely reflects poor germline expression of this transgene. We conclude that mg375 is an allele of dcr-1 and encodes a mutant variant of DCR-1 that harbors a G492R substitution within the N-terminal helicase domain. G492 is an evolutionarily conserved residue, which suggests an important function for this amino acid in RNAi-related processes (Figure 1D).
dcr-1(mg375Eri) and eri-9 animals exhibit defects in endo siRNA production and mRNA regulation:
Mutations in previously identified Eri genes result in a failure of cells to accumulate endo siRNAs (Lee et al. 2006; Duchaine et al. 2006; Yigit et al. 2006). The Argonaute protein NRDE-3 is expressed in most, if not all, somatic tissues and escorts a subset of endo siRNAs from the cytoplasm to the nucleus. In the absence of endo siRNAs, NRDE-3 resides in the cytoplasm, while, in the presence of endo siRNAs, NRDE-3 localizes to the nucleus (Guang et al. 2008). Thus, the subcellular distribution of NRDE-3 is reflective of endo siRNA abundance in somatic cells. To begin to assess whether eri-9 or dcr-1(mg375Eri) regulate endo siRNA expression, we examined the subcellular distribution of NRDE-3 in eri-9(−) or dcr-1(mg375Eri) animals. In contrast to wild-type controls, where NRDE-3 was predominantly nuclear, NRDE-3 localized predominantly to the cytoplasm in eri-9(−) and dcr-1(mg375Eri) animals (Figure 2A). These results suggest that wild-type eri-9 and dcr-1 are required for expression of the majority of endo siRNAs that associate with NRDE-3 in somatic tissues. We tested this hypothesis by performing Northern analysis of the expression of specific endo siRNAs in eri-9(−) or dcr-1(mg375Eri) animals. Endo siRNAs have been identified for the ORF E01G4.5 and an ∼3-kb noncoding region of the X chromosome (termed the X-cluster) (Ambros et al. 2003). eri-1(−), eri-3(−), and rrf-3(mg373) animals fail to express these endo siRNAs (Figure 2B and Figure S3). eri-9(−) and dcr-1(mg375Eri) animals also failed to express detectable E01G4.5 and X-cluster endo siRNAs (Figure 2B), supporting the hypothesis that these genes function in the eri-1 genetic pathway and indicating that ERI-9 and DCR-1 are required for generating and/or stabilizing these endo siRNAs.
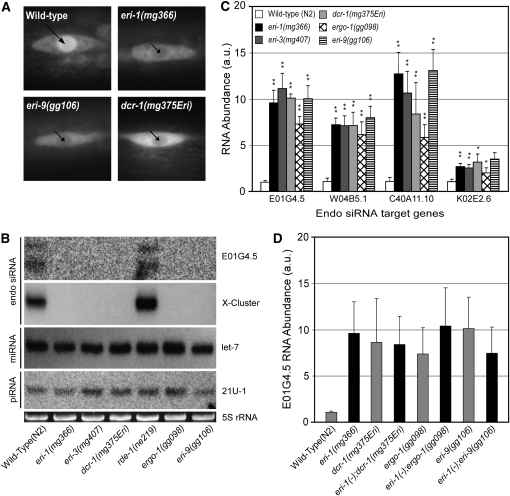
Figure 2.—
eri-9(−) and dcr-1(mg375Eri) animals exhibit defects in endo siRNA expression. (A) Fluorescence microscopy of a seam cell from larval stage L2 animals of the indicated genotypes expressing a GFP-tagged NRDE-3. Long arrow and short arrows indicate strong and weak nuclear localization of GFP∷NRDE-3, respectively. (B) Total RNA isolated from mixed-stage animals of the indicated genotype was subjected to Northern blot analysis to detect the E01G4.5, X-cluster, let-7, and 21U-1 (piRNA) small regulatory RNAs. (Bottom) 5S RNA loading control stained with ethidium bromide. (C) cDNA generated from RNA isolated from animals of the indicated genotype was subjected to qRT–PCR analysis. mRNA levels of E01G4.5, W04B5.1, C40A11.10, and K02E2.6 were normalized to a control mRNA, eft-3. Data are expressed as the ratio of mRNA abundance in mutant animals relative to wild type (n = 6, ±SD). *P < 0.05; **P < 0.01. (D) E01G4.5 mRNA levels were measured by qRT–PCR as described in C. (n = 3, ±SD).
In addition to endo siRNAs, C. elegans expresses at least two additional classes of small regulatory RNAs, the miRNAs, and the 21U-RNAs (the worm equivalent of the Piwi interacting RNAs). dcr-1(mg375Eri) and eri-9 animals retained the ability to express the miRNA let-7 and the 21U-1 piRNA at levels similar to that of wild-type animals (Figure 2B). In addition, dcr-1(mg375Eri) and eri-9 animals respond more robustly to dsRNA exposure (RNAi) than wild-type animals (Figure 1A and Table 1), strongly suggesting that these mutant animals are able to produce siRNAs from exogenous dsRNA substrates. Thus, the effect of the dcr-1(mg375Eri) and eri-9 mutations is restricted to the production of a subset of cellular small regulatory RNAs. Since DCR-1 is known to be involved in the biogenesis of miRNAs (Ketting et al. 2001; Lee et al. 2002, 2004; Lund et al. 2004), our results imply that the G492R substitution within the DCR-1 helicase domain preferentially impairs the endo siRNA-related function of DCR-1.
Endo siRNAs that map to the K02E2.6 ORF have been identified (Ambros et al. 2003; Ruby et al. 2006). Animals lacking components of the ERI/DICER complex fail to express K02E2.6 endo siRNAs and concomitantly overexpress the K02E2.6 mRNA (Duchaine et al. 2006). Consequently, it has been proposed that endo siRNAs negatively regulate mRNAs that exhibit sequence homology to their cognate endo siRNAs, which is known as endogenous RNAi. We found that K02E2.6 mRNA is likewise overexpressed in dcr-1(mg375Eri), ergo-1, rrf-3(mg373), and eri-9 animals (Figure 2C and Figure S3). We extended this analysis to three additional mRNAs (E01G4.5, W04B5.1, and C40A11.10) for which complementary endo siRNAs have previously been identified but transcript levels have not been characterized (Figure 2C and Ruby et al. 2006). All three mRNAs were overexpressed in eri-1, eri-3, dcr-1(mg375Eri), ergo-1, and eri-9 animals relative to wild-type controls (Figure 2C). Thus, the dcr-1, ergo-1, and eri-9 gene products (and previously identified components of the ERI/DICER complex) are required for negatively regulating these mRNAs. Animals harboring mutations in both eri-1 and eri-9, ergo-1, or dcr-1(mg375Eri) exhibit similar levels of mRNA misregulation to animals harboring the individual mutations (Figure 2D). Taken together, these data argue that ergo-1, eri-9, and the dcr-1 helicase domain are required for endo RNAi processes mediated by the eri-1 genetic pathway.
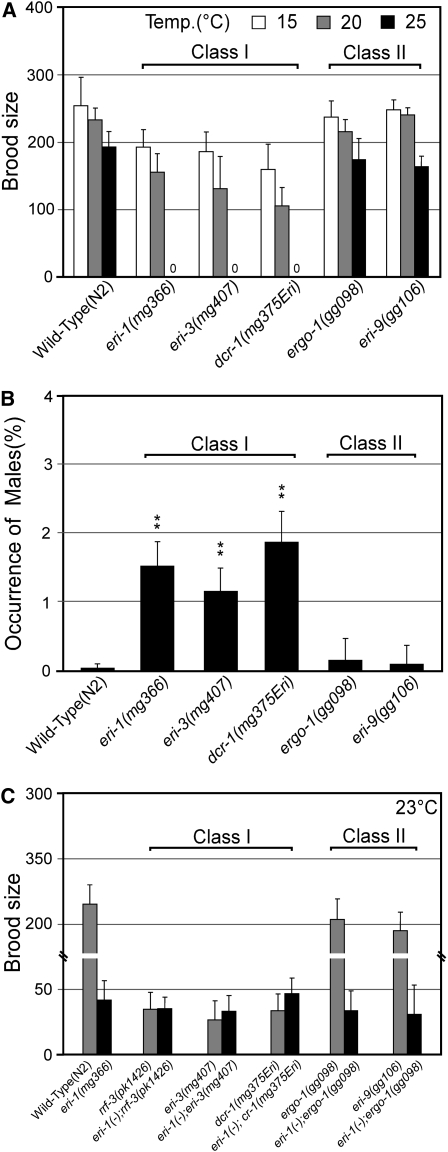
A subset of Eri animals exhibit sperm-specific endo RNAi defects:
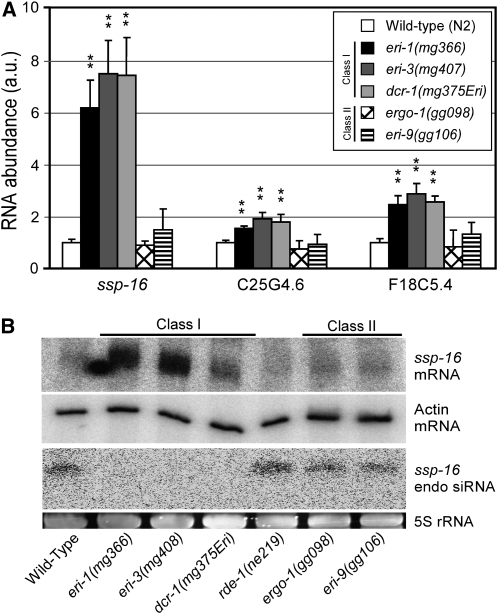
Endo siRNAs are thought to negatively regulate cellular mRNAs. Large-scale sequencing of endo siRNAs indicates that endo siRNAs are enriched for sequences with homology to mRNAs expressed in sperm (Ruby et al. 2006). We selected three sperm mRNAs [sperm-specific protein (ssp)-16, C25G4.6, and F18C5.4], for which endo siRNAs have been identified, for further investigation (Ruby et al. 2006). We confirmed that these mRNAs are expressed predominantly, if not exclusively, in the sperm-producing germline (Figure S4). We then investigated whether the Eri genes were required for regulating these mRNAs. qRT–PCR analysis demonstrated differential genetic requirements for the regulation of these sperm-enriched mRNAs. For example, eri-1, eri-3, rrf-3, and dcr-1(mg375Eri) mutant animals overexpressed these sperm-specific mRNAs while eri-9 and ergo-1 mutant animals expressed these mRNAs at levels similar to that of wild-type animals (Figure 3A and Figure S3). Henceforth, we will refer to eri-1, eri-3, rrf-3, and dcr-1(mg375Eri) as class I Eri genes and eri-9 and ergo-1 as class II Eri genes. Northern analysis of ssp-16 revealed a lack of ssp-16 endo siRNAs and a concomitant increase in ssp-16 transcript levels among class I, but not class II, mutant animals (Figure 3B). Interestingly, we found that mRNAs encoding class I Eri factors are expressed at similar levels in the sperm- and oocyte-producing germline, while mRNAs encoding class II Eri factors are enriched in the oocyte-producing germline, suggesting that the differential requirement for the class I and II Eri factors in sperm-specific mRNA regulation may be due to differential expression patterns of the class I and II Eri factors (Figure S5). Thus, the class I, but not class II, Eri gene products regulate sperm-enriched RNAs via endo RNAi.
Figure 3.—
Eri genes can be classified into two groups on the basis of defects in endo RNAi processes targeting sperm-enriched transcripts. (A) qRT–PCR analysis (as described in Figure 2C) detecting the indicated sperm-enriched mRNAs. Data were normalized to the sperm-specific mRNA, msp-3 (n = 10, ±SD). **P < 0.01. (B) Total RNA was subjected to Northern blot analysis detecting ssp-16 mRNA, Actin (act-1) mRNA, and ssp-16 endo siRNAs. (Bottom) 5S RNA loading control.
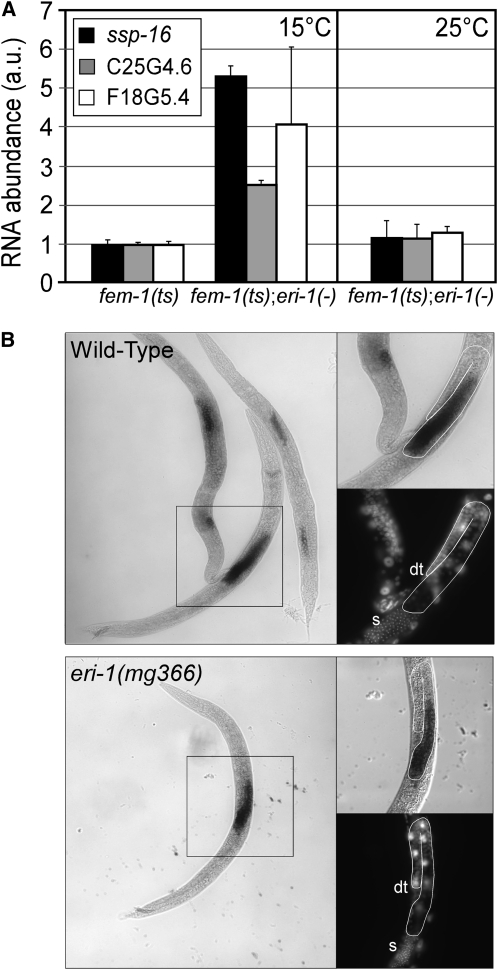
The class I Eri factors might negatively regulate sperm mRNAs specifically in the sperm-producing germline, or they might inhibit expression in tissues that normally do not produce these mRNAs. To address this question, we asked if class I Eri-dependent regulation of ssp-16, C25G4.6, and F18C5.4 mRNA occurs in the sperm-producing germline. Hermaphrodites that harbor a fem-1(ts) mutation produce both sperm and oocytes at 15°, but only oocytes at 25° (Nelson et al. 1978). Loss of eri-1 resulted in elevated levels of ssp-16, C25G4.6, and F18C5.4 in fem-1(ts) hermaphrodites when sperm were present, but not in the absence of sperm (Figure 4A). These results argue that the negative regulation of sperm gene expression by eri-1 requires the sperm-producing germline. In support of this idea, in situ hybridization detecting ssp-16 mRNA demonstrated that expression of ssp-16 is restricted to the sperm-producing gonad in wild-type animals and that eri-1(−) animals do not exhibit ectopic expression of ssp-16 in other tissues (Figure 4B). These results demonstrate that the class I Eri genes are required for endogenous RNAi processes within the sperm-producing germline.
Figure 4.—
Class I Eri-mediated RNA regulation occurs within the male germline. (A) cDNA was generated from total RNA isolated from age-synchronized L4 larvae of the indicated genotype reared at 15° or 25°. qRT–PCR detecting the indicated sperm-enriched transcripts as described in Figure 2C (n = 3, ±SD). (B) In situ hybridization of ssp-16 in wild-type and eri-1(mg366) animals reared at 25°. ssp-16 expression is restricted to the sperm-producing gonad. Insets are magnifications of boxed regions with the developing germlines of males outlined in white. dt, distal tip cell; s, condensed sperm nuclei. Surpisingly, we did not detect differences in ssp-16 mRNA staining intensity between eri-1(−) and wild-type animals in these experiments. As both our qRT–PCR and Northern analyses indicate a significant increase in ssp-16 mRNA levels in eri-1(−) animals, we hypothesize that our in situ hybridizations were not saturated and consequently were nonquantitative.
Class I Eri animals exhibit defects in sperm development:
Germline phenotypes have been reported for a few Eri mutants (Kennedy et al. 2004; Duchaine et al. 2006; Asikainen et al. 2007). eri-1, rrf-3, eri-3, and eri-5 hermaphrodites exhibit ts sterility; these animals are fertile at 15° and sterile at 25°. These animals also produce a higher-than-normal percentage of male progeny, referred to as a Him (high incidence of males) phenotype (Hodgkin et al. 1979). The Him phenotype of Eri mutants is due to an X-chromosome nondisjunction in the sperm-producing germline, which causes XX hermaphrodites to produce XO male offspring (Gent et al. 2009). We tested if these germline defects were shared by ergo-1(−), eri-9(−), rrf-3(mg373), or dcr-1(mg375Eri) animals. dcr-1(mg375Eri) and rrf-3(mg373) animals exhibited ts sterile and Him phenotypes (Figure 5 and Figure S3). In contrast, ergo-1(−) and eri-9(−) animals exhibited wild-type levels of fertility and normal frequencies of male progeny (Figure 5). Thus, the class I Eri genes, but not the class II Eri genes, are required for fertility at elevated temperatures. Finally, animals harboring both class I and class II Eri mutations are sterile at 25° (and exhibit brood sizes similar to class I Eri animals when reared at temperatures slightly below the nonpermissive temperature), indicating that class I Eri alleles are epistatic to class II alleles with regards to sterility (Figure 5C and data not shown).
Figure 5.—
Class I, but not class II, Eri animals exhibit germline defects. (A) The number of progeny from hermaphrodites of the indicated genotypes. Data are expressed as the mean number of progeny per adult (n ≥ 32, ±SD). (B) The percentage of progeny exhibiting male morphology (n = 12, ±SD). **P < 0.01. (C) The number of progeny from animals of the indicated genotype at a temperature (23°) slightly less than the nonpermissive temperature of class I Eri animals (n ≥ 32).
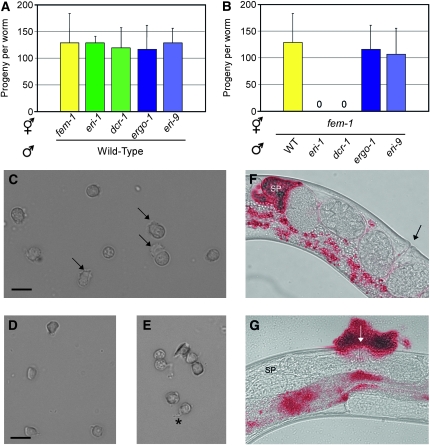
Sterility in hermaphrodites can arise from defects in sperm or oocyte function. Mating with wild-type males rescued the ts sterility of class I Eri hermaphrodites, a result suggestive of a sperm-specific defect (Figure 6A). Mating assays with fem-1 hermaphrodites (which produce only oocytes) crossed to class I Eri males confirmed this hypothesis; sperm from eri-1(−) and dcr-1(mg375Eri) males, reared at the nonpermissive temperature, were unable to fertilize eri-1(+) (fem-1) oocytes (Figure 6B). Sperm from class II Eri mutant males were functional in these assays (Figure 6B). Together, these data establish a role in sperm function for the class I, but not class II, Eri genes.
Figure 6.—
Sterility of class I Eri animals is attributable to defects in sperm function. (A) Wild-type males were crossed with hermaphrodites of the indicated genotype reared at the nonpermissive temperature (25°), and the number of cross-progeny were scored (n = 8, ±SD). (B) The number of progeny from hermaphrodite [fem-1(hc17ts)] animals mated with males of the indicated genotype at the nonpermissive temperature (25°) (n = 8, ±SD). (C–E) Spermatids dissected from wild-type (C) or dcr-1(mg375Eri) (D and E) adulthood males (3 days after final molt) were treated with the in vitro activator monensin and visualized by DIC Nomarski. Arrows indicate pseudopods of crawling spermatozoa. The dcr-1(mg375Eri) spermatids are misshapen and fail to activate (D) or extend thin immotile projections (asterisk in E). (F and G) Wild-type or dcr-1(mg375Eri) mid-adulthood males were stained with a vital fluorescent dye and then mated with fem-1(hc17ts) females. Mated females were visualized by both DIC Nomarski and fluorescence microscopy to assess sperm transfer and localization. The fluorescent and DIC images were overlaid to generate a composite image. Arrows indicate vulva. SP, spermatheca. (F) Sperm (in red) from wild-type males migrate to the spermatheca. (G) dcr-1(mg375Eri) sperm either remain at the vulva at insemination or are expelled from the uterus by passing oocytes. Intestinal autofluorescence seen in these images is also observed in the absence of dye staining.
We examined the mature gametes of males harboring class I Eri alleles for defects in morphology or motility that might underlie sperm-specific sterility. Wild-type adult males store round, immotile spermatids in the seminal vesicle that, upon insemination, are activated by an extracellular signal to generate amoeboid crawling spermatozoa with extended pseudopods (Wolf et al. 1978; Ward et al. 1981). The in vivo activation signal can be mimicked in vitro by proteases or compounds that increase intracellular pH (Nelson and Ward 1980; Ward et al. 1983; Shakes and Ward 1989). We assessed the ability of dcr-1(mg375Eri) spermatids to extend pseudopods and form crawling spermatozoa by treatment with the in vitro activators monensin or triethanolamine. In vitro activation of wild-type spermatids was successful 82% of the time (Figure 6C). In contrast, only 7% of dcr-1(mg375Eri) spermatids were capable of forming pseudopods, and the majority of dcr-1(mg375Eri) spermatids exhibited an aberrant morphology (Figure 6, D and E). Motile spermatozoa localize to the spermatheca, the site of fertilization, in the hermaphrodite reproductive tract. In vivo activation of class I Eri animal spermatids was also defective, as indicated by a failure of dcr-1(mg375Eri) spermatids to localize to the spermatheca. Vital staining of male sperm prior to mating indicated that dcr-1(mg375Eri) sperm can be transferred successfully. Temporal analysis following sperm transfer argued that dcr-1(mg375Eri) sperm are rapidly displaced from the hermaphrodite reproductive tract by passing oocytes and expelled through the vulva (compare Figure 6F to 6G). Similar results were observed for eri-1, eri-3, and rrf-3 sperm (data not shown). Thus, class I Eri males are defective in spermatid activation and cell motility, which are essential for fertility (L'Hernault 2006).
C. elegans hermaphrodites undergo spermatogenesis prior to oogenesis, while male animals continue to produce sperm throughout adult life (Wolf et al. 1978). Class I Eri hermaphrodites were sterile if reared at the nonpermissive temperature during spermatogenesis, but fertile if the temperature was raised during oogenesis (after spermatogenesis was completed) (Table 2). Class I Eri males were sterile if reared at the nonpermissive temperature, but eventually regained fertility following a downshift in temperature, indicating that the temperature-sensitive period for fertility associated with class I Eri mutants coincides with spermatogenesis (Table 2).
TABLE 2.
Temperature-sensitive period for class I Eri animals coincides with spermatogenesis
| ♀ fem-1a × ♂ N2 |
♀ fem-1a × ♂ eri-1 |
eri-1 (hermaphrodite self-cross) |
||||||
|---|---|---|---|---|---|---|---|---|
| Embryo L4b | Adultc | Progenyd | Embryo L4b | Adultc | Progenyd | Embryo L4e | Adultf | Progenyd |
| 15° | 25° | + | 15° | 25° | + | 15° | 25° | + |
| 25° | 25° | + | 25° | 25° | − | 25° | 25° | − |
| 25° |
15° |
+ |
25° |
15° |
+ |
25° |
15° |
− |
Feminized animals used to prevent self-fertilization.
Temperature of male animals during development and early spermatogenesis.
Temperature of male animals during mating and later spermatogenesis.
The presence (+) or absence (−) of progeny 4 days after temperature shift.
Temperature of hermaphrodite animals during development and spermatogenesis.
Temperature of hermaphrodite animals during oogenesis and fertilization.
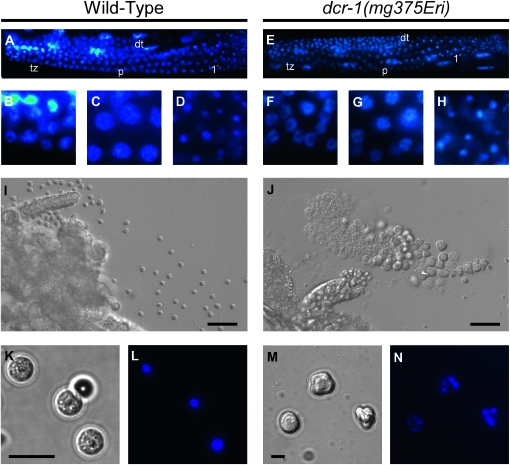
We next examined the gonads of class I Eri males for defects in sperm development. In wild-type males, a mitotically proliferating population of syncytial stem cells in the distal end of the germline gives rise to a transition zone at the onset of meiosis. Further progression through meiosis I produces pachytene nuclei that condense and cellularize into primary spermatocytes. Completion of the two meiotic divisions produces four haploid spermatids that separate from the residual body (L'Hernault 2006). Early events during spermatogenesis of dcr-1(mg375Eri) mutant males appeared to be unaffected, as indicated by the normal number and morphology of germ cells in the mitotic and meiotic regions of the germline (Figure 7, A–H). The first visible defect that we observed was a change in cytology at the spermatid stage. Wild-type animals produce small spherical spermatids that each contains a single, highly condensed nucleus (Figure 7, I, K, and L). In contrast, young adult males harboring the dcr-1(mg375Eri) mutation accumulated sperm cells in the seminal vesicle that were large and misshapen and often contained multiple nuclei (Figure 7, J, M, and N; note the difference in scale in K and M). Similar defects have been reported for sperm mutants (e.g., spe-26) that exhibit aberrant chromosome segregation (Varkey et al. 1995). The observed defects in the dcr-1(mg375Eri) mutant may reflect delayed progression through the sperm developmental program, as older males possessed spermatids that were more similar in size and shape to wild-type spermatids (see Figure 6, C–E). Defects similar to those described above were found in sperm for all class I Eri animals, whereas sperm of class II Eri animals exhibited normal morphology and function (data not shown). Taken together, the data indicate that class I Eri mutations produce multiple aberrations in spermatogenesis, beginning with cytokinesis at the meiotic divisions and extending through terminal differentiation into crawling spermatozoa. These defects likely explain the ts sterility associated with class I Eri animals.
Figure 7.—
Class I Eri animals exhibit defects in sperm development. (A–H) Early germline development of dcr-1(mg375Eri) animals is indistinguishable from wild type. (A) Germline of wild-type young adult male. DAPI staining indicates changes in nuclear morphology during development. dt, distal tip; tz, transition zone; p, pachytene of meiosis I; 1°, primary spermatocytes. (B–D) Higher magnification images of A showing characteristic crescent-shaped transition zone (B), pachytene (C), or highly condensed nuclei in primary spermatocytes (D). (E) Germline of dcr-1(mg375Eri) young adult male stained with DAPI. (F–H) Higher magnification images of transition zone, pachytene, and primary spermatocytes, respectively. (I–N) Defects in cytokinesis produce terminal spermatocyte arrest. (I and J) Dissected gonads of wild-type (I) or dcr-1(mg375Eri) (J) young adult male visualized by DIC Nomarski. Gametes are considerably larger in dcr-1(mg375Eri) animals. Bar, 10 μm (K–N) Terminal gametes dissected from wild-type (K and L) or dcr-1(mg375Eri) (M and N) young adult males visualized by DIC Nomarski (K and M) and DAPI (L and N). Wild-type spermatids are symmetrical and possess a centrally located single nucleus. dcr-1(mg375Eri) terminal spermatocytes are asymmetrical and contain multiple nuclei. Bar, 5 μm.
DISCUSSION
Here we report the identification and characterization of genes required for the production of endogenous siRNAs, small regulatory RNAs that are thought to mediate silencing of cellular transcripts. We establish that eri-9 encodes a novel component of the endo RNAi machinery and that eri-4 encodes a mutant variant of DCR-1 harboring a mutation within a conserved region of the DCR-1 helicase domain. We demonstrate that ERI-9 and the helicase domain of DCR-1 play a role in endo RNAi processes in C. elegans. Finally, we show that the class I Eri factors (including dcr-1), but not the class II Eri factors (including eri-9), are required for development of, and endo RNAi processes in, sperm.
We show that animals harboring a mutation in dcr-1 exhibit defects in endo siRNA production, mRNA regulation, X-chromosome segregation, and sperm development that are indistinguishable from phenotypes exhibited by eri-1(−), rrf-3(−), and eri-3(−) animals. ERI-1, RRF-3, and ERI-3 coprecipitate with DCR-1 (Duchaine et al. 2006). This latter observation has led to the hypothesis that the ERI proteins assemble into a protein complex with DCR-1. Our results provide genetic and biochemical evidence supporting the existence of a functional multi-protein complex composed of DCR-1 and the Eri factors and support the model that this complex is important for generating and stabilizing endo siRNAs. Interestingly, the N-terminal helicase domain of Dicer is well conserved throughout eukaryotes; however, the function of this domain remains enigmatic. Our identification and characterization of DCR-1(G492R) establishes that, in C. elegans, the helicase domain of Dicer is required for endo siRNA expression and spermatogenesis.
We have shown that eri-9 is a component of the endo RNAi machinery: ERI-9 functions in the eri-1 genetic pathway, encodes a DCR-1 coprecipitating protein (Duchaine et al. 2006), and is required for endo siRNA expression and for cognate mRNA regulation. Taken together, these data argue strongly that ERI-9 is a component of ERI/DICER complexes. Interestingly, we have shown that components of the ERI/DCR complex are not all functionally equivalent. Our division of the Eri genes into two groups on the basis of the presence [class I: eri-1, eri-3, rrf-3, and dcr-1(mg375Eri)] or absence (class II: ergo-1 and eri-9) of germline pleiotropies and endogenous RNAi processes in the sperm-producing germline indicate that these factors can have distinct biological function(s). In the simplest model, the class I proteins form a core ERI/DICER complex that is required for endo siRNA production and endogenous RNAi in both the soma and germline, whereas the class II proteins serve as accessory factors that modify core complex activity in tissues other than the male germline. In support of this hypothesis, expression analysis of Eri genes indicates that class I Eri mRNAs are expressed at equivalent levels during sperm and oocyte production, while the class II Eri mRNAs are enriched in the oocyte-producing germline (Figure S5). Future analyses that focus on endo RNAi defects in the sperm-producing germline may identify sperm equivalents of ergo-1 and eri-9.
Animals harboring class I Eri mutations exhibit defects in both sperm-specific endo RNAi and sperm development. The failure of class I Eri mutants to engage in sperm endo RNAi may directly cause spermatogenesis defects. Alternatively, it is possible that the sperm defects that we have observed result from non-RNAi and siRNA related functions of the class I Eri factors. We favor the former model for the following reasons: First, we have observed a one-to-one correlation between those Eri genes required for sperm-specific endo RNAi and those Eri genes required for normal sperm development. Second, large-scale sequencing of small RNAs has shown that endo siRNAs are enriched for sequences with the ability to target mRNAs expressed in sperm (Ruby et al. 2006). Third, several class I Eri factors, such as the RNA-dependent RNA polymerase RRF-3 and the RNase III enzyme DCR-1, are proteins whose only known biochemical function relates to small regulatory RNA biogenesis. Finally, our genetic screens have identified missense alleles of both dcr-1 and rrf-3 [encoding DCR-1(G492R) and RRF-3(G187E)], which do not affect expression of their encoded proteins or the ability of these mutant proteins to assemble into ERI/DCR complexes (data not shown). DCR-1(G492R) and RRF-3(G187E) animals do, however, exhibit fully penetrant endo RNAi defects and ts sperm defects, strongly suggesting that it is the small RNA products of DCR-1 and RRF-3 that are important for spermatogenesis. Taken together, these data argue that loss of endo siRNA expression in the male germline triggers defects in sperm development.
We have shown that mutant animals lacking Eri gene function overexpress mRNAs that exhibit sequence homology to Eri-dependent endo siRNAs. The most parsimonious explanation for these observations is that endo siRNAs negatively regulate their cognate mRNAs (and/or are the end product of this regulation). We consider two possible models for the role of endo siRNAs and endo RNAi during spermatogenesis.
In model 1, spermatogenesis defects are triggered by overexpression of sperm mRNAs, the negative regulation of which is important for sperm development. In such a case, either the target mRNA may be grossly overexpressed or the temporal expression may be precocious and/or persistent. In support of model 1, microarray analysis comparing wild-type animals to eri-1(−) or rrf-3(−) animals indicates that sperm RNAs are negatively regulated by ERI-1 and RRF-3 (Asikainen et al. 2007). We revisited the eri-1/rrf-3 microarray data sets and compared these data to existing microarray data sets, which identified genes preferentially expressed in the sperm (Reinke et al. 2000, 2004). This analysis showed that 96% of eri-1/rrf-3-regulated RNAs are sperm-enriched RNAs (Table S3). This remarkable degree of overlap between these two microarray data sets indicates that a large percentage of transcript misregulation observed in both eri-1 and rrf-3 mutants at this stage of development occurs among genes with elevated expression during spermatogenesis. At present, it is unclear which, if any, of these RNAs are responsible for the spermatogenesis defects that we have observed. We conducted RNAi-mediated knockdown of these 68 RNAs (individually) and failed to identify any suppressors of class I Eri sterility (data not shown). In addition, we screened several million genomes (following EMS mutagenesis) for suppressors of Eri-mediated sterility and failed to identify a single suppressor (data not shown). Taken together, these data hint that, if model 1 is correct, the sterility of class I Eri animals is unlikely to be due to overexpression of a single sperm mRNA.
In model 2, spermatogenesis defects result from loss of endo siRNA-directed heterochromatin in sperm. Some Eri-dependent endo siRNAs, such as those complementary to the C. elegans X-cluster, map to regions of the genome not predicted to encode functional mRNAs. In Schizosaccharomyces pombe, the RNAi machinery plays a role in heterochromatin formation at centromeres (Reinhart and Bartel 2002; Bühler et al. 2006; Colmenares et al. 2007). S. pombe mutants that lack components of the RNAi machinery exhibit chromosome nondisjunction phenotypes (Hall et al. 2003). We have observed an X chromosome nondisjunction phenotype in the class I Eri mutant animals. Thus, model 2 posits that C. elegans endo siRNAs function analogously to S. pombe siRNAs to regulate heterochromatin formation in such a way as to permit chromosome segregation and sperm function at elevated temperatures.
One puzzling aspect of our results is the temperature sensitivity of class I Eri germline phenotypes. The class I Eri mutants appear to be nonconditional for loss of endo siRNA accumulation and misregulation of target RNAs, as these defects occur at all temperatures (data not shown). Increased X chromosome nondisjunction and sperm-specific sterility of the class I Eri mutants, however, are manifested more robustly at elevated temperatures. Most of the class I Eri mutants that we characterized carry predicted null alleles, indicating that the ts sterility phenotype associated with the class I Eri mutants is very likely not due to temperature-sensitive proteins. Rather, data from wild-type animals suggest that both chromosome segregation and sperm development are inherently sensitive to perturbation by temperature. For example, brood size is reduced by ∼25% in wild-type animals reared at elevated temperature (Hirsh and Vanderslice 1976). Since fecundity is sperm limited in C. elegans hermaphrodites, this observation indicates that wild-type sperm production or function might be impaired at elevated temperatures. Similarly, the frequency of male progeny arising from X chromosome nondisjunction increases several-fold in wild-type hermaphrodites raised at higher temperature (Rose and Baillie 1979). Thus, the sperm defects associated with class I Eri animals may result from enhancement of inherently temperature-sensitive processes.
C. elegans expresses a diverse array of small regulatory RNAs. Our genetic screens continue to identify novel factors required for the biogenesis of endo siRNAs. To date, we have identified two classes of Eri factors, one of which promotes thermotolerance in sperm. Interestingly, animals lacking the C. elegans PIWI homologs (and consequently lacking piRNAs) exhibit temperature-sensitive spermatogenesis defects, hinting that other small RNAs may play important roles in spermatogenesis (Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008). Furthermore, we have identified differential germline functions among the Eri factors. Taken together, these results indicate that the mechanisms of small RNA biogenesis and function in C. elegans are complex and may vary in a tissue-dependent manner. Our genetic screens have not yet reached saturation. For example, two other Eri factors, eri-10 and eri-11, identified in our screens exhibit class II Eri phenotypes, are defined by single alleles, and are not allelic with any of the known endo RNAi genes, indicating that additional components of the cellular endo RNAi machinery remain to be identified. The identification and characterization of the full complement of endo RNAi machinery will likely facilitate the unraveling of the remarkably complex world of small regulatory RNAs.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.108134/DC1.
References
- Ambros, V., R. C. Lee, A. Lavanway, P. T. Williams and D. Jewell, 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13 807–818. [DOI] [PubMed] [Google Scholar]
- Asikainen, S., M. Storvik, M. Lakso and G. Wong, 2007. Whole genome microarray analysis of C. elegans rrf-3 and eri-1 mutants. FEBS Lett. 581 5050–5054. [DOI] [PubMed] [Google Scholar]
- Batista, P. J., J. G. Ruby, J. M. Claycomb, R. Chiang, N. Fahlgren et al., 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler, M., A. Verdel and D. Moazed, 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125 873–886. [DOI] [PubMed] [Google Scholar]
- Cam, H. P., T. Sugiyama, E. S. Chen, X. Chen, P. C. FitzGerald et al., 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37 809–819. [DOI] [PubMed] [Google Scholar]
- Colmenares, S. U., S. M. Buker, M. Buhler, M. Dlakic and D. Moazed, 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell 27 449–461. [DOI] [PubMed] [Google Scholar]
- Czech, B., C. D. Malone, R. Zhou, A. Stark, C. Schlingeheyde et al., 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, P. P., M. P. Bagijn, L. D. Goldstein, J. R. Woolford, N. J. Lehrbach et al., 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine, T. F., J. A. Wohlschlegel, S. Kennedy, Y. Bei, J. D. Conte et al., 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 343–354. [DOI] [PubMed] [Google Scholar]
- Gent, J. I., M. Schvarzstein, A. M. Villeneuve, S. G. Gu, V. Jantsch et al., 2009. Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNAi. Genetics 183 1297–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal, M., H. Seitz, M. D. Horwich, C. Li, T. Du et al., 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang, S., A. F. Bochner, D. M. Pavelec, K. B. Burkhart, S. Harding et al., 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I. M., G. D. Shankaranarayana, K.-i. Noma, N. Ayoub, A. Cohen et al., 2002. Establishment and maintenance of a heterochromatin domain. Science 297 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hall, I. M., K.-i. Noma and S. I. S. Grewal, 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A., O. Voinnet, L. Chappell and D. Baulcombe, 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. L., and S. W. L'Hernault, 2001. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev. Biol. 232 105–114. [DOI] [PubMed] [Google Scholar]
- Hirsh, D., and R. Vanderslice, 1976. Temperature-sensitive developmental mutants of Caenorhabditis elegans. Dev. Biol. 49 220–235. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Kawamura, Y., K. Saito, T. Kin, Y. Ono, K. Asai et al., 2008. Drosophila endogenous small RNAs bind to Argonaute[thinsp]2 in somatic cells. Nature 453 793–797. [DOI] [PubMed] [Google Scholar]
- Kennedy, S., D. Wang and G. Ruvkun, 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427 645–649. [DOI] [PubMed] [Google Scholar]
- Ketting, R. F., S. E. J. Fischer, E. Bernstein, T. Sijen, G. J. Hannon et al., 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V. N., 2005. Small RNAs: classification, biogenesis, and function. Mol. Cells 19 1–15. [PubMed] [Google Scholar]
- Knight, S. W., and B. L. Bass, 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. C., R. L. Feinbaum and V. Ambros, 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, R. C., C. M. Hammell and V. Ambros, 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., K. Jeon, J. T. Lee, S. Kim and V. N. Kim, 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He et al., 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117 69–81. [DOI] [PubMed] [Google Scholar]
- L'Hernault, S. W., 2006. Spermatogenesis, in WormBook, edited by The C. elegans Research Community. WormBook, http://www.wormbook.org.
- Llave, C., K. D. Kasschau, M. A. Rector and J. C. Carrington, 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg and U. Kutay, 2004. Nuclear export of microRNA precursors. Science 303 95–98. [DOI] [PubMed] [Google Scholar]
- Mochizuki, K., and M. A. Gorovsky, 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14 181–187. [DOI] [PubMed] [Google Scholar]
- Mochizuki, K., N. A. Fine, T. Fujisawa and M. A. Gorovsky, 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110 689–699. [DOI] [PubMed] [Google Scholar]
- Motohashi, T., H. Tabara and Y. Kohara, 2006. Protocols for large scale in situ hybridization on C. elegans larvae, in WormBook, edited by The C. elegans Research Community. WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Nelson, G. A., and S. Ward, 1980. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19 457–464. [DOI] [PubMed] [Google Scholar]
- Nelson, G. A., K. K. Lew and S. Ward, 1978. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev. Biol. 66 386–409. [DOI] [PubMed] [Google Scholar]
- Plasterk, R. H. A., 2002. RNA silencing: the genome's immune system. Science 296 1263–1265. [DOI] [PubMed] [Google Scholar]
- Reinhart, B. J., and D. P. Bartel, 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297 1831. [DOI] [PubMed] [Google Scholar]
- Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren et al., 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6 605–616. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Rose, A. M., and D. L. Baillie, 1979. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, J. G., C. Jan, C. Player, M. J. Axtell, W. Lee et al., 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127 1193–1207. [DOI] [PubMed] [Google Scholar]
- Shakes, D. C., and S. Ward, 1989. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev. Biol. 134 189–200. [DOI] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet et al., 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12 1317–1319. [DOI] [PubMed] [Google Scholar]
- Tam, O. H., A. A. Aravin, P. Stein, A. Girard, E. P. Murchison et al., 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., B. J. Reinhart, D. P. Bartel and P. D. Zamore, 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103–112. [DOI] [PubMed] [Google Scholar]
- van der Keyl, H., H. Kim, R. Espey, C. V. Oke and M. K. Edwards, 1994. Caenorhabditis elegans sqt-3 mutants have mutations in the col-1collagen gene. Dev. Dyn. 201 86–94. [DOI] [PubMed] [Google Scholar]
- Varkey, J. P., P. J. Muhlrad, A. N. Minniti, B. Do and S. Ward, 1995. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 9 1074–1086. [DOI] [PubMed] [Google Scholar]
- Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi et al., 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., S. Kennedy, D. Conte, J. K. Kim, H. W. Gabel et al., 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436 593–597. [DOI] [PubMed] [Google Scholar]
- Wang, G., and V. Reinke, 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S., Y. Argon and G. A. Nelson, 1981. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91 26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S., E. Hogan and G. A. Nelson, 1983. The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev. Biol. 98 70–79. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., Y. Totoki, A. Toyoda, M. Kaneda, S. Kuramochi-Miyagawa et al., 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453 539–543. [DOI] [PubMed] [Google Scholar]
- Wianny, F., and M. Zernicka-Goetz, 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2 70–75. [DOI] [PubMed] [Google Scholar]
- Wolf, N., D. Hirsh and J. R. McIntosh, 1978. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J. Ultrastruct. Res. 63 155–169. [DOI] [PubMed] [Google Scholar]
- Yigit, E., P. J. Batista, Y. Bei, K. M. Pang, C.-C. G. Chen et al., 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747–757. [DOI] [PubMed] [Google Scholar]