Abstract
Short interfering RNAs (siRNAs) are a class of regulatory effectors that enforce gene silencing through formation of RNA duplexes. Although progress has been made in identifying the capabilities of siRNAs in silencing foreign RNA and transposable elements, siRNA functions in endogenous gene regulation have remained mysterious. In certain organisms, siRNA biosynthesis involves novel enzymes that act as RNA-directed RNA polymerases (RdRPs). Here we analyze the function of a Caenorhabditis elegans RdRP, RRF-3, during spermatogenesis. We found that loss of RRF-3 function resulted in pleiotropic defects in sperm development and that sperm defects led to embryonic lethality. Notably, sperm nuclei in mutants of either rrf-3 or another component of the siRNA pathway, eri-1, were frequently surrounded by ectopic microtubule structures, with spindle abnormalities in a subset of the resulting embryos. Through high-throughput small RNA sequencing, we identified a population of cellular mRNAs from spermatogenic cells that appear to serve as templates for antisense siRNA synthesis. This set of genes includes the majority of genes known to have enriched expression during spermatogenesis, as well as many genes not previously known to be expressed during spermatogenesis. In a subset of these genes, we found that RRF-3 was required for effective siRNA accumulation. These and other data suggest a working model in which a major role of the RRF-3/ERI pathway is to generate siRNAs that set patterns of gene expression through feedback repression of a set of critical targets during spermatogenesis.
REPRESSION of gene expression by small RNAs of ∼20–30 nt in length is important for many aspects of multicellular eukaryotic development. A variety of classes of small RNA with distinct structural features, modes of biogenesis, and biological functions have been identified (reviewed in Hutvagner and Simard 2008). We are particularly interested in a class of small RNAs, called endogenous short interfering RNAs (siRNAs), that are similar to intermediates in exogenously triggered RNA interference (RNAi) in their perfect complementarity to mRNA targets. High-throughput sequencing technology has provided a valuable tool for characterization of endogenous siRNA populations from many diverse sources, including mouse embryonic stem cells (Babiarz et al. 2008), Drosophila heads (Ghildiyal et al. 2008), and Arabidopsis pollen (Slotkin et al. 2009). These siRNAs have been proposed to function in the regulation of both cellular processes and genome defense through downregulation of gene expression. Caenorhabditis elegans, like plants and fungi, utilizes RNA-copying enzymes called RNA-directed RNA polymerases (RdRPs) as part of the RNAi machinery (Smardon et al. 2000; Sijen et al. 2001). While two of the C. elegans RdRPs are nonessential (RRF-1 and RRF-2), mutations in either of the remaining two (EGO-1 or RRF-3) lead to fertility defects (Smardon et al. 2000; Simmer et al. 2002). RRF-3 is functionally distinct from EGO-1 in that the RRF-3 requirement in fertility is temperature dependent. In addition, RRF-3 activity has an inhibitory effect on exogenously triggered RNAi (resulting in an ERI, or enhanced RNAi, mutant phenotype in rrf-3 mutants). Mutants lacking either RRF-3 or another ERI factor, ERI-1, have been used as experimental tools because of their enhanced sensitivity in RNAi-based screens. One proposed mechanism for the enhancement in RNAi in rrf-3 and eri mutants has been a competition for cofactors between the exogenously triggered RNAi pathway and an endogenous RNAi pathway. Consistent with this hypothesis, siRNAs corresponding to several genes have been shown by Northern analysis to depend upon RRF-3 and other ERI factors for their accumulation (Duchaine et al. 2006; Lee et al. 2006; Yigit et al. 2006). Global microarray analyses have also been undertaken to identify messenger RNAs whose expression is affected by RRF-3 and ERI-1 (Lee et al. 2006; Asikainen et al. 2007).
A functional significance of the RRF-3/ERI pathway has been inferred by the inability of rrf-3, eri-1, eri-3, and eri-5 mutant strains to propagate at a high growth temperature (Simmer et al. 2002; Duchaine et al. 2006). Rather than producing temperature-sensitive mutant protein effects, RRF-3 and other ERI proteins are thought to act in a temperature-sensitive process, as evidenced by the predicted truncated and presumed nonfunctional protein fragments that would result from the available deletion alleles and by their shared temperature-sensitive phenotypes. rrf-3 mutant animals have been observed to exhibit X-chromosome missegregation (Simmer et al. 2002) and an unusual persistence of a chromatin mark on the X chromosome during male spermatogenesis (Maine et al. 2005). X-chromosome missegregation and defective spermatogenesis have been referred to in previous studies of eri-1 (Kennedy et al. 2004) and eri-3 and eri-5 (Duchaine et al. 2006). Furthermore, eri-3 mutant sterility can be rescued by insemination with wild-type sperm (Duchaine et al. 2006).
Here we investigated the role of RRF-3 during spermatogenesis. We found defects evident at multiple stages, including after fertilization, where defects in rrf-3 mutant sperm can produce subsequent nonviable embryos. By using high-throughput sequencing, we characterized a large population of siRNAs present in spermatogenic cells and found a strong enrichment for antisense siRNAs from genes with known mRNA expression during spermatogenesis. While the majority of siRNA production during spermatogenesis does not require RRF-3, we found a set of genes for which siRNA production was dependent upon RRF-3. Existing data indicate increased expression for these genes in rrf-3 and/or eri-1 mutants. Taken together, our analyses suggest a working model in which the RRF-3/ERI pathway generates siRNAs that downregulate specific genes during spermatogenesis, with this regulation playing a key role in generating functional sperm.
MATERIALS AND METHODS
C. elegans strains, growth conditions, and spermatogenic cell isolation
Strains:
Except as noted, C. elegans Bristol N2 (Brenner 1974) was used as the wild-type strain for these studies. The other strains used in this study were the following: GE24—pha-1(e2123) I (Schnabel and Schnabel 1990); GR1378—eri-1(mg366) IV (Kennedy et al. 2004); JK816—fem-3(q20) IV (Barton et al. 1987); JK987—tra-2(q276)/mnC1dpy-10(e128) unc-52(e444) II (Okkema and Kimble 1991); NL2099—rrf-3(pk1426) II (Sijen et al. 2001); PD3303—rrf-3(pk1426) II; pha-1(e2123) III; PD3330—rrf-3(pk1426) II; him-8(e1489) IV; PD3331—him-8(e1489) IV; VC407—rrf-3(ok629)/mIn1[mIs14dpy-10(e128)] II (http://www.celeganskoconsortium.omrf.org); WM48—rde-4 (ne299) III (Tabara et al. 1999); and WM161—prg-1(tm872) I (Yigit et al. 2006), tra-2(q122) II (Schedl and Kimble 1988).
Growth conditions:
As stated in the description of each experiment, animals were grown at 16°, 23°, or 25°. The rrf-3 and eri-1 mutant fertility phenotypes were strongest at 25°, but certain experiments were carried out at the milder temperature of 23° due to the insufficient numbers of embryos produced at 25°. Self-fertilized rrf-3(pk1426) hermaphrodites had an approximate surviving brood size of 2 at 25°, but 30 at 23°. rrf-3(pk1426) males were also significantly more fertile at 23°. Self-fertilized rrf-3(ok629) hermaphrodites also showed temperature-sensitive decreased fertility (they also exhibited a temperature-independent egg-laying-defective phenotype). Animals used for RNA extraction and sequencing were grown and harvested at 25° (see below).
Spermatogenic cell isolation:
A population of mixed stage fem-3(q20) hermaphrodites grown at 16° was treated with a 1.2% sodium hypochlorite, 0.5 n NaOH solution with periodic vortexing to dissolve everything but eggs. After washing in M9 buffer to remove hypochlorite, eggs were hatched overnight in M9 solution and allowed to starve as L1 larva. These were put onto OP50-seeded, peptone-enriched plates at 25° to mature to early adult stage (55 hr), when animals were washed off plates with M9 buffer. After repeated washes to remove bacteria, the animals were diced with a razor blade in a 10-cm tissue culture dish to release spermatogenic cells into solution (other cells remained attached to carcasses). The mixture of carcasses and released spermatogonic cells was filtered through a double layer of 10 μm Nitex bolting cloth (Wildlife Supply) and washed three times in M9 buffer before flash freezing in liquid N2. Visual inspection of a portion that was transferred to a microscope slide prior to freezing indicated that the sample contained a mix of spermatogenic cell types, including primary spermatocytes, secondary spermatocytes, residual bodies, and spermatids (supporting information, Figure S5). This procedure is optimized for small-scale isolation of mixed-stage spermatogenic cells to be immediately frozen [we note that studies of mature sperm function optimally utilize a Percoll gradient and SM buffer (L'Hernault and Roberts 1995; Machaca et al. 1996) rather than M9 buffer (Klass and Hirsh 1981; Miller 2006)].
Male isolation:
Set 1:
After sodium hypochlorite treatment of mixed stage animals grown at 16°, released eggs were hatched overnight in M9 solution to starve as L1 larva. Fifty-one hours after being put onto OP50-seeded NGM plates at 25°, the starved L1's had reached adulthood, and males were separated from hermaphrodites by filtration through 35-μm Nitex bolting cloth (Wildlife Supply). Embryos that passed through the filter were separated from males by putting both males and embryos on an unseeded area of a plate and allowing males to crawl toward food. Adult males were washed off plates in EN buffer (0.1 m NaCl, 10 mm EDTA) and filtered a second time immediately prior to freezing in liquid N2, 53 hr after initially being put on seeded plates as L1's. Approximately 95% of the him-8(e1489) sample and 90% of the rrf-3(pk1426); him-8(e1489) sample was male.
Set 2:
After sodium hypochlorite treatment of mixed stage animals grown at 16°, released eggs were put on OP50-seeded NGM plates at 25° directly without being starved as L1's. Fifty-six hours later, when they had reach adulthood, males were separated from hermaphrodites by filtration through 35-μm Nitex bolting cloth (Wildlife Supply). Embryos that passed through the filter were separated from males by putting both males and embryos on an unseeded area of a plate and allowing males to crawl toward food. Adult males were washed off the plates in EN buffer and frozen in liquid N2 60 hr after initially being put on seeded plates as eggs. Approximately 90% of the him-8(e1489) and 95% of the rrf-3(pk1426); him-8(e1489) sample was male.
Assaying embryonic viability
Mated tra-2(q122) females were allowed to lay eggs for several hours on NGM plates with thin, clear lawns of OP50 (≤1 day after seeding). After removing the mothers from the plates, the newly laid embryos were immediately counted. The surviving progeny were counted when they had reached L4 or adult stage. For temperature-shift experiments, matings were set up between L4 stage tra-2(q122) females and L4 or adult males raised at the starting temperature, where they were allowed to mate overnight. The gravid females were then separated from the males and shifted to the new temperature, where they were left for sufficient time to flush out previously fertilized embryos (6 hr at 23° and 25° or 12 hr at 16°). After this period they were allowed to lay eggs for several more hours (3 hr at 23° and 25° or 6 hr at 16°). In each experiment, multiple matings were set up, and a minimum of four gravid hermaphrodites were evaluated at each growth temperature. Males were raised at 23° rather than at 25° [because matings with rrf-3(pk1426) males at 25° are inefficient in terms of embryo production]. Twenty-three degrees represents an intermediate temperature at which enough competent sperm are produced to allow for continued egg production many hours after mating.
Cytological techniques
RAD-51, REC-8, HIM-8, and SYP-1 immunostaining:
Hermaphrodite gonads were dissected and fixed as previously described for anti-RAD-51, anti-HIM-8, anti-SYP-1 (Martinez-Perez and Villeneuve 2005), and anti-REC-8 (Pasierbek et al. 2001) stainings. Animals were synchronized by bleaching to kill everything but embryos. Embryos were then allowed to hatch overnight on NGM plates seeded with Escherichia coli (OP50) at 16°, after which time embryos were shifted to grow at 25° until the animals were ready for dissection. After fixation, samples were washed three times in 1× PBS for 5 min and then blocked with 3% bovine serum albumin (BSA) in 1× PBS for 20 min at room temperature. Antibodies were diluted in 1× PBS containing 3% BSA as follows: 1:80 anti-RAD-51 (Alpi et al. 2003), 1:100 anti-REC-8 (Pasierbek et al. 2001), 1:500 anti-HIM-8 (Phillips et al. 2005), and 1:200 anti-SYP-1 (MacQueen et al. 2002). Samples were then incubated overnight at 4° in a humid chamber. After washing three times in 1× PBS plus 0.1% Tween 20, secondary antibodies were applied at the following dilutions: anti-rabbit DyLight 547 nm (1:200), anti-rabbit Alexa Fluor 568 (1:500), and anti-guinea pig Alexa Fluor 488 (1:500). After 2 hr at room temperature, slides were washed three times in 1× PBS plus 0.1% Tween 20 and mounted in Vectashield antifading medium (Vector Laboratories, Burlingame, CA) containing 2 μg/ml DAPI. Samples were examined with a Zeiss Axio Imager M1 microscope. Images were recorded with a Spot camera (Diagnostics Instruments). For multicolor immunostaining, monochrome stacks of images were captured separately for each emission wavelength (MetaVue software; Molecular Devices, Sunnyvale, CA). Three-dimensional stacks of images were deconvolved (AutoQuant software; AutoQuant Imaging, Troy, NY) and projected (Helicon Focus software; http://helicon.com.ua/heliconfocus/).
α-Tubulin immunostaining:
We used fixation procedures optimized for preservation of microtubules (Gonczy et al. 1999; Oegema et al. 2001), with modifications as previously described (Martinez-Perez et al. 2008). FITC-conjugated anti-α-tubulin monoclonal antibody DM 1A (Sigma) was used at a concentration of 1 μg/ml. Images were acquired as stacks of optical sections at 0.2-μm intervals using a Deltavision deconvolution microscopy system. We quantified the incidence of microtubule wreaths surrounding sperm nuclei in reproductive tracts (gonads and spermathecae) dissected either from self-fertilizing hermaphrodites or from tra-2(q122) females that had been mated with either wild-type or rrf-3(pk1426) mutant males. Quantification was performed using animals that had been raised and maintained at 23°. Microtubule wreaths were frequent in sperm from both rrf-3 self-fertilizing hermaphrodites (15 worms; 237/287 sperm nuclei with microtubule wreaths) and tra-2(q122) females that had been mated with rrf-3 males (1 worm; 17/20 nuclei with microtubule wreaths). Self-fertilized eri-1 hermaphrodite sperm had microtubule wreaths in 59% of their sperm (5 worms; 152/256 nuclei with tubulin wreaths). Microtubule wreaths were not detected in either control N2 self-fertilizing hermaphrodites (2 worms; 0/93 nuclei with tubulin wreaths) or control tra-2(q122) females mated with N2 males (4 worms; 0/86 nuclei with tubulin wreaths). Although discrete tubulin wreaths were not detected in control sperm, some hazy/diffuse tubulin staining was detected in 4/179 control sperm. For experiments analyzing wild-type sperm, successful microtubule staining in mitotic germ cells from the same gonad served as an internal positive control.
Preparation of libraries of small RNAs for sequencing
Small RNA was extracted from frozen tissue samples with the mirVana microRNA (miRNA) isolation kit (Ambion). Libraries of small RNAs were prepared using a protocol similar to one previously described for miRNA cloning (Lau et al. 2001), in which small RNA species were flanked by adapter sequences to allow for PCR and sequencing. We modified the original protocol at several key steps to allow for capture of triphosphorylated species and for sequencing with the Illumina genome analyzer system. To remove the bias for 5′ monophosphorylated species but preserve the bias toward 3′ hydroxylated species, we removed both monophosphates and polyphosphates from the 5′-end of the small RNA with Antarctic Phosphatase (New England Biolabs) following the addition of the 3′ adapter with T4 RNA ligase 1 (New England Biolabs). The resulting hydroxylated 5′-ends were phosphorylated by treatment with T4 polynucleotide kinase (New England Biolabs) and ATP to form a subtrate for T4 RNA ligase 1-mediated addition of the 5′ adapter. T4 polynucleotide kinase treatment after addition of the 3′ adapter is important because of its 3′ phosphatase and 2′,3′ cyclic phosphodiesterase activity, which can convert unwanted RNA degradation products into a substrate for the 3′ adapter ligation reaction. After ligation of both the 3′ and 5′ adapters, the adapters were extended during the reverse transcription and PCR steps to include complete primer binding sites for subsequent amplification and sequencing reactions. To protect against cross-contamination and to allow for pooling of samples, we modified the 5′ adapter sequence to include the Illumina genomic sequencing primer immediately followed by a 4-nt barcode. The pair of barcodes used in the set 1 wild-type and mutant samples was switched in the set 2 wild-type and mutant samples to guard against any potential biases introduced by adapter sequences. The samples were PAGE purified at three stages: after initial RNA extraction and after each ligation step (except with the isolated spermatogenic cell sample, where the first PAGE purification was skipped to conserve material). Additional details for each sample are provided in Table S1. The following oligos were used:
3′ adapter (IDT linker-1 with 5′ adenylation and 3′ dideoxyC): rAppCTGTAGGCACCATCAATC;
5′ adapter (i.e., AF-PP-341 DNA/RNA hybrid oligo with a 5′ amino modifier and 4-nt barcode at the 3′-end): ACGCTCTTCCGATCTrGrUrUrA;
Dual reverse transcription primer and PCR primer (AF-JIG-37, complementary to IDT linker-1): CAAGCAGAAGACGGCATACGAattgatggtgcctacag;
PCR primer (AF-JIG-40, containing 5′ adapter sequence): AATGATACGGCGACCACCGACACTCTTTCCCTACACGACGCTCTTCCGATCT.
Sequence analyses
Thirty-six-nucleotide sequencing reads were generated using the Illumina Genome Analyzer system. The 3′ adapter sequences were trimmed off in silico by scanning from the 3′-end of the sequence for the first instance of the first 4 nt of the adapter: CTGT. Sequence reads that lacked CTGT in the 3′ most 13 nt were excluded from further analysis. After barcode sorting, the 4 nt of barcode were trimmed off the 5′-ends to yield 19- to 28-nt reads corresponding to the captured small RNAs. siRNA alignment counts were produced by aligning the reads to a cDNA reference set using a local installation of BLAT (Kent 2002), with default parameters, except tileSize was set to 10 and stepSize to 5. The term “siRNA alignment count” (above) refers to the number of matches to an individual cDNA obtained from the set of reads. The cDNA data set, derived from the C. elegans WS190 genome assembly, was downloaded from http://www.wormbase.org:80/biomart/martview. The data set of known transposons sequences was downloaded from http://www.sanger.ac.uk/Projects/C_elegans/REPEATS/elegans.lib. Read counts and alignment counts are listed in Table S1.
Statistical methods
P-value calculations for identification of genes with RRF-3-dependent siRNAs:
Gene-by-gene siRNA alignment counts were tallied and compared for each sample. To measure fold-changes in alignment count between samples for a particular gene, the alignment count was first converted to a proportion by dividing the alignment count of that gene by the sum of the alignments of the complete cDNA reference set. To obtain a measure of the statistical significance of differences between samples, we assumed that there was no difference in population proportions for each gene and then calculated the probability (P-value) of measuring a difference in sample proportions at least as extreme as the one observed. A one-tailed test was used since we had prior expectations that the proportions for certain genes would decrease in the rrf-3(pk1426) samples (see File S1 for a representative calculation). The standard errors of the means for sample proportion differences were calculated on the basis of predicted normal distributions of the sample proportions, which is justified by the large sample size and the central limit theorem (the proportions here are equivalent to means). The sample proportion differences were standardized to Z-scores by dividing by the standard errors of the means for the proportion differences.
P-values and fold-enrichment calculations for overlap between sets of genes with RRF-3-dependent siRNAs and genes with elevated mRNA levels in rrf-3 and eri-1 mutants:
P-values were derived using Fisher's exact test from the following numbers: 16,887 of the genes represented in the cDNA data set had probes on the microarray used by Asikainen et al. (2007). Of the 21 genes that we identified as spermatogenesis candidate RRF-3 targets, 18 were included in this set. Of the 16,887 total genes, 178 had increased mRNA levels in rrf-3(pk1426), while 46 had decreased levels. Of the 18 spermatogenesis candidate RRF-3 targets, 3 had increased mRNA levels in rrf-3(pk1426) (15.8-fold enrichment; P-value = 8.8 × 10−4); none had decreased levels. Of the 16,887 genes, 706 had increased mRNA levels in eri-1(mg366), while 263 had decreased levels. Of the 18 spermatogenesis candidate RRF-3 targets, 8 had increased mRNA levels in eri-1(mg366) (10.6-fold enrichment; P-value = 2.9 ×10−7). These 8 included the 3 with elevated mRNA levels in rrf-3(pk1426). None of the 18 had decreased levels in eri-1(mg366). When we considered all 44 candidate RRF-3 targets rather than just the 18 with siRNAs present in the spermatogenic cell sample, the numbers were still highly significant: fold enrichments of 6.5 and 4.4 with P-values of 1.5 × 10−2 and 4.4 × 10−4 for rrf-3(pk1426) and eri-1(mg366), respectively.
RESULTS
Compromised spermatogenesis accounts for reduced brood size and X-chromosome loss from rrf-3(pk1426) hermaphrodites:
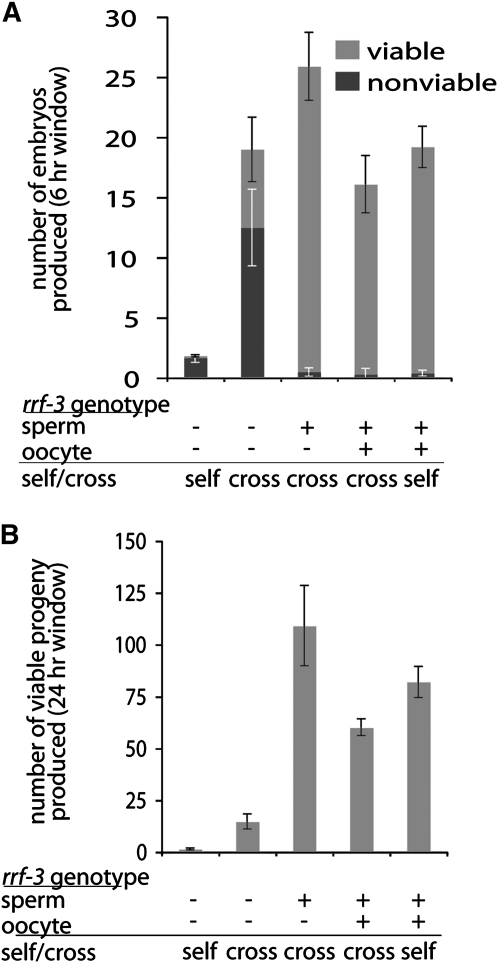
We first set out to determine whether the rrf-3(pk1426) reduced brood size and X-chromosome missegregation phenotypes were attributable to defects in sperm or oocytes. C. elegans exists in two sexes: males, which produce only sperm, and hermaphrodites, which produce first sperm and then oocytes in the same gonad. Defects in either gamete line can result in sterility of self-fertilizing hermaphrodites. rrf-3(pk1426) self-fertilizing hermaphrodites produced very few fertilized embryos at 25°, with 90% of these nonviable (Figure 1A). Here and in subsequent experiments, we defined “nonviable” as failure to give rise to L4 stage larva or adults. We found that a preponderance (or all) of the nonviability was accounted for by embryonic lethality prior to hatching. To determine the source of the reduced embryo count and viability, we set up matings between rrf-3(pk1426) hermaphrodites and wild-type N2 males at 25°. Fertilization by wild-type sperm rescued the rrf-3(pk1426) brood size reduction in terms of both numbers of embryos produced and embryonic viability (Figure 1, A and B), indicating that both phenotypes can be attributed to defects during spermatogenesis.
Figure 1.—
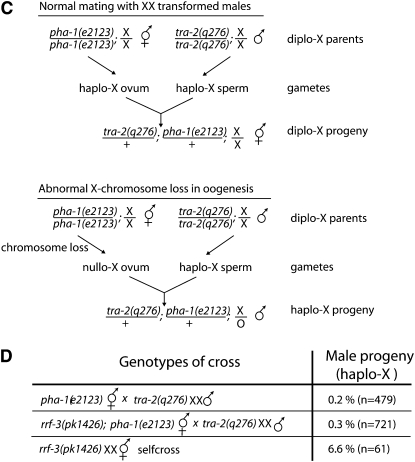
Lack of embryos, nonviable embryos, and X-chromosome loss are attributable to defective spermatogenesis in rrf-3 mutants. Individual hermaphrodites either were allowed to self-fertilize or were crossed with males at 25° and then allowed to lay eggs during a 6-hr window followed by an 18-hr window. The progeny from both periods were counted when they had reached L4 and adult stage. Cases where the matings were unsuccessful, as evidenced by few embryos or a lack of male progeny, were not counted. Multiple matings were set up and progeny from at least five gravid hermaphrodites were counted in every case. (A) Rescue of rrf-3(pk1426) embryo production and viability of wild-type sperm (6-hr window).The embryos laid during the first 6 hr were counted in addition to the surviving progeny to estimate the number of nonviable embryos. Error bars denote standard errors of the means. (B) Rescue of the rrf-3(pk1426) brood size (24-hr window). The number of surviving progeny from both the 6- and 18-hr windows were combined. Error bars denote standard errors of the means. (C) Diagram of assay for loss of the X chromosome during oogenesis. pha-1(e2123) is a temperature-sensitive, recessive, embryonic lethal mutation that ensures that only outcrossed progeny survive at 23°. The tra-2(q276) mutation causes XX animals, which would normally be hermaphrodites, to develop into partially fertile males. (D) Wild-type levels of X-chromosome loss in rrf-3(pk1426) oogenesis. Multiple matings were set up, each with five hermaphrodites and at least five males, with the exception of the rrf-3(pk1426) self-fertilizations, which were done with individual hermaphrodites. The number of male progeny was scored when they had reached L4 or adult stage. The total number of progeny counted, n, is smaller than would be expected from wild-type matings due to the inefficiency of tra-2(q276) males. These crosses were set up at 23° rather than 25° for increased success.
To determine whether X-chromosome missegregation is linked to spermatogenesis in rrf-3(pk1426) hermaphrodites, we utilized a genetic test that takes advantage of the C. elegans sex determination system. The ratio of X-chromosomes to autosomes in C. elegans determines the sex: animals with two X chromosomes and two sets of autosomes are hermaphrodites; animals with one X and two sets of autosomes are male (Nigon 1951; Madl and Herman 1979). Hence, in a normal self-fertilization, a hermaphrodite contributes one X from each sperm and from each ovum so that the resulting embryos are virtually all (∼99.8%) XX hermaphrodites. Generation of XO males from XX hermaphrodite self-fertilization is indicative of X-chromosome loss or nondisjunction in either the sperm or oocyte lineage. To test for X-chromosome loss in rrf-3(pk1426) oogenesis, we set up matings with tra-2(q276) XX-transformed males, which contribute an X from each sperm (like a hermaphrodite). Unless the X chromosome is lost during oogenesis, hermaphrodites mated with tra-2(q276) males give rise to almost entirely XX hermaphrodite progeny (Figure 1C) (Okkema and Kimble 1991). We set up matings between either rrf-3(pk1426) or wild-type hermaphrodites with tra-2(q276) males. To select for outcrossed progeny, the hermaphrodites also carried a temperature-sensitive mutation in pha-1 that causes self-fertilized progeny to die as embryos (Schnabel and Schnabel 1990). The progeny from this cross exhibited extremely low numbers of spontaneous males (wild type: 1 of 479; rrf-3(pk1426): 2 of 721) (Figure 1D). These results indicate that oogenesis is not the source of X-chromosome loss in rrf-3(pk1426) propagation, implicating spermatogenesis as the affected process. While RRF-3 may have additional functions in oogenesis, these results indicate that most or all of the fertility and X-chromosome segregation mutant phenotypes can be accounted for by roles for RRF-3 during spermatogenesis.
RRF-3 is required for spermatocyte cell division:
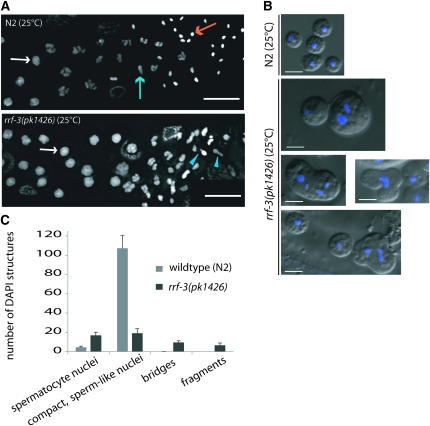
To further investigate the nature of the sperm defect, we examined germlines of rrf-3(pk1426) hermaphrodites prior to and during completion of spermatogenesis at 25°. We investigated chromosome organization during meiotic prophase using antibodies labeling RAD-51, which functions in recombination and decorates chiasmata (Alpi et al. 2003; Colaiacovo et al. 2003); the synaptonemal complex components REC-8 and SYP-1 (Pasierbek et al. 2001; MacQueen et al. 2002); and HIM-8, which marks a specific domain on the X chromosome (Phillips et al. 2005). This analysis did not reveal any overt defects in meiotic prophase (Figure S1 and Figure S2). In addition, DAPI-stained nuclei in the process of transitioning between prophase and the first meiotic division appeared normal in rrf-3 mutants (Figure 2A). Cytological abnormalities in rrf-3 mutant germ cells first started to become evident after the primary spermatocyte stage, where DAPI staining revealed nuclear abnormalities such as small or bridged structures (Figure 2A). Further analysis of unfixed late-stage spermatocytes by DIC and Hoechst staining revealed both unusual cell morphology and the presence of multiple nuclear structures within individual mutant spermatocytes (Figure 2B). Other spermatocytes within the same gonads appeared to develop without obvious morphological abnormalities and to produce residual bodies and sperm of the normal size and shape, although in smaller numbers than wild type. Additional DAPI staining of released gonads revealed that the production of compact sperm-like nuclei was both delayed and decreased. At the expected time of spermatogenesis completion (prior to ovulation but after appearance of oocytes), rrf-3 mutants had a fivefold reduction in compact, sperm-like nuclei relative to N2 (Figure 2C). By the time of the first ovulation, the number in rrf-3 mutants of sperm-like nuclei relative to N2 increased to half of the N2 levels (data not shown), but these gonads still contained cells with multiple or misshapen nuclei. Together, these phenotypes indicate defects in developmental progression and cell division processes during spermatogenesis in rrf-3(pk1426).
Figure 2.—
RRF-3 is required for normal spermatocyte cell division. (A) Abnormal nuclear structures and lack of sperm visible by DAPI staining of fixed hermaphrodite gonads undergoing spermatogenesis at 25°. Nuclei in both wild-type and mutant spermatocytes (white arrows) appeared visibly normal, but nuclei in subsequent stages often appeared fragmented or bridged in the mutant (blue arrowheads). A wild-type dividing spermatocyte nuclei (blue arrow) and spermatid nuclei (orange arrow) are also indicated. Bar, 10 μm. (B) Multiple and misshapen Hoechst-staining structures in mutant spermatocytes at 25°. Unfixed sperm and spermatocytes released from rrf-3(pk1426) dissected gonads and stained with Hoescht 33342 show multiple nuclear structures within individual cells (bottom four panels), often of inconsistent sizes and shapes compared to N2 control sperm nuclei (top). Mutant worms also produced sperm that were visibly similar to the N2 (e.g., upper left corner of bottom panel). (C) Quantification of DAPI-staining structures in fixed hermaphrodite gonads at 25°. Nuclear structures were counted in the proximal gonad at the expected timing of spermatogenesis completion (after the appearance of the first oocyte but prior to ovulation; at this stage the sperm have not yet moved into the spermatheca). Compact, sperm-like nuclei are not equivalent to cell counts because of the presence of multiple DNA structures within some individual cells in the mutant. Values shown are averages obtained from 9 N2 gonads and 10 rrf-3(pk1426) gonads. Error bars denote standard errors of the means.
An rrf-3 paternal effect on embryogenesis:
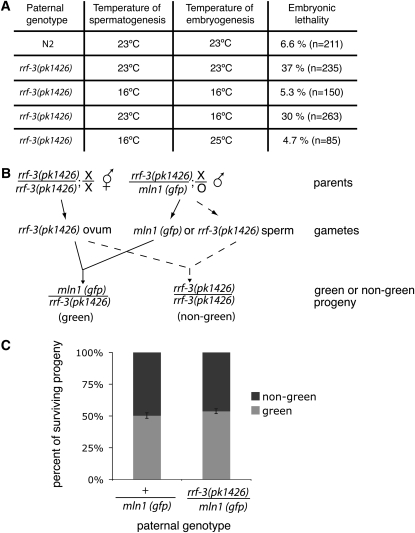
Since rrf-3(pk1426) animals at elevated temperatures yield a high percentage of nonviable embryos, we asked whether RRF-3 functions directly in embryos or in a spermatogenesis process linked to embryogenesis. To test this, we set up matings between rrf-3(pk1426) males and tra-2(q122) females, which genetically are XX hermaphrodites but have feminized germlines that produce only oocytes (Schedl and Kimble 1988). We found that rrf-3(pk1426) males produced sperm that were capable of fertilization, but many of the resulting embryos were nonviable (∼37% at 23°). This effect on embryos depended on the temperature at which spermatogenesis occurred but was independent of the temperature at which embryogenesis occurred (Figure 3A). The paternally derived lethality was not due to the presence of a second, unrelated recessive mutation because trans-heterozygous males bearing rrf-3(pk1426) and another deletion allele, rrf-3(ok629), also produced nonviable embryos. Such males should be heterozygous for any extraneous mutations outside rrf-3, so the contribution of any such mutations, if any, should be minimized. In setting up the test for the rrf-3 requirement, we utilized trans-heterozygote rather than ok629 homozygotes because of a male-tail-defective phenotype in the strain (which we have not definitively assigned to ok629). In matings between tra-2(q122) females and ok629/pk1426 trans-heterozygote males at 25°, the resulting embryos showed similar levels of nonviability as embryos sired by pk1426 homozygous males: 84% nonviability from pk1426/pk1426 and 72% nonviability from ok629/pk1426, contrasted with 2% nonviability from N2 (Figure S3).
Figure 3.—
RRF-3 is involved in a temperature-sensitive spermatogenic process with effects on embryonic viability. (A) The viability of embryos sired by rrf-3(pk1426) males is determined by temperature of spermatogenesis. rrf-3(pk1426) or N2 males raised at the indicated temperatures were mated with tra-2(q122) females. The level of embryonic viability was estimated by counting both the total number of eggs laid (n) and the number of surviving progeny (as described in materials and methods). (B) Diagram of assay for an embryonic requirement for a paternally delivered, wild-type copy of rrf-3. rrf-3(pk1426) homozygous hermaphrodites were mated with males that were heterozygous for rrf-3(pk1426) and the mIn1 inversion (which carries the wild-type rrf-3 locus and a transgene driving pharyngeal GFP expression). Inheritance of the mutant and wild-type rrf-3 allele could be tracked by GFP expression. No marker for outcrossed progeny was necessary because of the sperm defect in rrf-3(pk1426) hermaphrodites. Animals were raised and matings set up at 25°. (C) A wild-type copy of rrf-3 is dispensable for embryogenesis. GFP- and non-GFP-expressing progeny were counted at L4 or adult stage to determine the proportion with paternally inherited mIn1. To provide a control for the potential effects of the inversion and the dominant transgene marker, +/mIn1 males were used. A total of 526 progeny resulting from five matings were counted for +/mIn1, and a total of 675 progeny resulting from four matings were counted for rrf-3(pk1426)/mIn1. Error bars denote standard error of the means.
To distinguish a paternal effect from an embryonic requirement for a paternally delivered rrf-3(+) allele, we used the scheme in Figure 3B, setting up matings between rrf-3(pk1426) homozygous hermaphrodites and rrf-3(pk1426)/mIn1 heterozygous males. mIn1 is a chromosome inversion of chromosome II containing the wild-type rrf-3 locus and a transgene insertion that drives pharyngeal GFP expression (Edgley and Riddle 2001), enabling us to determine which paternal allele is inherited by the progeny. We found that survival was not affected by physical inheritance of the wild-type rrf-3 allele (on mIn1), indicating that rrf-3 expression is not required after separation of joined secondary spermatocytes into spermatids nor in embryogenesis (Figure 2 and Figure 3C). Together with the early temperature-sensitive period (prior to fertilization of the embryo as shown in Figure 3A), this result implicates spermatogenesis as the crucial timing for both RRF-3 expression and activity.
Roles for rrf-3 and eri-1 in preventing microtubule accumulation in sperm and abnormal microtubule structures in one-cell embryos:
To examine the basis of the embryonic lethality, we stained wild-type and rrf-3(pk1426) self-fertilized embryos with an antibody against α-tubulin. One particularly notable defect was the presence of abnormal microtubule structures in a subset of the mutant embryos at the one-cell stage (i.e., from formation of the maternal meiotic spindle through metaphase of the first mitotic division). Specifically, we detected supernumerary microtubule asters associated with the sperm pronucleus or tripolar spindles in 5 of 35 rrf-3 mutant one-cell embryos (Figure 4A), whereas such structures were never observed in control embryos (0 of 34; P-value: 0.029).
Figure 4.—
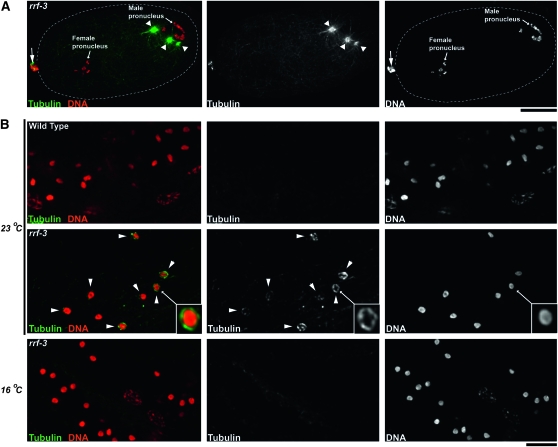
Abnormal microtubule structures in rrf-3 mutant embryos and sperm. An embryo (A) and sperm (B) stained with Hoechst to label chromosomes (red) and immunostained with antitubulin antibody to label microtubules (green). (A) One-cell embryo (prior to meeting of male and female pronuclei) laid by an rrf-3(pk1426) hermaphrodite at 23°. Although two asters are normally associated with the male pronucleus at this stage in wild-type embryos, this mutant embryo has three asters (marked by arrowheads) associated with the male pronucleus. An arrow marks the polar bodies, which are located at the anterior end of the embryo. Bar, 10 μm. (B) Sperm from either a wild-type hermaphrodite (top row) or rrf-3(pk1426) hermaphrodites (bottom three rows), raised at the indicated temperatures. Arrowheads highlight sperm nuclei surrounded by tubulin wreaths that were observed in rrf-3 mutant sperm at 23°. The insets in the middle row show an enlarged view of the tubulin wreath encasing the nucleus of an rrf-3 mutant sperm. Bar, 5 μm.
Immunofluorescence analysis also revealed striking microtubule-based abnormalities in rrf-3 mutant sperm. At 23°, we observed bright wreaths of microtubules surrounding the nuclei of both hermaphrodite and male-derived sperm (Figure 4B and Figure S4). These microtubule wreaths were clearly visible in 83% (n = 307) of rrf-3 mutant sperm. In contrast, only 2% of wild-type sperm exhibited any detectable tubulin staining (which had a diffuse appearance) other than the sperm centrioles. Microtubule wreath structures were never observed in wild-type sperm (n = 179). The fact that tubulin was largely undetectable is expected on the basis of previous observations that microtubules in secondary spermatocytes are effectively segregated to an enucleated cytoplast called the residual body (Roberts et al. 1986; Ward 1986; Ward et al. 1986). At 16°, where we did not see dead embryos, the mutant sperm lacked the microtubule wreath phenotype.
Because meiotic DNA damage resulting from compromised small RNA machinery in Drosophila results in microtubule polarization defects (Chen et al. 2007; Klattenhoff et al. 2007; Pane et al. 2007), we tested the possibility that the microtubule wreaths in rrf-3 mutant sperm might be a consequence of unrepaired meiotic DNA double-strand breaks (DSBs). (Although the normal kinetics of RAD-51 foci in rrf-3 mutants suggests successful DSB repair, our data could not exclude the possibility of persistent meiotic DNA damage.) Specifically, we looked for microtubule wreaths around sperm nuclei at 23° in double-mutant worms lacking SPO-11 (the enzyme required for inducing meiotic DSBs) (Keeney et al. 1997; Dernburg et al. 1998) and RRF-3. Microtubule wreaths were present in sperm from the double mutant, indicating that these structures do not depend on meiotic DSBs induced by SPO-11 (data not shown).
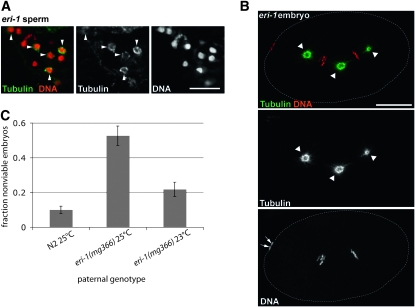
Given the similarities between the fertility and RNAi phenotypes of rrf-3 and eri-1 mutants, we examined tubulin distribution in sperm and embryos from eri-1(mg366) hermaphrodites raised at 23°. We found both microtubule wreaths in sperm (59%, n = 256) and microtubule/spindle defects in embryos (4/20) produced by eri mutants (Figure 5, A and B). We further confirmed a role for ERI-1 in producing functionally normal sperm by showing that mating eri-1(mg366) males with tra-2(q122) females resulted in nonviable embryos at elevated temperatures (Figure 5C).
Figure 5.—
Abnormal microtubule structures in eri-1 mutant embryos and sperm and paternal-effect nonviability from eri-1 mutant males. (A) eri-1(mg366) sperm and (B) embryo from hermaphrodites grown at 23° and stained with Hoechst to label chromosomes (red) and antitubulin antibody to label microtubules (green). (A) Some eri-1(mg366) sperm contain abnormal microtubule wreath structures (arrowheads) similar to those observed in rrf-3 mutant sperm. Bar, 5 μm. (B) One-cell eri-1 mutant embryo with a tripolar spindle. Arrowheads mark each of the three spindle poles. Arrows mark the polar bodies, which indicate the anterior of the embryo. Bar, 10 μm. (C) eri-1(mg366) males sire nonviable embryos at elevated temperatures. The fraction of nonviable embryos was estimated by counting both the number of eggs laid and the resulting number of surviving progeny. Matings were set up at both 23° and 25°. Error bars denote standard errors of the means.
Identification of siRNAs present in spermatogenic cells:
During the progression through the distinct stages of spermatogenesis from a premeiotic syncitium to a mature sperm, a large number of genes must be choreographed to ensure that the proper genes are expressed at the appropriate level and developmental stage (e.g., Johnston et al. 2008). Since several RNAi-related factors have been implicated in spermatogenesis through mutant studies, we expected that RNA-based regulatory machinery might contribute at several levels to this choreography. Consequently, we characterized the small RNAs present in the male germline. We took a sequencing-based approach to this, adapting sperm isolation and optimizing high-throughput sequencing methods to generate an extensive data set of small RNA sequences from isolated spermatogenic cells. Spermatogenic cells were isolated from adult fem-3(q20) XX animals using a technique that yields a large proportion of sperm precursors in addition to mature sperm (adapted from Lamunyon and Ward 1995, which describes a dicing method, and from Miller 2006, which uses M9 buffer and does not require a Percoll step to obtain sperm for biochemical analysis). fem-3(q20) animals produce an excess of sperm and no oocytes or embryos at 25° (Barton et al. 1987). Functionality of fem-3(gf) XX sperm has been demonstrated previously by their ability to sire live embryos following artificial insemination (Lamunyon and Ward 1994). The population of cells isolated and used for sequencing is shown in Figure S5.
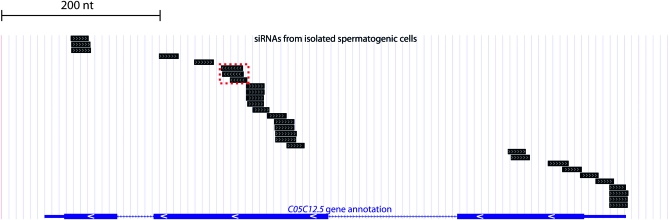
We prepared sequencing libraries using a method that allows for capture of small RNAs independently of the 5′ phosphorylation status. In C. elegans, the majority of RdRP products have a 5′ triphosphate (Ruby et al. 2006; Pak and Fire 2007; Sijen et al. 2007), but standard high-efficiency library preparation methods (e.g., Lau et al. 2001) rely on a 5′ monophosphate, which is characteristic of microRNAs and other DCR-1 products. The ability to capture 5′-triphosphorylated siRNAs was critical for this analysis, since this terminus structure is expected to be present on any initial RdRP transcript and would be retained unless there is covalent 5′-end processing. In materials and methods, we describe our procedure for preparing such libraries using a modification of Lau et al. (2001) with the addition of a dephosphorylation and a single phosphate addition step following 3′ adapter ligation. The resulting libraries of small RNAs were of diverse sequence composition, and we obtained large numbers of distinct small RNA species including microRNAs, piwi-interacting RNAs (also called piRNAs or 21U-RNAs), and siRNAs. These sequences are available at the NCBI short read archive (GEO accession number GSE18429). We aligned the small RNA sequences to a cDNA data set downloaded from http://www.wormbase.org to generate a list of siRNA alignment counts for each annotated gene (where “siRNA alignment count” for each gene is the number of alignments that it produced from the complete set of small RNA sequences) These data are tabulated in File S1. Also included in the NCBI short read archive are alignment files in psl format, which can be directly uploaded onto the UCSC Genome Browser (http://www.genome.ucsc.edu) for viewing. Two notable features of the siRNA alignments are a bias toward antisense orientation relative to the annotated mRNA and a diversity of positions within the individual target genes, as illustrated by siRNAs of the gene C05C12.5 (Figure 6). The antisense orientation of the spermatogenesis siRNAs suggests synthesis by RdRP and is consistent with potential modulatory roles of these small RNAs through a “standard” gene-silencing model in which an Argonuate-bound short antisense RNA segment catalytically locates and silences mRNAs with extended regions of homology (reviewed in Hutvagner and Simard 2008).
Figure 6.—
siRNA alignments to an mRNA: antisense predominance and diversity of intragenic locations. A modified UCSC Genome Browser Display illustrates the C05C12.5 annotation and siRNAs from the spermatogenic cell sample. For the C05C12.5 annotation, thin solid end segments denote untranslated regions, thick solid segments denote exons, and line segments denote introns. White arrows within exons indicate the gene's orientation. siRNA alignments are represented by short black segments. All but the three siRNA alignments outlined by a dashed red box are antisense to C05C12.5. Note that siRNAs that span exon–exon boundaries do not produce genomic alignments.
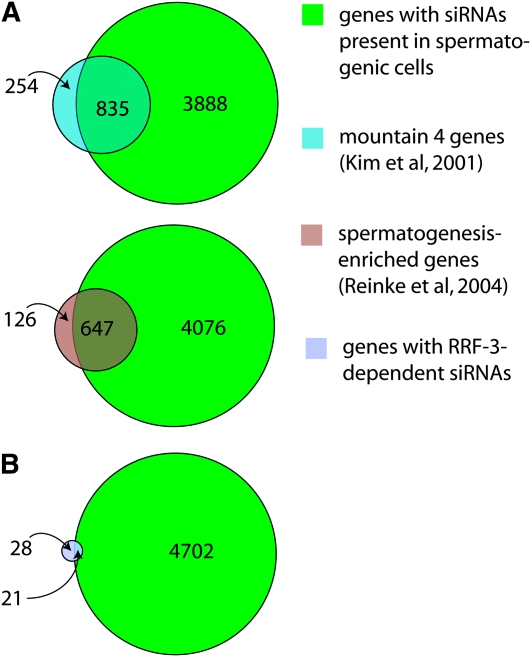
The full small RNA-ome from sperm is large, complex, and likely to reflect numerous distinct regulatory processes. We began by analyzing small RNAs with likely roles in the regulation of specific mRNAs by compiling siRNA alignment counts for each annotated mRNA-encoding gene in this data set. While a comprehensive list of spermatogenesis-expressed mRNAs is unavailable for C. elegans, several studies have categorized mRNAs with higher relative expression in spermatogenic cells than in other tissues. We compared the list of genes with siRNAs present in the spermatogenic cell sample with two gene lists, “spermatogenesis-enriched” and “mountain 4.” The spermatogenesis-enriched category was derived by comparison of mRNA expression from mutant hermaphrodites that were either masculinized to produce only sperm or feminized to produce only oocytes (Reinke et al. 2004). Mountain 4 was derived from a study that grouped genes according to correlated expression patterns over multiple growth conditions, developmental time points, and mutant backgrounds (Kim et al. 2001). For a gene to be categorized as having siRNAs in spermatogenic cells, we required that the number of alignments to its mRNA from our isolated spermatogenic cell siRNAs be at least three. A total of 4723 of the 21,133 genes in our cDNA reference set met this criterion. These 4723 genes show a 3.7- and 3.5-fold greater-than-expected overlap with the spermatogenesis-enriched and mountain 4 expression category, respectively (accounting for 84% of the total spermatogenesis-enriched category and 77% of the mountain 4 category, Figure 7A). Conversely, genes from the spermatogenesis-enriched and mountain 4 categories were significantly underrepresented among the group of mRNAs identified in our isolated spermatogenic cell sample with siRNA alignment counts of fewer than 3 (4.8- and 3.3-fold less-than-expected, respectively). These results indicate that siRNA abundance in spermatogenic cells correlates with mRNA expression during spermatogenesis and that siRNA production is a general feature of gene activity during spermatogenesis, involving the majority of genes with enriched expression during spermatogenesis.
Figure 7.—
Overlap between siRNA- and mRNA-based gene expression categories. (A) The genes with siRNAs present in spermatogenic cells are the majority of genes whose mRNAs have enriched expression during spermatogenesis. Venn diagrams display the numbers of genes either shared between two categories or exclusive to either category. Genes with siRNAs present in spermatogenic cells had an siRNA count of at least three from the isolated spermatogenic cell sample. Genes in mountain 4 (Kim et al. 2001) and spermatogenesis-enriched (Reinke et al. 2004) categories are associated with enriched mRNA expression during spermatogenesis as measured by comparative microarray analyses. Only genes that had probes on the microarrays and were present in the cDNA data set used for aligning small RNA sequences were counted in each category. (B) The candidate RRF-3 regulatory targets are enriched for genes with siRNAs present in spermatogenic cells. Relative siRNA expression levels were measured by siRNA sequencing from two sets of him-8(e1489) adult males (mutant and wild type for rrf-3). Forty-nine genes whose siRNA alignment counts were decreased by at least threefold with a P-value of <0.005 in rrf-3(pk1426) in both sets were categorized as candidate RRF-3 regulatory targets.
Identification of RRF-3-dependent siRNAs:
To identify siRNAs that depend upon RRF-3 for their production, we analyzed siRNA expression profiles from rrf-3 mutant samples. To avoid any possible confounding effects of the fem-3 mutant background (since FEM-3 affects the control of spermatogenesis), we carried these experiments out using populations of karyotypically normal XO males. For both rrf-3 mutant and wild-type control samples, a him-8(e1489) mutation that compromises X-chromosome pairing primarily in oocytes was used to generate populations with large numbers of males (Hodgkin et al. 1979).
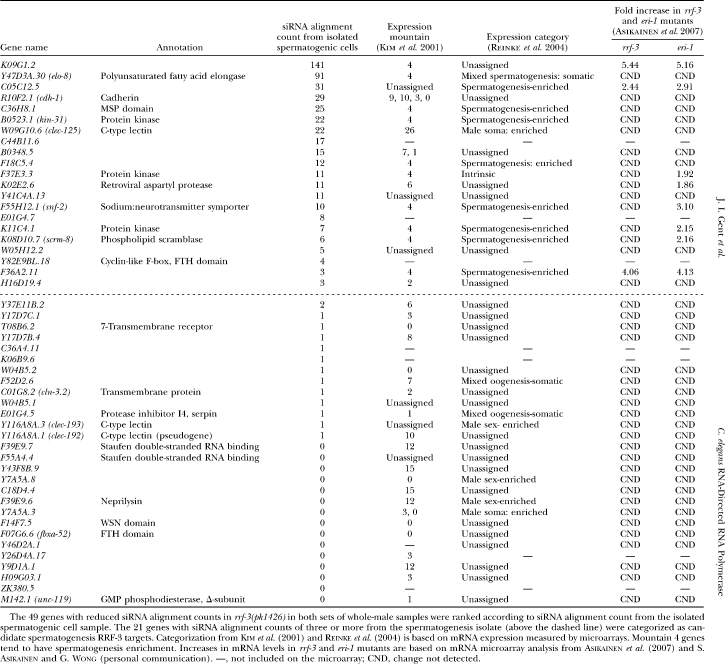
siRNA profiles from the him-8(e1489) whole males and from the isolated fem-3(q20) hermaphrodite spermatogenic cells were generally similar in that genes with abundant siRNAs in one sample tended to be abundant in the other, but a number of genes did show genotype/tissue specificity (Figure S6). To increase the stringency of our analysis, we prepared two sets of males of each genotype at 25° using different population synchronization methods for each set. We aligned the sequences from each sample to the cDNA data set and compared the siRNA alignment count for each gene in the rrf-3 mutant and control male sample. For the vast majority of genes, we saw little or no effect of rrf-3(pk1426) on siRNA levels in these assays: <5% of the genes had reproducible, greater-than-twofold decreases in siRNAs levels. A subset of genes showed dramatic effects, however, and we chose to track 49 genes whose expression is likely dependent upon RRF-3 by the criterion of a threefold-reduced alignment count with a P-value of <0.005 in both sets of experiments (Table 1; materials and methods).
TABLE 1.
Candidate RRF-3 regulatory targets
This list of 49 genes is intended as exemplary rather than complete: other genes appear to have similar properties without quite reaching our arbitrary statistical threshold criteria, while still more may produce siRNAs that are expressed at levels insufficient for statistical significance in these analyses. In addition, alignment of the small RNA sequences to the genome without regard to predicted coding status revealed additional unannotated loci with small RNAs dependent upon RRF-3, such as the previously identified RRF-3-dependent locus on the X chromosome called the X cluster (Ambros et al. 2003; Duchaine et al. 2006). The two genes on the list of 49 with the highest siRNA levels, C44B11.6 and K02E2.6, have previously been shown by siRNA Northern analysis to require RRF-3 for siRNA accumulation, providing an independent confirmation of our experimental approach (Duchaine et al. 2006; Lee et al. 2006).
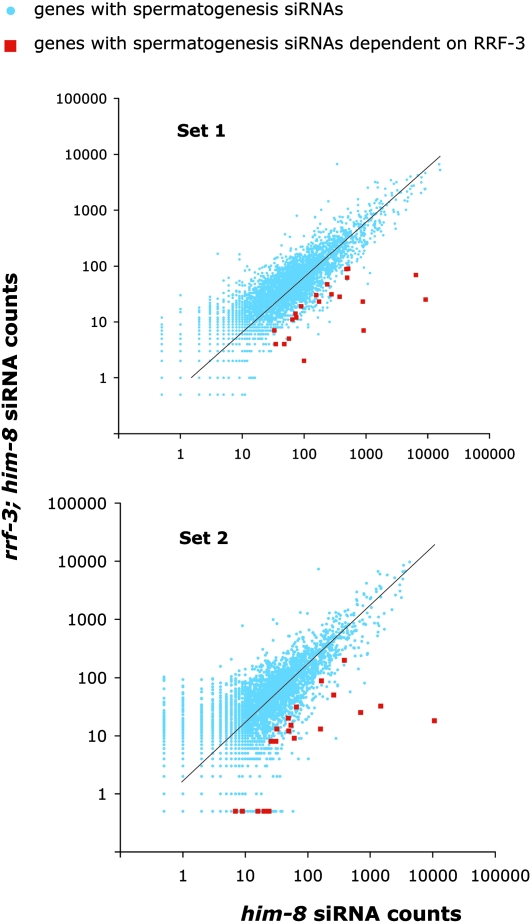
For any of these genes to be regulated by an RRF-3-driven mechanism during spermatogenesis, one would expect that both their mRNAs and siRNAs would be expressed during spermatogenesis. We found 5.2-fold enrichment for the mountain 4 expression category and 4.6-fold enrichment for the spermatogenesis-enriched category in our list of 49 candidate RRF-3 target genes, indicating that a substantial fraction of RRF-3-regulated genes are expressed during spermatogenesis (Table 1; Figure 7B). We also used an siRNA alignment count from spermatogenic cells as a means of ranking the candidate gene list in terms of the likelihood of spermatogenesis gene regulation. As before, we split the list into two categories using a spermatogenesis siRNA alignment count of 3 as the threshold, resulting in 21 genes with spermatogenesis siRNAs and 28 without (Table 1). These 21 genes with spermatogenesis siRNAs and dependency on RRF-3 represent a sample of candidate RRF-3 regulatory targets during spermatogenesis. An important result of this analysis was that RRF-3 had little or no effect on gene-specific siRNA counts for the majority of the 4723 genes with spermatogenesis siRNAs (Figure 8): <4% of the genes had reproducible, greater-than-twofold decreases in siRNA levels in rrf-3(pk1426) relative to control him-8(e1489) males (69 of 1784 genes; to exclude background noise from genes with low siRNA alignment counts, only genes with counts of at least 25 in both sets of control males were included). This result is consistent with the existence of many other RNAi-related factors with substantial germline phenotypes (Cox et al. 1998; Smardon et al. 2000; Knight and Bass 2001; Tops et al. 2005; Chen et al. 2007).
Figure 8.—
Global RRF-3 effects on spermatogenesis siRNA accumulation. Scatter plots depict the siRNA alignment count per gene from him-8(e1489) control males (x-axis) and from rrf-3(pk1426); him-8(e1489) (y-axis). Blue dots correspond to the 4723 genes with spermatogenic cell siRNAs. Red dots correspond to the 21 genes with both spermatogenic siRNAs and dependency on RRF-3. Genes with siRNA alignment counts of zero were assigned a value of 0.5 to avoid exclusion from the logarithmically scaled plots. The trendlines indicate the expected siRNA alignment counts per gene if both samples were derived from the same population. On a linear scaled plot, the slopes deviate from 45° because of differences in sample sizes.
In addition to mRNA-encoding genes, we examined small RNAs corresponding to a large number of repeated and/or transposable elements (http://www.sanger.ac.uk/Projects/C_elegans/REPEATS/elegans.lib). From this analysis we found that many transposons, especially Helitron (264 siRNAs aligned to a HELICOP1 sequence) and Tc1/mariner transposons (87 siRNAs aligned to a Tc1 sequence), were associated with high siRNA alignment counts from the spermatogenic cell sample. We did not find any transposons that required RRF-3 by the criteria of a threefold reduction in siRNA alignment count and a P-value of <0.005 between mutant and control from both sets of samples. Intriguingly, we found three cases in which siRNAs corresponding to a transposon were significantly more abundant in rrf-3 mutants: CELE1, LINE2, and TURMOIL2. Of these, only TURMOIL2 had siRNAs present in the spermatogenic cell sample. The increased siRNAs from these transposons could reflect an activation of RNAi-like processes as has previously been observed in exogenous RNAi and transgene silencing (Sijen et al. 2001; Simmer et al. 2002).
siRNAs produced by RRF-3 and ERI-1 function to downregulate mRNA levels during spermatogenesis:
In the absence of RRF-3 or ERI-1, at least one putative target, K02E2.6, has been shown to have increased mRNA levels (Duchaine et al. 2006). To test whether RRF-3 and ERI-1 generally act to downregulate their spermatogenesis targets, we compared the RRF-3 regulatory candidates with a global microarray analysis of RRF-3 and ERI-1 effects on mRNA levels in hermaphrodites undergoing spermatogenesis (Asikainen et al. 2007; S. Asikainen and G. Wong, personal communication). We first note some limitations in the comparison with the microarray analysis: Three of the 21 genes that we identified as differentially regulated in males were not included in the microarrays used by Asikainen et al. (2007). Furthermore, Asikainen et al. (2007) reported data for whole L4 hermaphrodites, in which sperm or sperm precursors may have been a relatively rare component (so that substantial differences in spermatogenic mRNA abundance could have been masked by pools of unaffected somatic RNA). Similarly, spermatogenic cell-type-specific silencing could be hard to detect in the background of other sperm precursor cells still expressing the transcript at normal levels. Despite these limitations, it is of considerable interest to compare our siRNA data with the mRNA abundances determined by Asikainen et al. (2007; Table 1). Of the 18 genes that were included in the microarray analysis and with RRF-3-dependent spermatogenesis siRNAs, 3 showed increased mRNA levels in both rrf-3 and eri mutants, and an additional 5 showed increased mRNA levels in eri-1 mutants alone (15.8-fold enrichment with a P-value of 8.8 × 10−4 for the rrf-3 mutant; 10.6-fold enrichment with a P-value of 2.9 × 10−7 for the eri-1 mutant; see materials and methods).
DISCUSSION
Evidence for a developmental role for RRF-3 during spermatogenesis:
Small RNA pathway functions are commonly categorized as defensive (e.g., suppression of transposable elements and viruses) or regulatory (e.g., downregulation of mRNA levels during development). Either function can be essential, as exemplified by the critical roles of microRNA developmental regulators (reviewed in Flynt and Lai 2008) and by the roles of piRNAs in transposon control (reviewed in Klattenhoff and Theurkauf 2008). These developmental/defense roles of small RNAs can overlap, as illustrated by a human cellular microRNA that targets a viral RNA (Lecellier et al. 2005) and by the recent description of a C. elegans small RNA family (21U- or piRNAs) that apparently modulates both transposons and cellular transcripts during spermatogenesis (Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008). Although products of cellular RdRPs apparently differ from piRNAs in both structure and biosynthetic mode, some RdRPs can participate in responses to foreign nucleic acids, including quelling in Neurospora crassa (Cogoni and Macino 1999) and viral defense in Arabidopsis (Mourrain et al. 2000). On the basis of developmental phenotypes in mutant strains, other RdRP components have been hypothesized to carry out endogenous regulatory roles (Smardon et al. 2000; Shiu and Metzenberg 2002).
Several characteristics of RRF-3 (the temperature-dependent character and the requirement for RRF-3 in a defined developmental pathway such as spermatogenesis) facilitate a detailed analysis of siRNAs associated with RRF-3 function and their potential regulatory roles. Using high-throughput sequencing as a tool for characterizing RNA populations, we found that antisense siRNAs corresponding to a variety of both transposable elements and cellular mRNAs are present in spermatogenic cells. Among the large and diverse set of cellular mRNAs reflected in the pool of siRNAs, we found a strong enrichment for those with expression during spermatogenesis (in particular, with mRNAs known to have elevated expression during spermatogenesis; Kim et al. 2001; Reinke et al. 2004). By comparing whole males either mutant or wild type for rrf-3, we have identified exemplary cellular mRNAs for which siRNA production during spermatogenesis depends on RRF-3. These cellular mRNAs show strong enrichment for previously identified genes whose mRNA levels are elevated in populations of whole animals in both rrf-3 and eri-1 mutants (Asikainen et al. 2007).
RRF-3 and paternal contributions to embryogenesis:
In principle, paternal-effect lethality associated with RRF-3 and ERI-1 mutants could result either from a lack of sperm-provided factors essential for normal embryogenesis or from pathological effects of inappropriate sperm content. We favor the latter possibility for several reasons of parsimony. First, RNAi pathways are known to function broadly in downregulation of gene expression, such that loss-of-function mutations result in overexpression of spermatogenic factors. Second, the presence of ectopic microtubule structures that we observed in mutant sperm (and later defective microtubule structures in embryos) suggests failure to eliminate at least one structure (microtubules) during budding of spermatids. Whether due to sequestration defects or ectopic microtubule polymerization after budding, a plausible interpretation is that microtubules or associated factors in mutant sperm could have a pathological effect in embryos.
Production of functionally normal sperm requires a highly specialized cellular differentiation program to generate cells that are motile, fertilization competent, and capable of contributing centrioles (which will nucleate the first mitotic spindles) and other polarity cues. Numerous genes whose expression is enriched or specific to spermatogenesis indicate that sperm production involves induction of gene expression programs (Kim et al. 2001; Reinke et al. 2004; Johnston et al. 2008). As with virtually any biological process, it would be expected that biological functions will also need to be downregulated during sperm differentiation (Lamitina and L'Hernault 2002; Muhlrad and Ward 2002; Khalil et al. 2004). In systems with interacting and potentially cooperating controls, loss of a single regulatory modality frequently leads to a conditional (rather than absolute) effect. For example, null mutations in certain regulators produce specific temperature-sensitive somatic phenotypes during larval development in C. elegans (Golden and Riddle 1984; Melendez and Greenwald 2000). Spermatogenesis in C. elegans is particularly temperature-sensitive even in wild-type worms (which become sterile if spermatogenesis is attempted at 27° (Shapira and Tan 2008). Null mutants of other C. elegans small RNA-related genes, including the Piwi-related gene prg-1, share an apparent null-mutant temperature-sensitive spermatogenesis character (Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008).
Models for negative regulation of gene expression by RRF-3:
A “simple” model for RRF-3 function during spermatogenesis would entail RRF-3 association with a set of transcripts, specific synthesis of small RNA pools from these transcripts, subsequent downregulation of new synthesis for the corresponding protein products, and a contribution of this downregulation in providing balanced protein product levels to allow for an efficient developmental process (spermatogenesis) under diverse conditions.
Despite the attractiveness of such a model, we note that our data do not establish a direct connection between RRF-3, the production of siRNAs from specific mRNA transcripts, the observed RRF-3-dependent downregulation, and spermatogenic phenotypes resulting in the mutant background. Certainly, the observation of similar sets of template mRNAs, siRNA products, mRNA modulation, and spermatogenic/embryonic phenotypes in at least one other member (ERI-1) of the ERI protein supercomplex where RRF-3 is known to reside argues for concerted participation of this complex in a defined regulatory process in sperm. We note that small RNA biogenesis and regulatory mechanisms in C. elegans and other organisms are sufficiently complex and multimodal, so that assuming that the siRNAs identified here are the immediate products of RRF-3 may be an oversimplification.
From the observed defects in sperm form and function, it was somewhat surprising to observe only a small number of genes for which siRNA production requires RRF-3. Given a requirement for the alternative RNA-directed RNA polymerase EGO-1 during spermatogenesis (Smardon et al. 2000), it seems plausible that two distinct classes of the cis-acting RNA signal (one for EGO-1 and one for RRF-3) are responsible for functional RdRP recruitment and subsequent genetic repression. Incomplete loss of RRF-3 target siRNAs in rrf-3 mutants suggests redundancy with other RdRPs or with other mechanisms that trigger siRNA production. EGO-1, RRF-3, and the other C. elegans RdRPs thus may overlap in their recognition of target RNAs due to either overlapping signal specificity of RdRP recruitment or differential timing of RdRP expression relative to developmental events. We stress that our lists of regulated genes (at the siRNA and mRNA levels) are exemplary and not complete: in addition to the genes that missed our detection due to siRNA expression that is redundantly triggered, we would have failed to detect unannotated genes, genes where siRNA expression was limited to hermaphrodites, or genes where siRNAs were at a level too low or too transient to be detected in the analysis.
Despite the incompleteness of the identified regulated gene set, it is important to note that the majority of genes showed no difference in either mRNA or siRNA expression in rrf-3 mutant animals. Thus the RRF-3 system appears somewhat exclusive in its choice of “client.” Misregulation of a small set of genes, or even of a single gene, could certainly have drastic effects on spermatogenesis. Single-gene requirements in C. elegans spermatogenesis have been extensively investigated in the literature, with hundreds of conditional and numerous nonconditional spermatogenesis mutants in several dozen genes (reviewed in L'Hernault 2009). While a single-gene process would be tempting to propose, multigenic requirements for developmental regulation have also been demonstrated in many small RNA-regulated systems; in the case of RRF-3 regulation, the existence of at least several dozen RRF-3-dependent siRNA templates suggests that the observed phenotypic syndrome could involve more than one target.
Our work, combined with other recent analyses, suggests the potential for at least four negative regulatory mechanisms that use small RNA effectors during spermatogenesis and that involve microRNAs (Ro et al. 2007; Marcon et al. 2008), piRNAs (reviewed in Klattenhoff and Theurkauf 2008), RRF-3-dependent siRNAs (Pavelec et al. 2009), and RRF-3-independent siRNAs. Each of these four mechanisms appears poised to control a specific set of RNA targets in a manner that would depend on the expression patterns for specific mRNAs and relevant populations of small RNA effectors.
RdRPs as providers of inexpensive feedback regulation:
For conventional feedback regulation (e.g., employing a transcription factor), a biological system must be devised such that a specific repressive interaction between a gene product (or a metabolite that depends on the gene product) and the corresponding RNA or protein occurs. RdRP-based mechanisms provide a much less costly mechanism to generate a self-regulated system. A key requirement for such a system is a negative regulator whose accumulation at any given time depends on the concentration of a product. An RdRP system would allow any gene to engage in feedback regulation by addition of an RdRP recruitment signal to its transcript. This would also provide the capacity to convert any “off-to-on” system into a temporal spike, since strong RdRP recruitment would result in a system in which an initial burst of gene expression could be self-silencing. Importantly for this potential contribution to gene regulation, the RdRP system could be present as a relatively ubiquitous component of cellular machinery, with any message to be autoregulated simply needing to make docking sites available for RdRP to acquire negative regulation following any burst of synthesis.
Acknowledgments
We thank Anton Gartner for anti-RAD-51, Josef Loidl for anti-REC-8, and Abby Dernburg for anti-HIM-8; Ayelet Lamm for flow cell preparation; Cheryl Smith, Ziming Weng, Phil Lacroute, Anton Valouev, and Arend Sidow for flow cell preparation, sequencing, and data processing; Weng-Onn Lui, Julia Pak, Poornima Parameswaran, and Jay Maniar for ideas on the preparation of small RNA libraries for sequencing; Virginia Walbot for comments on the manuscript; the Caenorhabditis Genetics Center for strains; and Suvi Asikainen, Gary Wong, Derek Pavelec, and Scott Kennedy for sharing data. This work was supported by National Institutes of Health (NIH) grant R01GM37706 to A.Z.F.; NIH grant R01GM53804 to A.M.V.; and a Vienna Science and Technology Fund WWTF grant LS05009 and an Austrian Science Fund FWF Elise Richter grant to V.J. J.I.G. was supported by the Stanford Department of Biological Sciences and by the Stanford Genome Training Program (NHGRI-T32-HG00044), and M.S. was supported by a Canadian Institutes of Health Research postdoctoral fellowship.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.109686/DC1.
References
- Alpi, A., P. Pasierbek, A. Gartner and J. Loidl, 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112 6–16. [DOI] [PubMed] [Google Scholar]
- Ambros, V., R. C. Lee, A. Lavanway, P. T. Williams and D. Jewell, 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13 807–818. [DOI] [PubMed] [Google Scholar]
- Asikainen, S., M. Storvik, M. Lakso and G. Wong, 2007. Whole genome microarray analysis of C. elegans rrf-3 and eri-1 mutants. FEBS Lett. 581 5050–5054. [DOI] [PubMed] [Google Scholar]
- Babiarz, J. E., J. G. Ruby, Y. Wang, D. P. Bartel and R. Blelloch, 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M. K., T. B. Schedl and J. Kimble, 1987. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista, P. J., J. G. Ruby, J. M. Claycomb, R. Chiang, N. Fahlgren et al., 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., A. Pane and T. Schupbach, 2007. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 17 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399 166–169. [DOI] [PubMed] [Google Scholar]
- Colaiacovo, M. P., A. J. MacQueen, E. Martinez-Perez, K. McDonald, A. Adamo et al., 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5 463–474. [DOI] [PubMed] [Google Scholar]
- Cox, D. N., A. Chao, J. Baker, L. Chang, D. Qiao et al., 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, P. P., M. P. Bagijn, L. D. Goldstein, J. R. Woolford, N. J. Lehrbach et al., 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94 387–398. [DOI] [PubMed] [Google Scholar]
- Duchaine, T. F., J. A. Wohlschlegel, S. Kennedy, Y. Bei, D. Conte, Jr. et al., 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 343–354. [DOI] [PubMed] [Google Scholar]
- Edgley, M. L., and D. L. Riddle, 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266 385–395. [DOI] [PubMed] [Google Scholar]
- Flynt, A. S., and E. C. Lai, 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal, M., H. Seitz, M. D. Horwich, C. Li, T. Du et al., 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1984. A pheromone-induced developmental switch in Caenorhabditis elegans: temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc. Natl. Acad. Sci. USA 81 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., S. Pichler, M. Kirkham and A. A. Hyman, 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and M. J. Simard, 2008. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9 22–32. [DOI] [PubMed] [Google Scholar]
- Johnston, D. S., W. W. Wright, P. Dicandeloro, E. Wilson, G. S. Kopf et al., 2008. Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc. Natl. Acad. Sci. USA 105 8315–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kennedy, S., D. Wang and G. Ruvkun, 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427 645–649. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, A. M., F. Z. Boyar and D. J. Driscoll, 2004. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc. Natl. Acad. Sci. USA 101 16583–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. K., J. Lund, M. Kiraly, K. Duke, M. Jiang et al., 2001. A gene expression map for Caenorhabditis elegans. Science 293 2087–2092. [DOI] [PubMed] [Google Scholar]
- Klass, M. R., and D. Hirsh, 1981. Sperm isolation and biochemical analysis of major sperm protein from Caenorhabditis elegans. Dev. Biol. 84 299–312. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C., and W. Theurkauf, 2008. Biogenesis and germline functions of piRNAs. Development 135 3–9. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C., D. P. Bratu, N. McGinnis-Schultz, B. S. Koppetsch, H. A. Cook et al., 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12 45–55. [DOI] [PubMed] [Google Scholar]
- Knight, S. W., and B. L. Bass, 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamitina, S. T., and S. W. L'Hernault, 2002. Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development 129 5009–5018. [DOI] [PubMed] [Google Scholar]
- LaMunyon, C. W., and S. Ward, 1994. Assessing the viability of mutant and manipulated sperm by artificial insemination of Caenorhabditis elegans. Genetics 138 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon, C. W., and S. Ward, 1995. Sperm precedence in a hermaphroditic nematode (Caenorhabditis elegans) is due to competitive superiority of male sperm. Experientia 51(8): 817–823. [DOI] [PubMed] [Google Scholar]
- Lau, N. C., L. P. Lim, E. G. Weinstein and D. P. Bartel, 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem et al., 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308 557–560. [DOI] [PubMed] [Google Scholar]
- Lee, R. C., C. M. Hammell and V. Ambros, 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault, S. W., 2009. The genetics and cell biology of spermatogenesis in the nematode C. elegans. Mol. Cell. Endocrinol. 306 59–65. [DOI] [PubMed] [Google Scholar]
- L'Hernault, S. W., and T. M. Roberts, 1995. Cell biology of nematode sperm. Methods Cell Biol. 48 273–301. [DOI] [PubMed] [Google Scholar]
- Machaca, K., L. J. DeFelice and S. W. L'Hernault, 1996. A novel chloride channel localizes to C. elegans spermatids and chloride channel blockers induce spermatid differentiation. Dev. Biol. 176 1–16. [DOI] [PubMed] [Google Scholar]
- MacQueen, A. J., M. P. Colaiacovo, K. McDonald and A. M. Villeneuve, 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl, J. E., and R. K. Herman, 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine, E. M., J. Hauth, T. Ratliff, V. E. Vought, X. She et al., 2005. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr. Biol. 15 1972–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon, E., T. Babak, G. Chua, T. Hughes and P. B. Moens, 2008. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 16 243–260. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez, E., and A. M. Villeneuve, 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez, E., M. Schvarzstein, C. Barroso, J. Lightfoot, A. F. Dernburg et al., 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez, A., and I. Greenwald, 2000. Caenorhabditis elegans lin-13, a member of the LIN-35 Rb class of genes involved in vulval development, encodes a protein with zinc fingers and an LXCXE motif. Genetics 155 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. A., 2006. Sperm and oocyte isolation methods for biochemical and proteomic analysis. Methods Mol. Biol. 351 193–201. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon et al., 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Muhlrad, P. J., and S. Ward, 2002. Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics 161 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigon, V., 1951. Polyploïdie expérimentale chez un nematode libre. Bull. Biol. Fr. Belg. 85 187–225. [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham and A. A. Hyman, 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema, P. G., and J. Kimble, 1991. Molecular analysis of tra-2, a sex determining gene in C. elegans. EMBO J. 10 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, J., and A. Fire, 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241–244. [DOI] [PubMed] [Google Scholar]
- Pane, A., K. Wehr and T. Schupbach, 2007. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer et al., 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec, Derek M., J. Lachowiec, T. F. Duchaine, H. E. Smith and Scott Kennedy, 2009. Requirement for ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C. M., C. Wong, N. Bhalla, P. M. Carlton, P. Weiser et al., 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Ro, S., C. Park, K. M. Sanders, J. R. McCarrey and W. Yan, 2007. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 311 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, T. M., F. M. Pavalko and S. Ward, 1986. Membrane and cytoplasmic proteins are transported in the same organelle complex during nematode spermatogenesis. J. Cell. 102 1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, J. G., C. Jan, C. Player, M. J. Axtell, W. Lee et al., 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127 1193–1207. [DOI] [PubMed] [Google Scholar]
- Schedl, T., and J. Kimble, 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel, H., and R. Schnabel, 1990. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science 250 686–688. [DOI] [PubMed] [Google Scholar]
- Shapira, M., and M. W. Tan, 2008. Genetic analysis of Caenorhabditis elegans innate immunity. Methods Mol. Biol. 415 429–442. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K., and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish et al., 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476. [DOI] [PubMed] [Google Scholar]
- Sijen, T., F. A. Steiner, K. L. Thijssen and R. H. Plasterk, 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244–247. [DOI] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet et al., 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12 1317–1319. [DOI] [PubMed] [Google Scholar]
- Slotkin, R. K., M. Vaughn, F. Borges, M. Tanurdzic, J. D. Becker et al., 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin et al., 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10 169–178. [DOI] [PubMed] [Google Scholar]
- Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok et al., 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132. [DOI] [PubMed] [Google Scholar]
- Tops, B. B., H. Tabara, T. Sijen, F. Simmer, C. C. Mello et al., 2005. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., and V. Reinke, 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S., 1986. The Asymmetric Localization of Gene Products During the Development of Caenorhabditis elegans Spermatozoa. A. R. Liss, New York.
- Ward, S., T. M. Roberts, S. Strome, F. M. Pavalko and E. Hogan, 1986. Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J. Cell Biol. 102 1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit, E., P. J. Batista, Y. Bei, K. M. Pang, C. C. Chen et al., 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747–757. [DOI] [PubMed] [Google Scholar]