Abstract
Enhanced stress response has been suggested to promote longevity in many species. Calorie restriction (CR) and conserved nutrient-sensing target of rapamycin (TOR) and protein kinase A (PKA) pathways have also been suggested to extend life span by increasing stress response, which protects cells from age-dependent accumulation of oxidative damages. Here we show that deleting the yeast 14-3-3 protein, Bmh1, extends chronological life span (CLS) by activating the stress response. 14-3-3 proteins are highly conserved chaperone-like proteins that play important roles in many cellular processes. bmh1Δ-induced heat resistance and CLS extension require the general stress-response transcription factors Msn2, Msn4, and Rim15. The bmh1Δ mutant also displays a decreased reactive oxygen species level and increased heat-shock-element-driven transcription activity. We also show that BMH1 genetically interacts with CR and conserved nutrient-sensing TOR- and PKA-signaling pathways to regulate life span. Interestingly, the level of phosphorylated Ser238 on Bmh1 increases during chronological aging, which is delayed by CR or by reduced TOR activities. In addition, we demonstrate that PKA can directly phosphorylate Ser238 on Bmh1. The status of Bmh1 phosphorylation is therefore likely to play important roles in life-span regulation. Together, our studies suggest that phosphorylated Bmh1 may cause inhibitory effects on downstream longevity factors, including stress-response proteins. Deleting Bmh1 may eliminate the inhibitory effects of Bmh1 on these longevity factors and therefore extends life span.
RECENT studies in genetically tractable model systems including yeasts, worms, flies, and mice demonstrated that longevity could be modulated by single-gene mutations (Kenyon 2001; Tissenbaum and Guarente 2002; Dilova et al. 2007). In addition to genetic interventions, calorie restriction (CR) has also been shown to extend life span in many species (Weindruch and Walford 1998; Roth et al. 2001) and to ameliorate age-related diseases such as cancer and diabetes (Weindruch and Walford 1998). Identification and study of novel longevity genes may therefore provide insights into the molecular mechanisms underlying CR, longevity regulation, and age-associated diseases.
The budding yeast Saccharomyces cerevisiae is an efficient model for studying longevity regulation. Yeast life span has been studied in two distinct ways: replicative life span (RLS) and chronological life span (CLS). RLS measures the number of cell divisions that an individual yeast cell undergoes before senescence. CLS measures the length of time that cells remain viable at a nondividing state. Yeast cells enter a nondividing stationary phase when nutrients are exhausted. This quiescent state has been suggested to resemble the G0 state in higher eukaryotes (Werner-Washburne et al. 1993). Moderate CR can be imposed in yeast by reducing the glucose concentration from 2% to 0.5% in rich media (Lin et al. 2000; Easlon et al. 2007; Smith et al. 2007; Wei et al. 2008), which extends both CLS and RLS. CR is suggested to function through reducing the activities of conserved nutrient-sensing pathways to extend life span. Decreasing the activity of the Ras-cyclic AMP-activated protein kinase A (Ras-cAMP/PKA) pathway, which regulates cell growth and stress response, extends life span (Lin et al. 2000; Fabrizio et al. 2001). Deleting the nutrient-responsive kinases Sch9 (a homolog of mammalian S6K kinases) and Tor1 also promotes longevity (Fabrizio et al. 2001; Kaeberlein et al. 2005). The conserved Sir2 family proteins have been shown to play important roles in moderate CR-induced RLS extension (Lin et al. 2000; Lamming et al. 2005; Easlon et al. 2007). Interestingly, Sir2 appeared to be dispensable for CLS (Fabrizio et al. 2005; Smith et al. 2007) and for RLS (Jiang et al. 2002; Kaeberlein et al. 2004; Easlon et al. 2007) in certain CR models, which further underscored the complexity of CR and longevity regulation. Identification of novel longevity factors is therefore essential to help elucidate the underlying mechanisms.
An enhanced stress response has been shown to play important roles in extending longevity. An increased stress response may protect cells from age-dependent accumulation of damages caused by reactive oxygen species (ROS) generated as metabolic by-products. In yeast, downregulation of the nutrient-sensing target of rapamycin (TOR)- and PKA-signaling activities confers resistance to various stresses such as heat shock and oxidative stress (Fabrizio et al. 2001; Kaeberlein et al. 2005). Stress-response transcription factors including Msn2, Msn4, Rim15, and Gis1 have been reported to mediate life-span extension and stress resistance in these nutrient-sensing mutants (Wei et al. 2008). In worms, the stress-response transcription factors DAF-16 and HSF-1 have also been shown to be required for increased life span and thermo-tolerance in the daf-2 mutants (Lin et al. 1997; Hsu et al. 2003). In mammals, fibroblasts derived from long-lived Snell dwarf mice were more resistant to different forms of stresses than cells from normal mice (Salmon et al. 2005). Furthermore, mice carrying deletions in the P66shc redox-sensing protein were resistant to stress-induced cell death and exhibited longer life span (Migliaccio et al. 1999). These studies demonstrate a correlation between increased life span and enhanced stress resistance.
In this study, we employed an accelerated cell-death-based genetic screen to identify novel longevity genes. Our screen identified the yeast 14-3-3 proteins, which are evolutionarily conserved dimeric acidic proteins involved in cell signaling, cell cycle control, apoptosis, and transcription (Aitken 2006; van Heusden and Steensma 2006). Yeast 14-3-3 proteins are encoded by two highly homologous genes, BMH1 and BMH2, and deleting both genes is lethal in most strains (van Heusden and Steensma 2006). 14-3-3 proteins function as molecular chaperones and protein tethers, which have >200 interacting partners (Kakiuchi et al. 2007). Most binding partners of 14-3-3 proteins contain either of the consensus sequence motifs RSXpS/TXP and RXXXpS/TXP (pS/T: phospho-Ser or phospho-Thr; X: any amino acid). Interactions of 14-3-3 proteins and their binding partners are regulated through protein phosphorylation (Aitken 2006; van Heusden and Steensma 2006). Genomewide gene/protein expression studies of yeast 14-3-3 protein (Bmh) mutants revealed that Bmh proteins affect the expression of many genes/proteins associated with carbon and nitrogen metabolism (Ichimura et al. 2004; Bruckmann et al. 2007). 14-3-3 proteins have also been shown to interact with the TOR and PKA pathways; however, it remains unclear how TOR and PKA regulate 14-3-3 proteins. In this study, we characterized the roles of yeast 14-3-3 protein Bmh1 in stress response and in CR-, TOR-, and PKA-mediated longevity pathways.
MATERIALS AND METHODS
Yeast strains and media:
The yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 and the isogenic gene deletion collections were acquired from Open Biosystems (Brachmann et al. 1998). W303AR MATa ura3-1, leu2-3, 112 his3-11,15 trp1-1 ade2-1 RDN1∷ADE2 can1-100 were described previously (Kaeberlein et al. 1999; Lin et al. 2000). Rich media YPD and synthetic SD media were made as described (Burke et al. 2000). Media used for CLS analysis were minimal synthetic SD supplemented with 4× auxotrophic amino acids (leucine, histine, lysine, and uracil) and glucose to a final concentration of 2% or 0.5%. All gene deletions were generated and verified as described (Guldener et al. 1996). Strains used in this study are listed in Table 1. Strains overexpressing Bmh1 or Bmh2 were made by integrating the pADH1-BMH1 or pADH1-BMH2 plasmid into the genome.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303AR | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Lin et al. (2000) |
| RDN∷ADE2 can1-100 | ||
| LSC1041 | W303AR bmh1Δ∷Kanr | This study |
| LSC14 | W303AR cdc25-10 | Lin et al. (2000) |
| BY4742 | MATahis3Δ1 leu2Δ0 lysΔ0 ura3Δ0 | Easlon et al. (2007) |
| LSC722 | BY4742 bmh1Δ∷Kanr | This study |
| LSC716 | BY4742 bmh2Δ∷Kanr | This study |
| LSC622 | BY4742 pADH1-BMH1-LEU2 | This study |
| LSC623 | BY4742 pADH1-BMH2-LEU2 | This study |
| LSC89 | BY4742 pADH1-XXX-LEU2 (pPP81, vector control) | Easlon et al. (2007) |
| LSC586 | BY4742 tor1Δ∷Kanr | This study |
| LSC157 | BY4742 cdc25-10 | Easlon et al. (2007) |
| LSC837 | BY4742 cdc25-10 bmh1Δ∷Kanr | This study |
| LSC801 | BY4742 tor1Δ bmh1Δ∷Kanr | This study |
| LSC1021 | BY4742 bmh1Δ∷Kanr pADH1-Bmh1-S189A-LEU2 | This study |
| LSC1020 | BY4742 bmh1Δ∷Kanr pADH1-Bmh1-S238A-LEU2 | This study |
| LSC327 | BY4742 msn2Δmsn4Δ∷Kanr | This study |
| LSC971 | BY4742 msn2Δmsn4Δbmh1Δ∷Kanr | This study |
| LSC516 | BY4742 rtg3Δ∷Kanr | This study |
| LSC997 | BY4742 bmh1Δrtg3Δ∷Kanr | This study |
| LSC1246 | BY4742 gnc4Δ∷Kanr | This study |
| LSC1245 | BY4742 bmh1Δgcn4Δ∷Kanr | This study |
| LSC1244 | BY4742 tor1Δgcn4Δ∷Kanr | This study |
| LSC1218 | BY4742 rim15Δ∷Kanr | This study |
| LSC1219 |
BY4742 bmh1Δrim15Δ∷Kanr |
This study |
Genetic screen conditions:
About 20,000 colonies, which represented about five copies of the yeast genome, were screened. Colonies carrying the 2μ genomic library were replica plated onto two sets of YPD plates. One set (assay plates) was upshifted to nonpermissive temperature (38°) and the other set (master plates) was incubated at 25°. After 4 days at 38°, the assay plates were shifted to permissive temperature (25°) for an additional 2 days. Cell patches that grew on the assay plates at 38° were excluded because they were likely to carry cdc25-10-specific suppressors that simply rescued the growth defects of the cdc25-10 mutant at 38°. Cell patches on the assay plates that did not grow at 38° but grew after shifting to 25° were likely to carry genes extending survival. Four cell patches were identified under these conditions. Plasmid DNA conferring the strongest survival was recovered from the corresponding master plate and retransformed into the cdc25-10 mutant to confirm the phenotype. Sequencing analysis of this DNA fragment indicated that it contained both the NDJ1 and the BMH2 genes. Each gene was cloned into an integrative vector driven by the ADH1 promoter pPP81. Only BMH2 overexpression extended survival.
Plasmid constructions:
Bmh overexpression constructs pADH1-BMH1 and pADH1-BMH2 were made as follows: specific oligonucleotides were designed (with a Not1 site added to the 5′-end and a NheI site to the 3′-end) to amplify the BMH1 (or BMH2) coding region via PCR using Pfu polymerase. Amplified DNA was digested with NotI and NheI and then ligated to pPP81 (an integrative vector carrying a LEU2 auxotrophic marker and an ADH1 promoter).
Chronological life span:
Four colonies from each strain were analyzed in each experiment as described (Fabrizio and Longo 2003) with some modifications. Cells were grown in 10 ml SD (at a starting OD600 of 0.1) in 50-ml tubes on a roller drum set at the maximum speed to ensure proper aeration. Cells were continuously grown in SD or shifted to sterile water after entering stationary phase (typically after 48 hr). Cells shifted to water showed more significant CLS extension compared to cells maintained in SD (Fabrizio et al. 2004) (C. Wang and S.-J. Lin, unpublished results). However, a regrowth phenomenon was often observed: after ∼90–99% of cells died, the number of viable cells increased. This regrowth phenomenon has also been reported in several other strains (Fabrizio et al. 2004) and was likely due to nutrients released by dead cells and to increased adaptive mutations during prolonged culture (Fabrizio et al. 2004). We found that the regrowth problem could be alleviated by transferring 10-fold fewer cells to water at a final density of OD600 ∼1 (107 cells/ml; ∼108 cells were examined for each CLS assay in water). Cell viability was monitored every other day by plating a fraction of culture onto fresh YPD to determine the colony-forming units (CFU). The rate of cell survival was calculated by normalizing each CFU to the CFU obtained 48 hr after starting CLS in SD (when cells just entered the stationary phase).
Heat shock and oxidative stress resistance:
Cells were first grown in SD containing 2% glucose (normal) or 0.5% glucose (CR) for 2 days with a starting OD600 of 0.1 prior to analysis. For heat-shock studies, cells were spotted onto YPD plates (2% glucose) in fivefold serial dilutions (started at OD600 of 1) and then were incubated at 55° or 25° for 45 min or 60 min. After heat shock, plates were transferred to 30° and continued to incubate for 2–3 days. For the hydrogen peroxide toxicity test, cells were spotted onto YPD plates (with 0 or 3 mm H2O2) in fivefold serial dilutions and were allowed to grow for 2 days. For the paraquat toxicity test, cell growth was monitored in SD containing indicated concentrations of paraquat with a starting OD600 of 0.05 after incubation at 30° for 16 hr.
ROS detection:
Cells grown in minimal SD to stationary phase were washed twice with PBS buffer (pH 7.4) and then resuspended in PBS with 10 μm H2DCFDA following incubation at 4° in the dark for 45 min (Zuin et al. 2008). Next, cells were washed once with PBS and then added to 96-well fluorescence assay plates (∼5 × 106 cells/well). Fluorescence signals were detected using a plate reader with excitation at 485 nm, and emission was monitored at 535 nm.
Superoxide dismutase gel assay:
Cells grown in minimal SD were harvested at indicated time points. Total protein extract was obtained by agitations using glass beads and the FastPrep beads beater. About 15 μg protein was loaded in each lane of a 12% native polyacrylamide gel. After electrophoresis, gel was stained in a buffer with 0.025% Nitro Blue Tetrazolium, 0.01% riboflavin, 0.01% N,N,N′,N′-tetramethylethylenediamine for 45 min in the dark at room temperature (Raychaudhuri et al. 2003). Stained gel was then exposed to intensive light. The superoxide dismutase (SOD)-active bands appeared white in a dark-blue background on the gel. Results shown in Figure 2G are inverted images of the original gels.
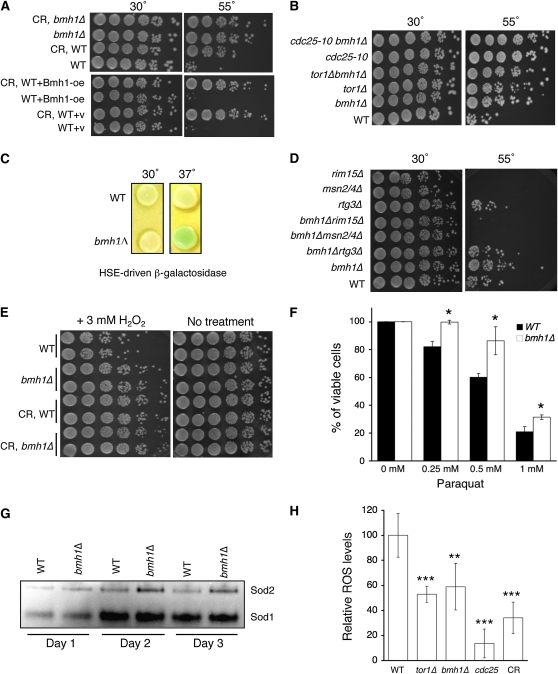
Figure 2.—
The bmh1Δ mutant shows increased stress response and decreased ROS levels. (A) bmh1Δ increases heat resistance, which is not enhanced by CR (top). Bmh1-oe does not increase heat resistance (bottom). Results show fivefold serial dilutions of cells grown on YPD with or without heat shock. (B) bmh1Δ does not further increase heat resistance in the tor1Δ or cdc25-10 mutants. (C) The bmh1Δ mutant shows increased HSE-driven transcription activity. Results show wild-type and bmh1Δ mutant cells (carrying an HSE-driven β-galactosidase reporter plasmid) grown on YPD containing 4 mg X-gal. The light green color shown in the bmh1Δ mutant grown at 37° indicates higher HSE-driven transcription activity. (D) bmh1Δ-induced heat resistance requires Msn2/4 and Rim15. (E) bmh1Δ confers resistance to H2O2-induced toxicity. (F) bmh1Δ confers resistance to paraquat-induced toxicity. (G) The bmh1Δ mutant has higher Sod2 activity in stationary phase using a SOD gel activity assay. (H) bmh1Δ decreases intracellular ROS levels. P-values are calculated using the Student's t-test (**P < 0.01; ***P < 0.005). WT, wild-type control; CR, cells pregrown in SD with 0.5% glucose prior to analysis.
Epitope tagging:
Bmh1 was tagged by the HA epitope tag in the genome using the pFA6a-3HA-KanMX6 plasmid as described (Longtine et al. 1998). Yeast strains expressing Myc-tagged Bmh1 proteins were made by introducing the pBMH1-13MycXX plasmid (Easlon et al. 2008).
Site-directed mutagenesis:
The Bmh1-S189A and Bmh1-S238A mutants were generated using the QuickChange kit (Stratagene) by PCR. The pADH1-Bmh1 plasmid was used as a template in a 50-μl reaction with 4 μl 2.5 mm dNTP, 5 μl 10× Pfu buffer, 1 μl pADH1-Bmh1 plasmid DNA, 2 μl Pfu Turbo polymerase, and 1 μl of each pair of oligos: S189A-f (5′-GAAATTCAAAACGCTCCAGAC-3′) + S189A-r (5′-GTCTGGAGCGTTTTGAATTTC-3′) or S238A-f (5′-CAGACATGGCCGAGTCCGGTC-3′ + S238A-r (5′-GACCGGACTCGGCCATGTCTG-3′). The PCR products were digested with DpnI and then introduced into XL1-Blue competent cells by electroporation. Plasmids with desired point mutations were verified by sequencing.
Antibody production:
Antibodies to total Bmh1 proteins (anti-Bmh1-total) and phosphorylated Bmh1 proteins (anti-Bmh1-pS238) were generated in rabbits using the keyhole-limpet-hemocyanin-conjugated phosphopeptides SVFYYEIQN(p)SPDKAC (flanking Ser189) or TLWTSDM(p)SESGQAEDQ (flanking Ser238) from Antagene. Antibodies were purified from the resulting antiserum by column chromatography on phosphopeptide-conjugated affinity resin.
Protein extraction and Western blot analysis:
Total protein extract was obtained as described (Easlon et al. 2008). About 15 μg of total proteins were loaded in each lane. After electrophoresis, proteins were transferred to nitrocellulose membranes (Whatman), which were then washed and blotted with anti-Bmh1-total, anti-Bmh1-pS238, anti-actin (Abcam), or anti-Myc antiserums (Covance). Proteins were visualized using anti-mouse or anti-rabbit antiserum conjugate to the horseradish peroxidase (Amersham) and the ECL reagents (Pierce). Chemiluminescent images were analyzed using the Alpha Innotech imaging system.
Calf intestinal alkaline phosphatase treatment:
A total of 250 ml cells (∼5 × 109) grown in SD to mid-log phase were harvested and resuspened in 500 μl breaking buffer: 50 mm Tris–HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 1 mm PMSF, and protease inhibitors (Roche). Cell suspensions were lysed by agitations using glass beads and the FastPrep beads beater. Monocolonal anti-Myc antibody (7 μl; Covance) was used to immunoprecipitate Myc epitope-tagged Bmh1 in 400 μl cell extract. For calf intestinal alkaline phosphatase (CIP) treatments (Ai et al. 2002), the immunocomplex was spun down, washed, and then resuspended in 200 μl CIP buffer (100 mm NaCl, 50 mm Tris–HCl, 10 mm MgCl2, 1 mm DTT, pH 7.9). A total of 100 μl of this suspension was incubated with CIP (20 units, Roche) for 20 min at 37°. About 30 μl of the resultant protein suspension was used in Western blot analysis.
Cloning and purification of recombinant Tpk1:
The coding region of TPK1 was cloned into the 6xHis-tag-containing pET28b expression vector using engineered BamHI and XhoI sites. This plasmid was then electroporated into BL21(DE3) cells using kanamycin selection. These cells were grown to OD600 ∼1 in a total volume of 100 ml and induced for 2 hr with 0.4 mm IPTG. Following induction, cells were collected, and recombinant Tpk1-6xHis was purified using the His Bind purification kit (Novagen). Purified Tpk1 was concentrated by the 5000 NMWL filter unit (Millipore).
Kinase assay:
Cells (15 ml) were harvested at OD600 ∼10 and resuspended in 500 μl breaking buffer: 50 mm Tris–HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, and 1 mm PMSF. Cell suspension was lysed by agitations using glass beads and the FastPrep beads beater. Monocolonal anti-HA antibody (5 μl; Covance) was used to immunoprecipitate HA epitope-tagged Bmh1 in 400 μl cell extract. After CIP treatment, beads were washed with buffer (50 mm Tris–HCL, pH 7.5, 100 mm NaCl, and 0.2% Triton X-100) to remove the CIP in the reaction and then resuspended in kinase buffer (20 mm Tris–HCl, pH 7.5, 20 mm β-glycerophosphate, 100 μm orthovanadate, 10 mm MgCl2, 1 mm DTT, 50 μm ATP, and 1 mm NaF). Thirty microliters of recombinant Tpk1 (0.1 μg/μl) was added to initiate the kinase reaction at 30° for 30 min.
Heat-shock-element reporter assay:
To monitor Hsf1-mediated transcription activity, we transformed yeast cells with the HSE4Ptt-CYC1-LacZ reporter plasmid (Bandhakavi et al. 2008). Cells carrying the plasmid were first grown in SD without URA at 30° with a starting OD600 of 0.1. After 2 days, cells were spotted onto YPD plates containing 4 mg X-gal. After incubation at 37° for 4 days, plates were transferred to room temperature for 2 days.
RESULTS
Deleting Bmh1 extends chronological life span:
To further understand the mechanisms of longevity regulation, we screened for factors that could extend the survival of cells at a nondividing state (CLS). Since yeast cells could survive up to several months in stationary phase, we utilized a cdc25-10 temperature-sensitive mutant to accelerate the screening process: an accelerated cell death system. CDC25 encodes a GTP–GDP exchange factor that activates Ras in the cAMP/PKA pathway in response to glucose. When arrested at 38°, the cdc25-10 mutant exhibited phenotypes similar to that of stationary phase cells (Gray et al. 2004) and survived only ∼3 days. Therefore, factors that extended the survival of the cdc25-10 mutant at 38° might also extend survival of wild type cells in stationary phase (CLS). We first introduced a 2μ-based yeast overexpression library into the cdc25-10 mutant to obtain genes that could extend survival (see materials and methods). As shown in Figure 1A, a genomic DNA fragment containing the BMH2 gene (clone 1) was identified, and its overexpression extended the survival of the cdc25-10 mutant cells at 38°.
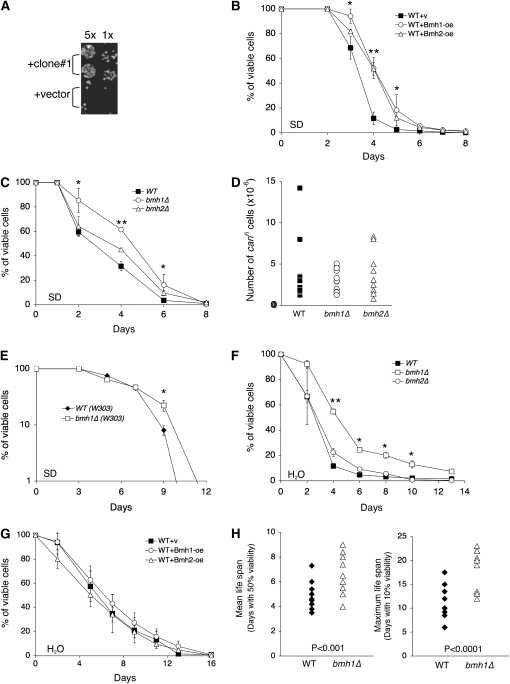
Figure 1.—
Characterization of yeast 14-3-3 proteins as novel longevity factors that regulate CLS. (A) Overexpression of clone 1 (containing BMH2) extends survival in an accelerated cell death assay. Temperature-sensitive cdc25-10 mutants carrying a control 2μ vector or clone 1 are first incubated at 38° for 4 days and then are incubated at 30° for 2 days. About 5 × 104 cells are spotted in the first column (“5×”). (B) Bmh1 and Bmh2 overexpressions extend the CLS of cells grown and kept in SD. (C) bmh1Δ extends the CLS of cells grown and kept in SD. (D) The bmh1Δ and bmh2Δ mutants do not show increased mutation rates in stationary phase. The mutation rates of wild-type and bmh1Δ and bmh2Δ mutant cells are determined by measuring the mutation frequency of the CAN1 gene. Stationary-phase cells (day 4 in SD) are collected and then plated onto both YPD and SD −ARG (without arginine, and containing 60 mg/liter l-canavanine sulfate). Mutation frequency is calculated by normalizing the number of colonies that appeared on SD −ARG to that of the corresponding YPD. For each strain, 12 independent colonies were examined. (E) bmh1Δ extends maximum the CLS in a different strain, W303. (F) bmh1Δ extends the CLS of cells shifted to water. Cells are first grown in SD to stationary phase and then are shifted to sterile water. (G) Bmh1 and Bmh2 overexpressions do not extend the CLS of cells shifted to water. (H) bmh1Δ extends both mean and maximum CLS. Results show statistics of 11 independent experiments, each of which was conducted in triplicate or quadruplicate. Each symbol represents the average viability of four samples, each containing 107 cells. For CLS analysis, one representative set of three independent experiments, each conducted in quadruplicate, is shown. “Days” denotes the number of days that cells were in SD or H2O. Error bars denote standard deviations. P-values were calculated using the Student's t-test (*P < 0.05; **P < 0.01). WT, wild-type control; v, vector control; oe, overexpression.
We then examined if both yeast 14-3-3 proteins indeed played important roles in longevity regulation. Wild-type (BY4742) cells overexpressing Bmh1 and Bmh2 (Bmh1-oe and Bmh2-oe) were subject to standard CLS analysis in minimal synthetic SD. As shown in Figure 1B, both Bmh1 and Bmh2 overexpressions significantly extended CLS. We then examined whether deleting BMH1 or BMH2 would also affect CLS. Surprisingly, the bmh1Δ mutant showed extended survival compared to wild-type cells (Figure 1C). This phenotype was not due to adaptive mutagenesis since the frequency of canavanine-resistant mutations did not increase significantly in the bmh mutants (Figure 1D). Deleting BMH1 also extended the maximum CLS in a different strain, W303 (Figure 1E), indicating that the observed bmh1Δ life-span phenotype was not specific to the BY4742 strain. To further understand the role of Bmh in CLS, we examined the effects of Bmh on CLS under more stringent and stressful growth conditions by shifting cells to water after they had entered stationary phase. Shifting cells to water has been suggested to mimic the adverse growth conditions that yeast cells frequently encounter in a natural environment (Fabrizio and Longo 2003; Gray et al. 2004). Interestingly, under this condition, bmh1Δ still extended CLS (Figure 1F) whereas Bmh1-oe and Bmh2-oe failed to extend CLS (Figure 1G). Since bmh1Δ extends CLS regardless of growth conditions (in both SD and H2O), it is more likely to function in conserved longevity pathways. Moreover, although bmh1Δ did not dramatically extend CLS, distribution of the mean and maximum life span from multiple experiments demonstrated that bmh1Δ indeed extended CLS consistently and significantly (Figure 1H).
bmh1Δ extends chronological life span by increasing stress response:
Many studies have associated increased CLS to activation of the stress response (Fabrizio and Longo 2003; Powers et al. 2006). As shown in Figure 2A (top), the bmh1Δ mutant exhibited increased resistance to heat stress at a level similar to that observed in CR cells and in the long-lived tor1Δ and cdc25-10 mutants (Figure 2B). Interestingly, Bmh1-oe did not show increased resistance (Figure 2A, bottom), which was consistent with the finding that Bmh1-oe failed to extend CLS under more stressful growth conditions (Figure 1G). These results suggested that bmh1Δ might extend CLS by activating the stress response.
The heat-shock factor HSF-1 has been shown to be required for starvation-induced life-span extension as well as for responding to heat shock and oxidative stress in worms (Hsu et al. 2003). Since the bmh1Δ mutant showed increased heat resistance, we examined whether Hsf1-mediated transcription activity was increased in the bmh1Δ mutant using a reporter assay (Bandhakavi et al. 2008). Hsf1 recognizes heat shock elements (HSEs) in promoters of target genes such as molecular chaperons and heat-shock proteins. As shown in Figure 2C, the bmh1Δ mutant displayed higher HSE-driven transcription activity at the LacZ (β-galactosidase) reporter gene. Yeast Bmh proteins have also been suggested to sequester the stress-sensing transcription factors Rim15 and Msn2/4 (van Heusden and Steensma 2006) and to inhibit the activity of the retrograde response that senses mitochondrial genome integrity (van Heusden and Steensma 2006). We therefore examined whether bmh1Δ required these stress-response transcription factors to confer heat resistance. As shown in Figure 2D, deleting Msn2/4 and Rim15 abolished bmh1Δ-induced resistance to heat stress. Furthermore, deleting Msn2/4 also abolished bmh1Δ-induced CLS extension (supporting information, Figure S1), suggesting that these stress-response factors played important roles in bmh1Δ-induced heat resistance and CLS extension.
Intracellular homeostasis of ROS has also been shown to affect the expectancy of life span (Reverter-Branchat et al. 2004). Increased intracellular ROS levels cause damages to macromolecules such as DNA, proteins, and lipids. To further understand the roles of Bmh1 in ROS homeostasis and life-span regulation, we first examined whether the bmh1Δ mutant was more resistant to oxidative stress by challenging cells with hydrogen peroxide (H2O2) and paraquat (generating superoxide anions). As shown in Figure 2E, the bmh1Δ mutant showed an ∼10-fold increased resistance to H2O2 compared to wild type. In addition, bmh1Δ also conferred resistance to paraquat-induced toxicity (Figure 2F). These results demonstrate that deleting Bmh1 protects cells from oxidative stress. We next compared the activities of SODs in cell extracts of the bmh1Δ mutant and wild-type cells using a SOD gel assay (Raychaudhuri et al. 2003). Both bmh1Δ and wild-type cells showed higher Sod1 activities (cytosolic Cu/Zn-SOD) after entering the stationary phase (days 2 and 3) (Figure 2G). Interestingly, wild-type cells failed to show increased Sod2 activities (mitochondrial Mn-SOD) whereas the bmh1Δ mutant showed slightly higher Sod2 activities, suggesting that Sod2 might play an important role in bmh1Δ-induced stress response. Finally, we determined the level of ROS in the bmh1Δ mutant using a ROS-specific fluorescence dye (Zuin et al. 2008). Similar to CR-treated cells and to the tor1Δ and cdc25-10 mutants, the bmh1Δ mutant showed a lower ROS level compared to wild type (Figure 2H). These data suggest that bmh1Δ activates the stress response, leading to increased stress resistance, decreased ROS level, and CLS extension.
BMH1 genetically interacts with CR, PKA, and TOR longevity pathways to regulate life span:
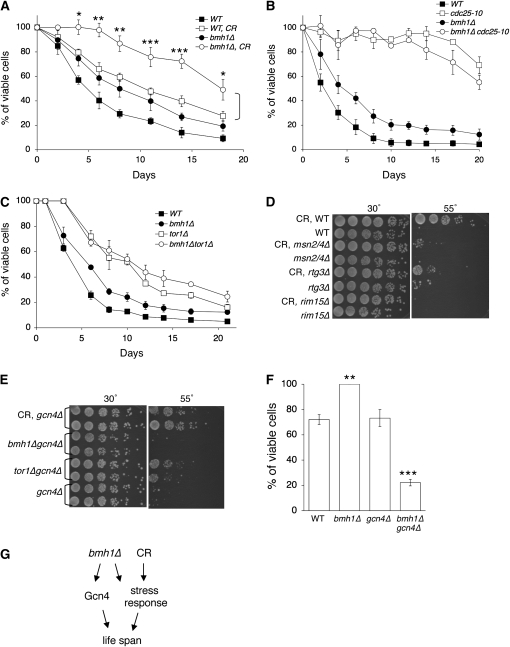
To further understand the roles of Bmh1 in longevity regulation, we examined the effects of bmh1Δ on CR, tor1Δ- and cdc25-10-induced CLS extension to determine whether bmh1Δ functioned in these pathways. Figure 3A showed that bmh1Δ further extended CR-induced CLS, suggesting that bmh1Δ and CR functioned in parallel or in partially overlapping pathways to extend CLS. Figure 3, B and C, showed that bmh1Δ did not significantly further extend CLS of the tor1Δ and cdc25-10 mutants, suggesting that bmh1Δ might function in the TOR and PKA pathways to extend CLS.
Figure 3.—
Analyses of genetic interactions between bmh1Δ, CR, and long-lived nutrient-sensing mutants. (A) bmh1Δ further extends CR-induced CLS. (B) bmh1Δ does not significantly affect cdc25-10-induced CLS. (C) bmh1Δ does not significantly affect tor1Δ-induced CLS. For CLS, cells are first grown in SD containing 2% (normal) or 0.5% glucose (CR) to stationary phase (48 hr) and then are shifted to sterile water. “Days” denotes the number of days that cells are in H2O. One representative set of three independent experiments, each conducted in quadruplicate, is shown. (D) CR-induced heat resistance requires the stress-response factors Msn2/4, Rtg3, and Rim15. (E) Deleting GCN4 totally abolishes bmh1Δ-induced heat resistance. (F) bmh1Δ-induced increase in survival requires Gcn4. Results show cell viability determined in day 6 cultures (4 days after shifting to H2O). (G) A proposed model for bmh1Δ- and CR-induced CLS extension. bmh1Δ and CR function in overlapping pathways to regulate CLS since they share common downstream stress-response factors such as Msn2/4 and Rim15. Gcn4 appears to be more specific to the bmh1Δ pathway. For simplicity, other factors/pathways are not shown. Error bars denote standard deviations. P-values were calculated using the Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.005). WT, wild-type control; CR, cells pregrown in SD with 0.5% glucose prior to analysis.
Since bmh1Δ further extended CR-induced CLS (Figure 3A), it was possible that bmh1Δ activated additional stress-response factors to extend the CLS of cells grown in CR. Deleting this bmh1Δ-specific stress-response factor should affect only bmh1Δ-induced but not CR-induced heat resistance. Since deleting Msn2/4, Rim15, and Rtg3 also abolished CR-induced heat resistance (Figure 3D), these factors were not only specific to the bmh1Δ pathway. Because our genetic studies suggested that bmh1Δ and tor1Δ might function in the same pathway to extend CLS (Figure 3C), we examined whether Gcn4, a downstream target of TOR, might play a role in bmh1Δ-induced heat resistance and CLS extension. Decreased TOR-signaling activities have been shown to enhance Gcn4 (a nutrient- and stress-sensing transcription factor) expression, which activates genes involved in nitrogen utilization (Valenzuela et al. 2001). Gcn4 was also required for the response and resistance to hydrogen peroxide (Mascarenhas et al. 2008). Figure 3E showed that gcn4Δ specifically abolished bmh1Δ- and tor1Δ-induced but not CR-induced heat resistance. Furthermore, Gcn4 was also required for bmh1Δ-induced CLS extension (Figure 3F). These results suggest that Gcn4 is an important factor in the bmh1Δ pathway, which works in concert with other factors such as Msn2/4 and Rim15 to regulate the stress response and CLS (Figure 3G).
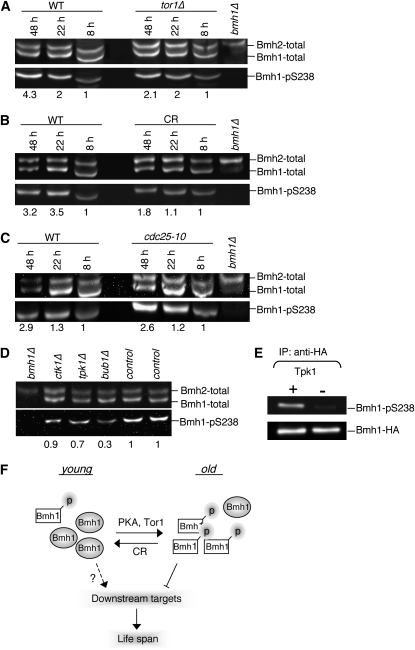
The level of phosphorylated Bmh1-Ser238 is increased during chronological aging:
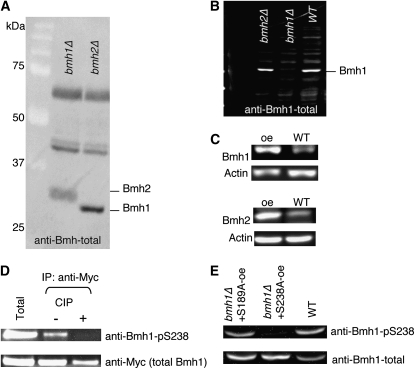
We next examined the roles of Bmh1 phosphorylation in chronological aging. Studies of mammalian 14-3-3 proteins have shown that phosphorylation of the Ser185 or Ser/Thr233 residues affect the interactions between 14-3-3 proteins and their binding partners (Aitken 2006). The equivalent phosphorylation sites on yeast Bmh1 are Ser189 and Ser238. We attempted to raise peptide antibodies that specifically targeted phosphorylated Ser189 and Ser238 on Bmh1 and total Bmh1 proteins. However, only anti-Bmh-total (specific to both Bmh1 and Bmh2 proteins; Figure 4A), anti-Bmh1-total (specific to total Bmh1 proteins; Figure 4B), and anti-Bmh1-pS238 (specific to phosphorylated Ser238 on Bmh1) antibodies showed specificity. The anti-Bmh1-pS238 antibody specifically recognized phosphorylated Ser238 on Bmh1 since it was unable to recognize Bmh1 after phosphatase (CIP) treatment (Figure 4D) or when Ser238 was mutated to alanine (Figure 4E).
Figure 4.—
Western blot analyses of anti-Bmh1 antibodies and Bmh1 and Bmh2 protein expression. (A) The anti-Bmh-total antibody recognizes both Bmh1 and Bmh2. Total protein extracts of the bmh1Δ and bmh2Δ mutants are analyzed. About 10 μl cell extract was loaded in each lane (A, B, C, and E). (B) The anti-Bmh1-total antibody specifically recognizes Bmh1 proteins. (C) Levels of protein expression in cells overexpressing Bmh1 or Bmh2 are analyzed using the anti-Bmh-total antibody. (D) The anti-Bmh1-pS238 specifically recognizes phosphorylated Ser238 on Bmh1. Results show CIP treatment of immunoprecipitated Myc-tagged Bmh1. CIP, calf intestinal alkaline phosphatase; IP, immunoprecipitation. (E) The anti-Bmh1-pS238 antibody is specific to phosphorylated Ser238 of Bmh1. Total protein extracts of wild-type and bmh1Δ cells expressing mutated Bmh1-S238A or Bmh1-S189A are analyzed. WT, wild-type control; oe, overexpression.
We next monitored the phosphorylation status of Bmh1-Ser238 during chronological aging using the anti-Bmh1-pS238 antibody. As shown in Figure 5, A–C, the level of phosphorylated Bmh1 proteins increased approximately three- to fourfold in wild-type cells upon entering stationary phase (48 hr). It was possible that phosphorylated Bmh1-Ser238 caused inhibitory effects on CLS because deleting Bmh1 extended CLS. In addition, both tor1Δ (Figure 5A) and CR (Figure 5B) largely prevented the increase in Bmh1 phosphorylation whereas the cdc25-10 mutation did not significantly affect Bmh1 phosphorylation (Figure 5C). These results suggested that phosphorylated Bmh1-Ser238 might impede the beneficial effects induced by CR and tor1Δ and that CR and tor1Δ extend CLS in part by preventing Bmh1-Ser238 phosphorylation.
Figure 5.—
Analyses of Bmh1-Ser238 phosphorylation levels using site-specific anti-Bmh1-pS238 antibody. (A) Phosphorylation of Ser238 on Bmh1 is increased in stationary phase, which is delayed by tor1Δ. Numbers below the bottom panels in A–E indicate the relative amount of Bmh1-pS238 normalized to the levels of Bmh1-pS238 at 8 hr. (B) CR reduces Bmh1-Ser238 phosphorylation in stationary phase. (C) cdc-25-10 does not decrease Bmh1-Ser238 phosphorylation in stationary phase. (D) Screening kinase mutants that show reduced Bmh1-Ser238 phosphorylation. (A–D) Results show duplicated blots, each loaded with the same amount of proteins from the same sample in each lane. (A–C) The amount of Bmh1-pS238 is first normalized to an internal loading control and then normalized to the levels of Bmh1-pS238 at 8 hr. WT, wild-type control; CR, 0.5% glucose. (E) Recombinant Tpk1 can phosphorylate Bmh1 at Ser238 in vitro. (F) A proposed model for the role of Bmh1 phosphorylation in life-span regulation. Bmh1 phosphorylation (at Ser238) is increased in old cells, which may inhibit the downstream longevity factors, including stress-response proteins. CR, low PKA, and low TOR activities can decrease the level of phosphorylated Bmh1 (at Ser238), which may help release its inhibitory effects on downstream longevity factors. It is possible that phosphorylation of Bmh1 at other residues also plays an important role in life-span regulation and that nonphosphorylated Bmh1 may also induce certain beneficial effects on life span.
Protein kinase A can directly phosphorylate Bmh1 at Ser238:
To gain further insight into the regulation of Bmh1, we screened 132 putative protein kinase mutants (defined in the Saccharomyces Genome Database) for kinases involved in the phosphorylation of Bmh1-Ser238 (Table S1) by using the yeast nonessential gene deletion collections. We found that the levels of phosphorylated Bmh1 were significantly decreased (70% decrease) in the bub1Δ mutant (Figure 5D). BUB1 encodes the protein kinase that plays a crucial role in the anaphase checkpoint control. Previous studies have linked 14-3-3 proteins to DNA checkpoint controls (Usui and Petrini 2007). Our findings supported a role for 14-3-3-mediated cell cycle checkpoint controls in life-span regulation. Interestingly, tpk1Δ also reduced the levels of phosphorylated Bmh1-Ser238 (30% decrease). Yeast protein kinase A is encoded by three different genes: TPK1, TPK2, and TPK3. Although these Tpk proteins are functionally redundant for viability, they have also been reported to play different roles in many processes. Since the decrease in Bmh1-Ser238 phosphorylation was only 30% in the tpk1Δ mutant, it was possible that all Tpk proteins contributed to Bmh1 phosphorylation. We next directly examined whether Tpk1 could phosphorylate Bmh1-Ser238. As shown in Figure 5E, recombinant Tpk1 was able to phosphorylate immunoprecipitated Bmh1 on the Ser238 residue in vitro. It remains highly possible that Tpk1 also phosphorylates other Ser/Thr residues on Bmh1. Phosphorylation of other Ser/Thr residues on Bmh1 by Tpk1 and/or other kinases may also play important roles in life-span regulation.
DISCUSSION
The yeast 14-3-3 proteins, Bmh1 and Bmh2, have been implicated in many cellular processes. Our studies demonstrate that yeast 14-3-3 proteins also play important roles in longevity regulation and stress response. In this study, 14-3-3 proteins were identified as longevity factors because their overexpressions extended cell survival in an accelerated cell death assay (Figure 1A) and extended CLS (Figure 1B). Interestingly, we discovered that Bmh1 deletion also extended CLS (Figure 1C). Since Bmh1-oe extended CLS only under certain growth conditions whereas bmh1Δ extended CLS regardless of growth conditions, bmh1Δ was more likely to function in a conserved pathway to extend CLS. Therefore, we focused on determining the mechanisms underlying bmh1Δ-induced CLS extension. We showed that the bmh1Δ mutant was more resistant to challenges of heat shock (Figure 2A) and oxidative stress-inducing reagents (Figure 2, E and F). Deleting Bmh1 also increased HSE-mediated transcription (Figure 2C). In addition, cells lacking Bmh1 had a lower intracellular ROS level (Figure 2H). Together, our results demonstrated that bmh1Δ-induced CLS extension was likely due to activation of stress-response mechanisms, which protects cells from ROS-induced damages during chronological aging.
In line with our findings, RNA-interference-mediated knockdown of specific 14-3-3 proteins in worms was also shown to increase the expression of antioxidant enzymes such as Sod3 (extracellular superoxide dismutase) (Li et al. 2007). It remained unclear why Bmh1-oe extended CLS only in cells grown and kept in SD but failed to extend CLS in cells shifted to H2O (Figure 1, B and G). It is possible that Bmh1-oe extends CLS only in cells that are more metabolically active. It has been shown that cells remain at a high metabolic state (post-diauxic phase) if they are maintained in SD media (Fabrizio and Longo 2003). Conversely, shifting cells to water induces a low metabolic state (stationary phase), which mimics the adverse natural environment that yeast cells frequently encounter (Fabrizio and Longo 2003; Gray et al. 2004). Overexpressions of 14-3-3 proteins have also been reported to extend life span in worms (Berdichevsky et al. 2006; Wang et al. 2006); however, the detailed mechanisms remain unclear. Since 14-3-3 proteins have many interacting partners and affect many cellular pathways, they are likely to regulate life span via multiple mechanisms. Understanding how Bmh1-oe extends life span in yeast may provide further insight into the roles that 14-3-3 proteins play in longevity regulation and cellular metabolism.
Phosphorylated Bmh1-Ser238 might cause inhibitory effects on CLS. The amount of phosphorylated Bmh1-Ser238 increased in stationary phase, which was significantly delayed by CR (Figure 5A) and tor1Δ (Figure 5B). Although cdc25-10 did not decrease the level of phosphorylated Bmh1-Ser238 (Figure 5C), our kinase screen (Figure 5D) and in vitro PKA kinase assay (Figure 5E) showed that PKA could directly phosphorylate Bmh1-Ser238. These results suggest that decreasing the level of phosphorylated Bmh1-Ser238 might promote longevity and that CR and low TOR and PKA activities might extend CLS in part by this mechanism (Figure 5F). In addition, Bmh proteins have been shown to interact with the retrograde response proteins (van Heusden and Steensma 2006), the stress-sensing transcription factors Msn2/4 and Rim15 (van Heusden and Steensma 2006), and the components of the autophagy pathway (Kakiuchi et al. 2007). Our data showed that bmh1Δ required these components for heat resistance (Figure 2D) and/or CLS extension (Figure S1), suggesting that the ability to respond to various types of metabolic and oxidative stresses was essential for bmh1Δ-mediated CLS extension. It is possible that during chronological aging, an increased amount of phosphorylated Bmh1-Ser238 might enhance the ability of Bmh1 to sequester and/or interfere with the interactions of these stress-response factors with their interacting partners. According to this model, deleting Bmh1 was sufficient to extend CLS. Therefore, bmh1Δ-induced stress resistance and CLS extension was likely due to elimination of the inhibitory effects of Bmh1 on downstream longevity factors, including stress-response proteins.
Our data also suggest that bmh1Δ might function in the same pathway as the tor1Δ and low PKA activity mutations to extend CLS. We showed that bmh1Δ did not further extend the long life span induced by tor1Δ and cdc25-10 (Figure 3, B and C). We also showed that both TOR and PKA could affect Bmh1-238 phosphorylation (Figure 5, A and E). Although bmh1Δ further extended CR-induced CLS (Figure 3A), bmh1Δ and CR appeared to share common downstream stress-response factors such as Msn2/4 and Rim15 (Figure 2D and Figure 3D). In addition, CR also decreased the level of Bmh1-Ser238 phosphorylation during chronological aging (Figure 5B). Together, these data suggest that Bmh1 is a novel downstream target of the TOR, PKA, and CR pathways and functions in accordance with other longevity factors to regulate CLS. Consistent with our studies, mammalian 14-3-3 proteins have also been reported to interact with the PKA and TOR pathways. For example, mammalian PKA has been shown to phosphorylate the binding motif of 14-3-3-interacting proteins, therefore altering the conformation of these proteins and their functions (Aitken 2006). A novel mTOR-binding partner, PRAS40, was also suggested to associate with 14-3-3 proteins, causing insulin to stimulate mTOR activities (Vander Haar et al. 2007).
14-3-3 proteins have been suggested to play a role in several aging-associated diseases, including cancers and neurodegenerative diseases (Wilker and Yaffe 2004; Darling et al. 2005). However, it remains unclear how and which types of 14-3-3 protein abnormalities contribute to the cause of these diseases. 14-3-3 protein levels are abundant in the neurofibrillary tangle in patients with Alzheimer's disease (Umahara et al. 2004). Elevated 14-3-3 expressions in lung and breast cancers have also been described. It would be interesting to determine whether phosphorylation of 14-3-3 proteins plays an important role in these diseases as well as in life-span regulation in mammals.
Acknowledgments
We thank T. Powers for providing the pFA6a-13Myc-kanMX6 plasmid and suggestions and members of the Lin laboratory for discussions and suggestions. The HSE4Ptt-CYC1-LacZ reporter plasmid was kindly provided by T. Griffin. We also thank members of the Parales laboratory for assistance with oxygen consumption assays and members of the Hunter laboratory for assistance with image analysis. This study was supported by the National Institute on Aging, the Ellison Medical Foundation, and an American Cancer Society Institutional Research Grant.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107797/DC1.
References
- Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan and X. F. Zheng, 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10 1295–1305. [DOI] [PubMed] [Google Scholar]
- Aitken, A., 2006. 14–3-3 proteins: a historic overview. Semin. Cancer Biol. 16 162–172. [DOI] [PubMed] [Google Scholar]
- Bandhakavi, S., H. Xie, B. O'Callaghan, H. Sakurai, D. H. Kim et al., 2008. Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis. PLoS One 3 e1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdichevsky, A., M. Viswanathan, H. R. Horvitz and L. Guarente, 2006. C. elegans SIR-2.1 interacts with 14–3-3 proteins to activate DAF-16 and extend life span. Cell 125 1165–1177. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Bruckmann, A., P. J. Hensbergen, C. I. Balog, A. M. Deelder, H. Y. de Steensma et al., 2007. Post-transcriptional control of the Saccharomyces cerevisiae proteome by 14–3-3 proteins. J. Proteome Res. 6 1689–1699. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Sterns, 2000. Methods in Yeast Genetics, pp. 171–174. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Darling, D. L., J. Yingling and A. Wynshaw-Boris, 2005. Role of 14–3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68 281–315. [DOI] [PubMed] [Google Scholar]
- Dilova, I., E. Easlon and S. J. Lin, 2007. Calorie restriction and the nutrient sensing signaling pathways. Cell. Mol. Life Sci. 64 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon, E., F. Tsang, I. Dilova, C. Wang, S. P. Lu et al., 2007. The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J. Biol. Chem. 282 6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon, E., F. Tsang, C. Skinner, C. Wang and S. J. Lin, 2008. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 22 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., and V. D. Longo, 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2 73–81. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron and V. D. Longo, 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292 288–290. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., L. Battistella, R. Vardavas, C. Gattazzo, L. L. Liou et al., 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., C. Gattazzo, L. Battistella, M. Wei, C. Cheng et al., 2005. Sir2 blocks extreme life-span extension. Cell 123 655–667. [DOI] [PubMed] [Google Scholar]
- Gray, J. V., G. A. Petsko, G. C. Johnston, D. Ringe, R. A. Singer et al., 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, A. L., C. T. Murphy and C. Kenyon, 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 1142–1145. [DOI] [PubMed] [Google Scholar]
- Ichimura, T., H. Kubota, T. Goma, N. Mizushima, Y. Ohsumi et al., 2004. Transcriptomic and proteomic analysis of a 14–3-3 gene-deficient yeast. Biochemistry 43 6149–6158. [DOI] [PubMed] [Google Scholar]
- Jiang, J. C., J. Wawryn, H. M. Shantha Kumara and S. M. Jazwinski, 2002. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp. Gerontol. 37 1023–1030. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., M. McVey and L. Guarente, 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., K. T. Kirkland, S. Fields and B. K. Kennedy, 2004. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., R. W. Powers, III, K. K. Steffen, E. A. Westman, D. Hu et al., 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kakiuchi, K., Y. Yamauchi, M. Taoka, M. Iwago, T. Fujita et al., 2007. Proteomic analysis of in vivo 14–3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46 7781–7792. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., 2001. A conserved regulatory system for aging. Cell 105 165–168. [DOI] [PubMed] [Google Scholar]
- Lamming, D. W., M. Latorre-Esteves, O. Medvedik, S. N. Wong, F. A. Tsang et al., 2005. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 309 1861–1864. [DOI] [PubMed] [Google Scholar]
- Li, J., M. Tewari, M. Vidal and S. S. Lee, 2007. The 14–3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev. Biol. 301 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K., J. B. Dorman, A. Rodan and C. Kenyon, 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 1319–1322. [DOI] [PubMed] [Google Scholar]
- Lin, S. J., P. A. Defossez and L. Guarente, 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 2126–2128. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Mascarenhas, C., L. C. Edwards-Ingram, L. Zeef, D. Shenton, M. P. Ashe et al., 2008. Gcn4 is required for the response to peroxide stress in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 19 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio, E., M. Giorgio, S. Mele, G. Pelicci, P. Reboldi et al., 1999. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402 309–313. [DOI] [PubMed] [Google Scholar]
- Powers, R. W., III, M. Kaeberlein, S. D. Caldwell, B. K. Kennedy and S. Fields, 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri, S., M. M. Reddy, N. R. Rajkumar and R. Rajasekharan, 2003. Cytosolic iron superoxide dismutase is a part of the triacylglycerol biosynthetic complex in oleaginous yeast. Biochem. J. 372 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter-Branchat, G., E. Cabiscol, J. Tamarit, J. Ros, E. Piulats et al., 2004. Oxidative damage to specific proteins in replicative and chronological-aged Saccharomyces cerevisiae: common targets and prevention by calorie restriction oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 279 31983–31989. [DOI] [PubMed] [Google Scholar]
- Roth, G. S., D. K. Ingram and M. A. Lane, 2001. Caloric restriction in primates and relevance to humans. Ann. NY Acad. Sci. 928 305–315. [DOI] [PubMed] [Google Scholar]
- Salmon, A. B., S. Murakami, A. Bartke, J. Kopchick, K. Yasumura et al., 2005. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am. J. Physiol. Endocrinol. Metab. 289 E23–E29. [DOI] [PubMed] [Google Scholar]
- Smith, D. L., Jr., J. M. McClure, M. Matecic and J. S. Smith, 2007. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 6 649–662. [DOI] [PubMed] [Google Scholar]
- Tissenbaum, H. A., and L. Guarente, 2002. Model organisms as a guide to mammalian aging. Dev. Cell 2 9–19. [DOI] [PubMed] [Google Scholar]
- Umahara, T., T. Uchihara, K. Tsuchiya, A. Nakamura, T. Iwamoto et al., 2004. 14–3-3 proteins and zeta isoform containing neurofibrillary tangles in patients with Alzheimer's disease. Acta Neuropathol. 108 279–286. [DOI] [PubMed] [Google Scholar]
- Usui, T., and J. H. Petrini, 2007. The Saccharomyces cerevisiae 14–3-3 proteins Bmh1 and Bmh2 directly influence the DNA damage-dependent functions of Rad53. Proc. Natl. Acad. Sci. USA 104 2797–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela, L., C. Aranda and A. Gonzalez, 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183 2331–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar, E., S. I. Lee, S. Bandhakavi, T. J. Griffin and D. H. Kim, 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9 316–323. [DOI] [PubMed] [Google Scholar]
- van Heusden, G. P., and H. Y. Steensma, 2006. Yeast 14–3-3 proteins. Yeast 23 159–171. [DOI] [PubMed] [Google Scholar]
- Wang, Y., S. W. Oh, B. Deplancke, J. Luo, A. J. Walhout et al., 2006. C. elegans 14–3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 127 741–747. [DOI] [PubMed] [Google Scholar]
- Wei, M., P. Fabrizio, J. Hu, H. Ge, C. Cheng et al., 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 4 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch, W., and R. L. Walford, 1998. The Retardation of Aging and Diseases by Dietary Restriction. Charles C Thomas, Springfield, IL.
- Werner-Washburne, M., E. Braun, G. C. Johnston and R. A. Singer, 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker, E., and M. B. Yaffe, 2004. 14–3-3 proteins: a focus on cancer and human disease. J. Mol. Cell. Cardiol. 37 633–642. [DOI] [PubMed] [Google Scholar]
- Zuin, A., N. Gabrielli, I. A. Calvo, S. Garcia-Santamarina, K. L. Hoe et al., 2008. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS ONE 3 e2842. [DOI] [PMC free article] [PubMed] [Google Scholar]