Abstract
The African fish Nothobranchius furzeri is the shortest-lived vertebrate species that can reproduce in captivity, with a median life span of 9–11 weeks for the shortest-lived strain. Natural populations of N. furzeri display differences in life span, aging biomarkers, behavior, and color, which make N. furzeri a unique vertebrate system for studying the genetic basis of these traits. We mapped regions of the genome involved in sex determination and tail color by genotyping microsatellite markers in the F2 progeny of a cross between a short-lived, yellow-tailed strain and a long-lived, red-tailed strain of N. furzeri. We identified one region linked with the yellow/red tail color that maps close to melanocortin 1 receptor (mc1r), a gene involved in pigmentation in several vertebrate species. Analysis of the segregation of sex-linked markers revealed that N. furzeri has a genetic sex determination system with males as the heterogametic sex and markedly reduced recombination in the male sex-determining region. Our results demonstrate that both naturally-evolved pigmentation differences and sex determination in N. furzeri are controlled by simple genetic mechanisms and set the stage for the molecular genetic dissection of factors underlying such traits. The microsatellite-based linkage map we developed for N. furzeri will also facilitate analysis of the genetic architecture of traits that characterize this group of vertebrates, including short life span and adaptation to extreme environmental conditions.
THE Nothobranchius fish species are present in eastern and southeastern Africa, where they populate ephemeral water pools that often undergo complete desiccation during the dry season (Terzibasi et al. 2008; Reichard et al. 2009; Wildekamp 2009). Nothobranchius species tend to live in extreme habitats and have evolved unique adaptations to harsh environmental conditions, including extremely short life cycles, resistance to a wide range of temperatures and water salinity, embryonic development that does not require the presence of water, and a developmental diapause that allows embryos to survive for months in dry conditions (Wourms 1972; Inglima et al. 1981; Genade et al. 2005).
Nothobranchius furzeri is the shortest-lived species of the Nothobranchius genus, with an intergeneration time of 40 days, a median life span of 9–11 weeks, and a maximum life span of 12–15 weeks for the shortest-lived strain GRZ (Valdesalici and Cellerino 2003; Genade et al. 2005; Valenzano et al. 2006; Terzibasi et al. 2008, 2009; Hartmann et al. 2009). Natural populations of N. furzeri can vary substantially in phenotypic traits. For example, N. furzeri strains derived from Zimbabwe and northern Mozambique (e.g., GRZ) exhibit a shorter life span than strains derived from more humid areas in southern Mozambique (e.g., MZM-0403) under controlled conditions (Terzibasi et al. 2008). The extremely short life cycle of N. furzeri and the presence of natural populations with phenotypic variations make this species a promising model system for studying aging and adult-specific traits, including color and behavior.
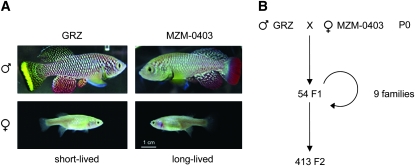
The color pattern of the adult male tail differs among N. furzeri strains. GRZ males show a yellow submarginal band and a black marginal band (yellow morph) whereas MZM-0403 males display a broad red band (red morph) in the caudal fin (Figure 1A). This dichromatism is present in natural populations of N. furzeri (Terzibasi et al. 2008; Reichard et al. 2009). Similar color polymorphism among males is also observed in other species of Nothobranchius (Wildekamp 2009) and in other fish species, including guppies and cichlids (Hughes et al. 1999; Brooks and Endler 2001; Maan et al. 2004). Differences in male color morphs within the same species are associated with sexual preference by females, different recognition by predators depending on the habitat, and differential susceptibility to pathogens (Price et al. 2008), which could all influence the evolution of this trait. Despite the widespread variation in Nothobranchius coloration, the genetic basis of this trait is unknown.
Figure 1.—
Cross between two strains of N. furzeri that differ in color and life span (A) Color phenotypes of GRZ and MZM-0403. (B) A yellow-tailed, short-lived male GRZ and a red-tailed, long-lived female MZM-0403 were the founders of cross 1.
Genetic information on N. furzeri is still limited. The N. furzeri genome is 1.6–1.9 Gb in size and is characterized by a high repeat content (45%) (Reichwald et al. 2009). N. furzeri has 19 chromosomes, but no morphologically discernible sex chromosomes (Reichwald et al. 2009). The sex determination system in N. furzeri has not been characterized yet. Sex can be determined either genetically or environmentally in fish (Volff 2005; Marshall Graves 2008). For example, medaka, platyfish, guppy, and sticklebacks all have recently evolved genetic sex-determining systems (Volff and Schartl 2002; Peichel et al. 2004; Shapiro et al. 2009; Tripathi et al. 2009a), whereas zebrafish do not have a clear genetic basis of sex determination (von Hofsten and Olsson 2005).
Genetic studies in N. furzeri would greatly benefit from the building of a linkage map in this species of fish. However, to date there is no linkage map available for N. furzeri or for any Nothobranchius species, although linkage maps have been generated for fish of the same order, e.g., Poecilia reticulata (guppy) and Xiphophorus maculatus (platyfish) (Khoo et al. 2003; Walter et al. 2004; Tripathi et al. 2009b), and of the same superorder, e.g., Oryzias latipes (medaka) (Wada et al. 1995).
Here, we report a microsatellite-based linkage map for N. furzeri using a genetic cross between the short-lived yellow-tailed GRZ strain and the long-lived red-tailed MZM-0403 strain. This N. furzeri linkage map allowed us to map the male-specific tail color trait on linkage group (LG) V. Synteny analysis revealed that LG V has homology to a region of the medaka genome that contains the melanocortin 1 receptor (mc1r) gene, which is known to play a key role in vertebrate pigmentation. We identified a sequence polymorphism in mc1r between the two strains of N. furzeri, allowing us to map mc1r on LG V. This analysis revealed that mc1r is located in close proximity to the color locus, but that the sequence polymorphism is probably not causative for the color difference. We also found that sex is genetically determined in N. furzeri, with males as the heterogametic sex. The sex-determining region is located on LG XIII and is characterized by male-specific suppression of recombination. Our findings will be pivotal for the identification of the genetic determinants of color in N. furzeri and for expanding our knowledge about sex-determination mechanisms in vertebrates. Due to the array of intraspecific phenotypic differences displayed by the various populations of N. furzeri, this linkage map will also be a key tool for mapping phenotypic variation in this short-lived vertebrate species, including differences in life span.
MATERIALS AND METHODS
Fish housing and husbandry:
Fish were grown at 25° in a centralized filtration water system at a density of two fish per gallon tank. Fish were fed freshly hatched Artemia brine shrimp until 3 weeks of age and then dried Chironomid bloodworms two times per day every day. Adults spawned freely in the system. Tanks were inspected daily, and freshly laid embryos were collected and stored in dry peat moss until they were ready to hatch, as indicated by spontaneous twitching inside the eggshell. Once ready to hatch, embryos were immersed in Yamamoto embryo solution (17 mm NaCl, 2.7 mm KCl, 2.5 mm CaCl2, 0.02 mm NaHCO3, pH 7.3) (Rembold et al. 2006). Fry were placed in 0.2-gallon tanks at the density of five fry per tank.
Cross between two strains of N. furzeri:
One male from the GRZ strain was crossed with one female from the MZM-0403 strain (cross 1). From the F1 generation (54 fish), nine independent “families” (eight tanks with a spawning pair in each tank and one tank with three males and four females spawning together) were formed, and fertilized eggs were collected. A total of 413 F2 individuals were produced. An independent, reciprocal cross was established between one male MZM-0403 and one female GRZ (cross 2). Two F1 individuals from cross 2 produced a total of 34 F2 individuals.
Color phenotyping:
Fish were removed from tanks at death, rinsed in tap water, and stored in 100% ethanol. Tail color (yellow vs. red) was scored in all 203 F2 adult males and all 23 F1 males from cross 1 by visual inspection, immediately preceding submersion in ethanol. N. furzeri male fish are extremely colorful, and thus male color at death was clearly visible. F1 and F2 fish with yellow tails overlaid with red spots were scored as “yellow.”
Sex phenotyping:
Sex was assessed at death on the basis of the presence or absence of the typical male tail coloration. Fish that had not yet reached sexual maturity at death were scored as “undetermined.”
Microsatellite identification by hybridization:
Genomic DNA was isolated from a 7-week-old MZM-0403 male and digested with RsaI. Fragments (1000–1650 bp) were cloned into the pCR-Blunt-TOPO vector (Invitrogen) and transformed into chemically competent Escherichia coli. Transformants were selected for the presence of CA/GT microsatellites using a [32P]dCTP end-labeled (CA)15 hybridization probe (Elim Biopharmeceuticals) following a described method (Peichel et al. 2001). Of the 773 positive clones analyzed, 318 contained microsatellites and 311 met the following criteria: (i) at least 15 repeat units and (ii) at least 100 bp of sequence flanking the microsatellite. To amplify these microsatellite repeats by PCR, primers that have melting temperatures between 57°–63° and that amplify fragments between 150 and 400 bp were designed using Primer3 (http://frodo.wi.mit.edu/). A 5′ M13 sequence (5′-TGTAAAACGACGGCCAGT-3′) was added to all forward primers to enable fluorescent labeling of fragments during PCR (Schuelke 2000).
Microsatellite identification by whole-genome sample sequencing:
Whole-genome sample sequencing of a male specimen of the GRZ strain resulted in 5540 sequences composing a total of 5.4 Mb (Reichwald et al. 2009). Through in silico analyses using Sputnik (http://cbi.labri.fr/outils/Pise/sputnik.html), 289 microsatellites that met the following criteria were identified: (i) one microsatellite per sequence, (ii) at least 20 repeat units for dinucleotide repeats, (iii) at least 100 bp of microsatellite flanking sequence, and (iv) perfect repeats. Primers were designed using the GAP4 module of the Staden Sequence Analysis Package, as described (Reichwald et al. 2009). One hundred and thirty-nine microsatellites were experimentally validated by PCR and subsequent genotyping using ABI3730xl analyzers and GeneMapper software v4 (Applied Biosystems).
Genotyping:
A total of 244 pairs of primers were used to amplify microsatellites from the grandparents and 246 F2 offspring (160 males and 86 females) of cross 1. One hundred and fifty-two microsatellite markers were informative for the grandparents, 148 of which were used to genotype F2 individuals. Of the 246 individuals genotyped to build the map, 234 were genotyped at all the markers whereas 12 were genotyped only at the 121 markers corresponding to the four terminal markers of each LG and to the singletons. PCRs were performed in 5 μl in 384-well plates with 0.2 units Taq DNA polymerase, 1× PCR buffer (50 mm KCl, 100 mm Tris–HCl, pH 9.0, 0.1% Triton-X), 0.5 ng/μl DNA, 0.25 mm dNTP (Invitrogen), 800 nm FAM56-labeled M13 primer, 10–20 nm M13-forward primer, and 400–800 nm reverse primer. Samples were heated to 94° for 5 min, followed by 30 cycles of 30 sec at 94°, 45 sec at 56°, and 45 sec at 72° and 8 cycles of 30 sec at 94°, 45 sec at 53°, and 45 sec at 72°. Amplicons were denatured by incubation in denaturation solution (1:1.15 Hi-Di Formamide (Applied Biosystems) and 1:300 GeneScan-500 LIZ Size Standard (Applied Biosystems)) at 95° for 5 min and electrophoresed on an ABI 3730 capillary sequencer. Chromatographs were analyzed manually using PeakScanner software v1.0 (Applied Biosystems).
Linkage map generation and map length calculation:
Genotypes were scored according to JoinMap 3.0 (Van Ooijen and Voorrips 2001). The Kosambi mapping function was used to convert recombination frequencies (REC = 0.4) to centimorgans (cM). The map was also calculated using Haldane's function to account for double crossing-overs, which gave similar results. The assignment of markers to LG was carried out with a LOD score threshold of 4 and a maximum linkage distance of 25 cM. The calculation of phase cannot be exploited in this particular case, since parental (F1) genotypes were tractable for only 96 of 246 F2 individuals. Raw data are available for download (supporting information, File S1).
The map length was computed by adding 2 × s to each LG length in centimorgans, where s is the average intermaker distance, to account for chromosome ends, as described (Tripathi et al. 2009b). This measure was averaged with the measure obtained by multiplying each LG's length in centimorgans by (m + 1)/(m − 1), where m is the number of markers in each LG, as described (Tripathi et al. 2009b).
Mapping color:
Color mapping was computed manually by scoring the recombination events in all the F2 red-tailed fish (n = 61) of cross 1, genotyped for all 148 microsatellites. Only red-tailed F2 fish were used to map color because the red phenotype is more reliable than the yellow phenotype, as yellow-tailed fish can develop red spots with advancing age (see below). These genotypes were then further included in the whole-genome map calculation. For each microsatellite marker, the presence of two, one, or none of the two alleles inherited from the red-tailed MZM-0403 grandparent was scored. The LOD score for each marker was calculated according to the following formula: LOD = M × LOD10(m) + (N − M) × LOD10(1 − m) − N × LOD10(0.5), where M is the number of recombination events, m is the fraction of recombination events over all the alleles (m = M/N), N is twice the number of genotyped individuals (corresponding to the total number of alleles genotyped), and LOD10 is the 10-base logarithm. Considering the color marker position as the position at which all red individuals have both MZM-0403 alleles, m × 100 corresponds to the distance in centimorgans of each marker from the color gene. This analysis was performed over all the markers in the map.
Synteny analysis:
BLASTn searches were performed using the flanking regions from all 11 microsatellites that cosegregated with tail color as query against the medaka genome (October 2005 MEDAKA1 assembly) at Ensembl (http://www.ensembl.org/Oryzias_latipes/Info/Index). The microsatellites flanking regions having a P value <10−5, and 100% sequence identity over >22 bp was considered a significant hit. Search sensitivity was set to “no optimization.”
Cloning of mc1r:
Total RNA was isolated from caudal fins of three male adult individuals (two GRZ and one MZM-0403) using Trizol (Invitrogen). cDNA was generated using MMLV reverse transcriptase (Clontech), according to the manufacturer's protocol. A 681-bp fragment of N. furzeri mc1r was generated by PCR using primers derived from conserved mc1r regions in medaka, stickleback, Takifugu, and Tetraodon (forward primer: 5′ GAA CCG CAA CCT GCA CTC 3′; reverse primer: 5′ GGG TCG ATG AGC GAG TTA CA 3′). A 1402-bp DNA fragment containing the mc1r open reading frame and 5′ and 3′ untranslated regions was amplified by RACE PCR (Clontech) and subcloned in the pCR 2.1-TOPO cloning vector (Invitrogen).
Identification and genotyping of mc1r sequence polymorphism:
The mc1r 1402-bp region was amplified from the grandparents of cross 1 (male GRZ and female MZM-0403), cloned, and entirely sequenced. A sequence polymorphism between GRZ and MZM-0403 was identified at nucleotide 67 of the mc1r coding sequence. This polymorphism was genotyped in 61 F2 fish from cross 1. For sequencing, a 312-bp region flanking nucleotide 67 of mc1r was amplified by PCR using the following primers (forward primer: 5′ GTG GAC CCC TGC TTT AAT GA 3′; reverse primer: 5′ TAG TAC ATG GGC GAG TGC AG 3′). The PCR products were purified using a PCR purification kit (Qiagen) and sequenced using Molecular Cloning Laboratories (http://www.mclab.com). Sequences were analyzed using Sequencher 4.7 (Gene Codes).
Mapping sex:
The genotypes at 148 microsatellites of 239 F2 individuals from cross 1 were sorted by sex (female, male, unknown) to isolate the markers carrying a significant sex-biased allele distribution. To confirm these results, F2 individuals from cross 2 (34 individuals) were also genotyped at 9 sex-linked microsatellites identified in cross 1 and sorted by sex (female, male, unknown). The presence of potentially suppressed recombination in the male vs. female sex-determining region was determined by scoring recombination events in two specific F1 families of cross 1 (family 3 and family 7), in which the two parental pairs were both heterozygous at all the sex-linked loci. The sex-specific recombination events were determined by assessing the sex-specific allele distribution in the offspring of these two F1 families (22 F2 individuals for family 3 and 46 F2 individuals for family 7—68 F2 individuals total) at the sex-linked markers that were heterozygous for both F1 parents and that had more than two alleles.
RESULTS
Microsatellite-based linkage map for N. furzeri:
To develop genetic markers for linkage mapping in N. furzeri, we identified microsatellites using two strategies of large-scale genomic library screening and sequencing. In the first approach, we screened a genomic library from MZM-0403, a wild-derived strain of N. furzeri with a broad red marginal band in the caudal fin (“red morph”), by using a (CA)15 probe because CA microsatellite repeats are frequent in fish genomes. We sequenced 773 positive clones and identified 318 clones containing microsatellites, 105 of which gave rise to a PCR product. In the second approach, we performed whole-genome sample sequencing of GRZ, which has a black marginal band and a yellow submarginal band in the caudal fin (“yellow morph”) (Reichwald et al. 2009). We identified 289 clones containing microsatellites by in silico analysis using Sputnik (http://cbi.labri.fr/outils/Pise/sputnik.html), 139 of which were experimentally validated.
To generate a linkage map, we set up a cross between one long-lived red-tailed MZM-0403 female and one short-lived yellow-tailed GRZ male (Figure 1A). We obtained 54 F1 progeny, which were used to form nine families (Figure 1B). A total of 413 F2 progeny from these F1 families developed into adulthood (Figure 1B). We genotyped 244 microsatellite markers in the grandparents and found that 152 (62%) were polymorphic and thus informative for building a genetic map (Table S1). All microsatellite markers were homozygous in the GRZ male grandparent, except those linked with sex (see below), confirming that GRZ is an inbred strain (Reichwald et al. 2009). Forty-one percent of the microsatellite markers (63 of 152) were heterozygous in the MZM-0403 female grandparent, which is consistent with this strain being recently derived from the wild and propagated in captivity for no more than seven generations (Terzibasi et al. 2008; Hartmann et al. 2009).
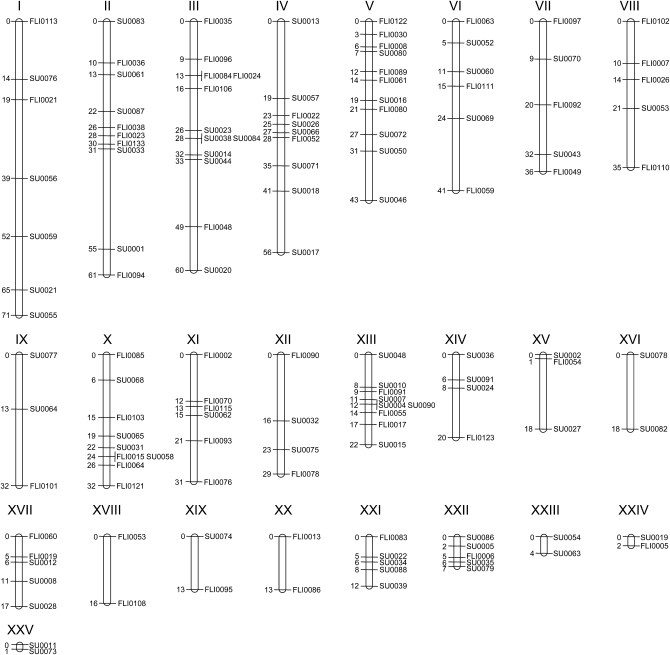
To build the linkage map, we genotyped 246 F2 fish at 148 of the 152 informative microsatellite markers (see materials and methods). Significant linkage was found for 138 (93%) microsatellite markers, and 10 markers were singletons. Six of the 138 linked markers were excluded from the map calculation because they gave incomplete genotypes in >50% of the individuals. The resulting N. furzeri linkage map consists of 25 LGs, with 2–12 markers per LG (Figure 2). Since N. furzeri has only 19 chromosomes (Reichwald et al. 2009), we anticipate that some of the linkage groups will coalesce when additional markers are included. The total calculated map length is 1012 cM, with an average intermarker distance of 5.3 cM. Considering 25 cM as the maximum intermarker distance, 10 singletons, and 6 more LGs than chromosomes, we estimate that up to 400 cM could still not be accounted for in this map (25 × 10 + 25 × 6 = 400 cM), corresponding to 28% of the N. furzeri genome [400/(400 + 1012)]. Thus, we have generated a first-generation microsatellite-based linkage map for N. furzeri that can be used to map phenotypic variation between the populations of this species.
Figure 2.—
Microsatellite-based genetic linkage map of N. furzeri. Each linkage group is designated by a Roman numeral and ordered on the basis of genetic length. The map distance in centimorgans is reported on the left side of each linkage group. Microsatellite loci identified at Stanford University were labeled as SU followed by a four-digit number based on their order of identification. Microsatellite loci identified at the Fritz Lipmann Institute were labeled as FLI followed by a four-digit number. In the text, these markers are termed NfuSU or NfuFLI followed by a four-digit number, with Nfu standing for N. furzeri.
Mapping tail color on LG V in N. furzeri:
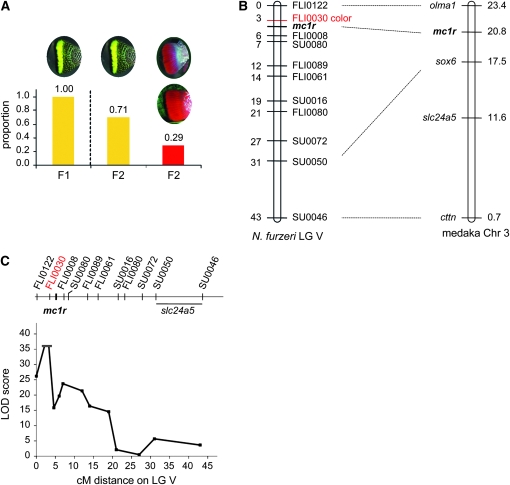
A conspicuous difference between the GRZ and MZM-0403 strains of N. furzeri is the color and pattern of the caudal fin in males (Figure 1A, Figure 3A). To examine the genetic basis of this dichromatism, we scored all males in the F1 and F2 generations for a “yellow” vs. a “red” caudal fin. All F1 males display a yellow morph, indicating that yellow is dominant over red (Figure 3A). We also observed the progressive appearance of red spots in the caudal fin of adult F1 male fish with advancing age (data not shown). In the 203 males of the F2 generation, the yellow/red tail-color trait segregated as 142 yellow (yellow and yellow with red spots) and 61 red (Figure 3A). The ratio between the two color morphs is close to 3:1 (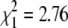 , P = 0.09), which suggests Mendelian transmission.
, P = 0.09), which suggests Mendelian transmission.
Figure 3.—
Genetics of the tail color in N. furzeri males. (A) Transmission of the color trait in the F1 and F2 generations. A χ2 test shows that the ratio is close to 3:1. Yellow tails always have black vertical bars. Red tails can be with (bottom) or without (top) black vertical bars. (B) LG V contains the color locus, which corresponds to marker NfuFLI0030 (FLI0030). The expected map location of N. furzeri mc1r is indicated in bold. The distance in centimorgans is reported on the left side of LG V. mc1r is represented between NfuFLI0030 and NfuFLI0008 and closer to NfuFLI0030 than to NfuFLI0008 because there is stronger linkage among mc1r, NfuFLI0030, and NfuFLI0122 than between mc1r and NfuFLI0008 and because mc1r is 4 cM from NfuFLI0122 and 2 cM from NfuFLI0030. Note that the distances in centimorgans between NfuFLI0122, NfuFLI0030, mc1r, and NfuFLI0008 do not add up because the distances between mc1r and each of these three microsatellite markers were calculated independently. Dashed lines indicate the synteny of microsatellite markers on N. furzeri LG V and annotated genes on medaka chromosome 3. The distance in megabases is reported on the right side of medaka chromosome 3. (C) LOD score plot of the color locus region. The LOD score is undetermined at marker NfuFLI0030 (FLI0030) due to lack of recombination between this marker and the color locus. Therefore the peak at NfuFLI0030 is represented by two short horizontal lines. slc24a5 was not directly mapped and therefore is represented by a horizontal line between marker NfuSU0050 (SU0050) and NfuSU0046 (SU0046).
To map the yellow/red tail-color trait, we genotyped all the red males of our cross (61 fish) at all 148 markers. We found a strong linkage signal on LG V (Figure 3B, Table 1). The peak LOD score corresponds to the NfuFLI0030 marker (Figure 3C, Table 1). At the current map resolution, the NfuFLI0030 marker is indistinguishable from the color locus; i.e., all red males in the F2 generation inherited both alleles from the red MZM-0403 grandparent. These results indicate that the yellow/red tail-color trait is determined mainly by one locus, although we cannot exclude that multiple linked loci contribute to this color trait or that other minor unlinked loci may influence the trait.
TABLE 1.
Genotype frequencies of microsatellite markers on the color locus containing LG V
| Genotype |
|||
|---|---|---|---|
| Markers | MZM-0403 | GRZ | GRZ/MZM-0403 |
| FLI0122 | 46 (97) | 0 (0) | 1 (2) |
| FLI0030a | 48 (97) | 0 (0) | 0 (0) |
| mc1r | 26 (85) | 0 (0) | 1 (3) |
| FLI0008 | 35 (93) | 0 (0) | 0 (0) |
| SU0080 | 50 (93) | 1 (2) | 3 (6) |
| FLI0089 | 41 (91) | 0 (0) | 3 (7) |
| FLI0061 | 40 (85) | 1 (2) | 6 (13) |
| SU0016 | 41 (73) | 1 (2) | 13 (23) |
| FLI0080 | 9 (64) | 1 (7) | 4 (29) |
| SU0072 | 5 (36) | 1 (7) | 8 (57) |
| SU0050 | 28 (56) | 3 (6) | 19 (38) |
| SU0046 |
25 (42) |
3 (5) |
31 (53) |
Genotype frequencies for 11 markers and for mc1r on LG V in 61 F2 red-tailed males. The number of red individuals is presented. Genotype frequencies are presented as percentages in parentheses. Only complete genotypes with both alleles present were reported; therefore, percentage values do not add up to 100.
Marker corresponding to the LOD peak.
We sought to identify candidate genes that underlie the tail-color trait in N. furzeri. Synteny with other species can be used to infer the position of genes on a linkage map of a species without a sequenced genome (Gross et al. 2008). We performed a BLASTn search in the medaka genome with the known sequences flanking the 11 microsatellite markers of LG V as queries, since medaka is the sequenced fish species most closely related to N. furzeri (Reichwald et al. 2009). Markers NfuFLI0122, NfuSU0050, and NfuSU0046 displayed significant sequence homology with, respectively, olma1 (cardiac muscle actin, orthologous to human ACTC1; P < 10−17), sox6 (sry box containing gene 6; P < 10−13), and cttn (cortactin; P < 10−11) on medaka chromosome 3 (Figure 3B). In addition, NfuFLI0122, NfuSU0050, and NfuSU0046 are in the same order on N. furzeri LG V as their respective counterparts on medaka chromosome 3 (Figure 3B). Together, these results suggest that there is synteny and colinearity between N. furzeri LG V and medaka chromosome 3.
We next asked whether genes known to mediate skin/hair pigmentation in other vertebrate species are present in the region that is syntenic to N. furzeri LG V on medaka chromosome 3 (Figure 3B). The genes that we specifically examined were the following: melanocortin 1 receptor (Mc1r) and its ligand (Asip) (Rees 2003; Shriver et al. 2003; Hoekstra 2006; Sulem et al. 2008; Le Pape et al. 2009), the Kit receptor and its ligand (Kitl) (Geissler et al. 1988; Miller et al. 2007; Sulem et al. 2007), CBD103 (Candille et al. 2007), MATP/SLC45A2 (Norton et al. 2007), SLC24A4 (Sulem et al. 2007), SLC24A5 (Lamason et al. 2005), IRF4 (Sulem et al. 2007; Han et al. 2008), TPCN2 (Bonilla et al. 2005; Sulem et al. 2008), Sly (Alizadeh et al. 2009), oca2 (Shriver et al. 2003; Protas and Patel 2008), and TYR (Shriver et al. 2003; Sulem et al. 2007). Of all these genes, only mc1r and slc24a5 were located on the medaka chromosome syntenic to N. furzeri LG V. Conversely, the annotated genes on medaka chromosome 3 do not include any other genes known to be associated with color determination, although this does not rule out their existence. In medaka, mc1r is located 2.6 Mb from olma1, the marker that is syntenic with NfuFLI0122, whereas slc24a5 is located outside of the medaka region that is syntenic to the region linked with color in N. furzeri (Figure 3B). Together, these results suggest that mc1r is a better functional candidate for tail color than slc24a5 in N. furzeri.
To determine if mc1r is the gene underlying male tail color, we cloned N. furzeri mc1r cDNA and mapped mc1r on the linkage map. Sequence comparison of GRZ and MZM-0403, the strains used in our cross, revealed a single nucleotide polymorphism at position 67 of the mc1r coding sequence (C in GRZ and G in MZM-0403) (Figure S1; GenBank GQ463613). This variation is a nonsynonymous substitution that results in a change from histidine (GRZ) to aspartic acid (MZM-0403) at amino acid 23 in the N-terminal region of the Mc1r protein (Figure S1). Sequencing three additional specimens of each strain confirmed that MZM-0403 individuals were homozygous for G, and GRZ individuals were homozygous for C at position 67 of the mc1r coding sequence. The segregation of this mc1r polymorphism in 61 F2 fish allowed us to place mc1r on LG V, the LG that contains the color locus (Figure 3B). However, there was one recombinant red-tailed F2 fish with a GC genotype, as well as three recombinant F2 yellow-tailed fish with a GG genotype at position 67 (data not shown), and these genotypes were independently confirmed. These results indicate that mc1r is closely linked to the color locus, but 1.7 cM away from it. The marker to which mc1r is most closely linked is NfuFLI0030 (Figure 3B). The distances between mc1r and NfuFLI0030 (2 cM) and between mc1r and the color locus (1.7 cM) are not the same, because the former was calculated in both male and female F2 individuals whereas the latter was calculated only in red F2 males. The distance between mc1r and the color locus indicates that the Mc1r amino acid difference is unlikely to be causal for the yellow/red color determination in N. furzeri, although we cannot rule out the involvement of cis-acting elements for the mc1r gene in the determination of color. An alternative possibility is that another gene in close proximity to mc1r is involved in color determination in N. furzeri.
A sex determination system on linkage group XIII in N. furzeri:
Sex is another phenotype that differs between the two grandparents of our cross. To establish the sex determination system in N. furzeri, we counted the number of males and females in the F1 and F2 generations. The F1 and F2 offspring displayed an even sex ratio (F1: 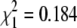 , P = 0.668; F2:
, P = 0.668; F2: 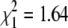 , P = 0.2) (Figure 4A), consistent with a genetic sex determination system. We sorted the genotypes by sex and searched for markers that show alleles that are present predominantly in males or in females. We identified nine microsatellite markers on LG XIII that were linked with sex. Interestingly, five of these makers (NfuSU0004, NfuSU0007, NfuSU0010, NfuSU0090, and NfuFLI0091) show a significant male-specific allelic bias in that most of the F2 males carry a combination of a fixed, male-specific allele and a nonfixed, non-sexually-biased allele (Table 2). Importantly, the male-biased markers are the only ones for which the male grandparent (GRZ) is heterozygous. These results strongly suggest that male is the heterogametic sex in N. furzeri and that the sex determination system is XY/XX. To analyze whether the male-specific allelic bias is shared by different N. furzeri strains, we genotyped the sex-linked markers in the F2 progeny (20 males and 14 females) of a reciprocal cross of a male MZM-0403 and a female GRZ (cross 2) and found a male-allelic bias at the same loci (Table 2). Together, these results indicate the presence in N. furzeri of a conserved haplotype for the male sex chromosome with marked sex-linkage disequilibrium.
, P = 0.2) (Figure 4A), consistent with a genetic sex determination system. We sorted the genotypes by sex and searched for markers that show alleles that are present predominantly in males or in females. We identified nine microsatellite markers on LG XIII that were linked with sex. Interestingly, five of these makers (NfuSU0004, NfuSU0007, NfuSU0010, NfuSU0090, and NfuFLI0091) show a significant male-specific allelic bias in that most of the F2 males carry a combination of a fixed, male-specific allele and a nonfixed, non-sexually-biased allele (Table 2). Importantly, the male-biased markers are the only ones for which the male grandparent (GRZ) is heterozygous. These results strongly suggest that male is the heterogametic sex in N. furzeri and that the sex determination system is XY/XX. To analyze whether the male-specific allelic bias is shared by different N. furzeri strains, we genotyped the sex-linked markers in the F2 progeny (20 males and 14 females) of a reciprocal cross of a male MZM-0403 and a female GRZ (cross 2) and found a male-allelic bias at the same loci (Table 2). Together, these results indicate the presence in N. furzeri of a conserved haplotype for the male sex chromosome with marked sex-linkage disequilibrium.
Figure 4.—
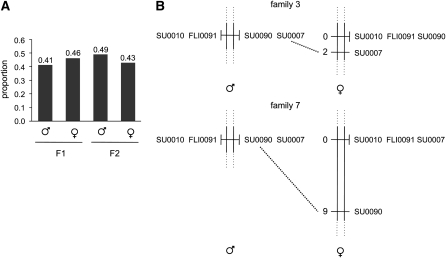
Sex determination in N. furzeri (A) The proportion of males and females in the F1 (23 males and 26 females) and F2 (203 males and 178 females) generations. The χ2 test shows that the ratio is 1:1 (F1 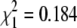 , P = 0.668; F2
, P = 0.668; F2 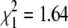 , P = 0.2). The individuals that could not be phenotyped as males or females (undetermined) are not reported; therefore the proportion of males and females does not add up to 1. (B) Recombination scores for males and females at LG XIII for two F1 families (family 3 = 22 individuals; family 7 = 46 individuals). In both families, F2 males do not show recombination at the four sex-linked markers NfuSU0010 (SU0010), NfuFLI0091 (FLI0091), NfuSU0007 (SU0007), and NfuSU0090 (SU0090). In contrast, F2 females show recombination at markers NfuSU0007 (SU0007) and NfuSU0090 (SU0090). Note that F2 females show recombination at different markers in family 3 and family 7, which is likely due to the low number of individuals that were genotyped. This figure displays only the markers on LG XIII that were heterozygous in the F1 parents of family 3 and family 7 and therefore allowed the analysis of male vs. female meioses. The partial representation of LG XIII is depicted by dotted lines. The map distance in centimorgans is reported on the left of the LGs.
, P = 0.2). The individuals that could not be phenotyped as males or females (undetermined) are not reported; therefore the proportion of males and females does not add up to 1. (B) Recombination scores for males and females at LG XIII for two F1 families (family 3 = 22 individuals; family 7 = 46 individuals). In both families, F2 males do not show recombination at the four sex-linked markers NfuSU0010 (SU0010), NfuFLI0091 (FLI0091), NfuSU0007 (SU0007), and NfuSU0090 (SU0090). In contrast, F2 females show recombination at markers NfuSU0007 (SU0007) and NfuSU0090 (SU0090). Note that F2 females show recombination at different markers in family 3 and family 7, which is likely due to the low number of individuals that were genotyped. This figure displays only the markers on LG XIII that were heterozygous in the F1 parents of family 3 and family 7 and therefore allowed the analysis of male vs. female meioses. The partial representation of LG XIII is depicted by dotted lines. The map distance in centimorgans is reported on the left of the LGs.
TABLE 2.
Genotype frequencies of sex-linked markers on the sex-determining region on LG XIII
| m1 allele (%) |
m2 allele (%) |
f allele (%) |
||||
|---|---|---|---|---|---|---|
| Markers | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ |
| Cross 1 | ||||||
| SU0048 | 0 | 0 | 77 | 27 | 23 | 73 |
| SU0010 | 50 | 0 | 23 | 24 | 27 | 76 |
| FLl0091 | 48 | 0 | 23 | 26 | 29 | 74 |
| SU0007 | 51 | 0 | 22 | 28 | 27 | 72 |
| SU0004 | 50 | 0 | 0 | 0 | 50 | 100 |
| SU0090 | 49 | 0 | 21 | 26 | 30 | 74 |
| FLl0055 | 0 | 0 | 80 | 28 | 20 | 72 |
| FLl0017 | 0 | 0 | 70 | 21 | 30 | 79 |
| SU0015 | 0 | 0 | 65 | 31 | 35 | 69 |
| Cross 2 | ||||||
| SU0010 | 50 | 0 | 15 | 25 | 35 | 75 |
| FLl0091 | 53 | 0 | 27 | 25 | 20 | 75 |
| SU0007 | 50 | 0 | 30 | 18 | 20 | 82 |
| SU0090 |
0 |
0 |
67 |
25 |
33 |
75 |
Genotype frequencies of sex-linked markers on LG XIII in 160 males and 86 females (cross 1) and in 20 males and 14 females (cross 2). m1 and m2 are alleles derived from the male grandparent. The m1 allele is exclusively present in F2 males. The f allele is derived from the female grandparent. Cross 1: GRZ male crossed with MZM-0403 female. Cross 2: MZM-0403 male crossed with GRZ female.
Chromosomal regions carrying a sex-determining locus are characterized by suppressed meiotic recombination (Charlesworth 2004; Marshall Graves 2008). To test whether recombination is suppressed at the sex-linked markers in N. furzeri, we independently calculated male and female meiotic recombination frequencies in 68 F2 offspring from two F1 families of our first cross (cross 1). We performed this analysis in F2 individuals instead of F1 individuals because the P0 MZM-0403 female was homozygous at all sex-linked loci. F1 females were heterozygous at the sex-linked loci, except at marker NfuSU0004, which was therefore excluded from the analysis. In F2 progeny from each of the two F1 families, there was no recombination event for markers NfuSU0090, NfuSU0007, NfuSU0010, and NfuFLI0091 in males, whereas females displayed a total of six recombination events, which account for a map distance of 2 cM for family 3 and 9 cM for family 7 at markers NfuSU0007 and NfuSU0090, respectively (Figure 4B). This result indicates that recombination is largely suppressed in males in the sex-linked region of LG XIII and further supports the conclusion that male is the heterogametic sex in N. furzeri.
To determine if the sex-linked markers in N. furzeri have similarities with sex-determining regions in other fish species, we searched for synteny between the nine microsatellite markers on LG XIII on N. furzeri and medaka chromosomes. Marker NfuSU0015 on LG XIII shows significant homology with a region on medaka chromosome 16 (BLASTn P = 1.5 × 10−7), although there is no annotated gene in this region. Interestingly, chromosome 16 corresponds to the sex chromosome in one medaka species (Oryzias javanicus) (Takehana et al. 2008), raising the interesting possibility that sex determination in N. furzeri and O. javanicus might have evolved from a common system. However, N. furzeri LG XIII is not syntenic with the sex-determining chromosome in another species of medaka (Oryzias latipes) (Matsuda et al. 2002; Kasahara et al. 2007) and with the sex-determining LGs in two stickleback species (Peichel et al. 2004; Ross and Peichel 2008; Shapiro et al. 2009), suggesting that the sex-determination system of N. furzeri probably arose independently of these species.
DISCUSSION
Linkage map in N. furzeri:
Our first-generation microsatellite-based linkage map consists of 25 LGs. The number of LGs is higher than the number of chromosomes visible in a metaphase spread (19) (Reichwald et al. 2009). First-generation linkage maps usually contain more LGs than chromosomes, and these LGs tend to collapse if more markers are added (Ohtsuka et al. 1999; Naruse et al. 2000). Thus, it is likely that the current LG number will eventually collapse to 19 if additional markers/meioses are added. Our analyses also show that the phylogenetic relationship between N. furzeri and medaka allows the use of sequence similarity and synteny to predict the location of specific genes on the N. furzeri map. The synteny between medaka and N. furzeri is likely to be high, given that medaka and the more distant stickleback genomes already exhibit remarkable synteny (http://oxgrid.angis.org.au/ensembl_grids/oxg__gacu_All__vs__olat_All__500.html). A high level of synteny will be particularly useful in identifying candidate genes underlying specific traits, as was recently described for the cave fish Astyanax mexicanus (Gross et al. 2008, 2009).
Color determination in N. furzeri:
Our linkage map allowed us to identify a locus linked with male yellow/red tail color on LG V in N. furzeri. Synteny analysis between N. furzeri and medaka revealed a potential candidate gene for color, mc1r (Rees 2003; Hoekstra 2006). The Mc1r protein is a G-protein-coupled receptor for the Agouti ligand Asip. In mice, mutations in either Mc1r or Asip result in changes in the pattern of melanogenesis and in coat color (Jackson 1993) and affect the relative amounts of eumelanin and pheomelanin in mammalian melanocytes (Andersson 2003). In humans, MC1R mutations are associated with red hair and fair skin (Valverde et al. 1995; Mundy 2009) and with the presence of freckles and skin sensitivity to sun (Sulem et al. 2007). However, while mc1r is important for the brown pigmentation in different populations of the cave fish A. mexicanus (Gross et al. 2009), whether mc1r plays any role in yellow/red pigmentation in fish is unknown. In fact, fish pigmentation is fundamentally different from that of mammals because it depends on several cell types, including xantophores and erythrophores, which can synthesize yellow pigments de novo (Parichy 2003; Braasch et al. 2007; Protas and Patel 2008).
The red and yellow grandparents of our cross have a nonsynonymous variation in the coding sequence of mc1r, leading to a change in amino acid in the Mc1r protein from an aspartic acid in the red strain to a histidine in the yellow strain. However, this nonsynonymous change in Mc1r does not map exactly at the color locus, but 1.7 cM from it, likely ruling out its direct functional implication in male coloration. This amino acid change is in the extracellular N-terminal domain of Mc1r, a region poorly conserved between different teleost species and not essential for ligand binding (Selz et al. 2007). This region does not contain any of the known Mc1r mutations that have previously been found to be causative for color differences in other species (Selz et al. 2007). Thus, at this point, we consider unlikely an implication of mc1r in fish yellow/red color determination, although we cannot exclude a potential involvement of mc1r via cis-regulation. It is more likely that another gene located in the same region is responsible for color determination in N. furzeri. Fine mapping will reveal whether color determination in N. furzeri is mediated by mc1r or by another gene.
Mapping the color locus will provide important cues for the evolution of a trait under strong sexual selection. In the genus Nothobranchius, coloration is specific to males and is likely shaped by female mate selection. For example, in Nothobranchius guentheri, females prefer conspecific males on the basis of color (Haas 1976). More intensely colored males are preferred over less intensely colored ones (Haas 1976). However, a bright coloration also renders males more conspicuous to predators and likely comes with a survival cost, in line with the observation that, in the wild, the sex ratio for this species is biased toward females, which are not colored (Haas 1976; Reichard et al. 2009). The balance between the fitness advantage due to mate preference and the fitness costs due to predation may underlie the rate of evolution of male coloration in this species. When we identify a gene or regulatory region underlying the difference in color in N. furzeri, it will be interesting to test whether natural populations of N. furzeri, as well as different species of the genus Nothobranchius, harbor polymorphisms in this region.
Sex determination in N. furzeri:
Sex can be determined by mechanisms that are genetic, environmental, or a combination of both (Volff 2005; Marshall Graves 2008). Environmental factors that control sex determination in fish species include water temperature, density, and social interactions. Genetic control of sex determination is governed by the presence of sex chromosomes (visible sex chromosomes or heteromorphic chromosomes) that can be present either in males (XY) or in females (ZW). We found that N. furzeri has a genetic sex-determination system, with males as the heterogametic sex, indicative of an XY/XX system. The male sex-determining region in N. furzeri harbors a nonrecombining region, similar to that in medaka, guppies, platyfish, and sticklebacks (Kondo et al. 2001; Matsuda et al. 2002; Nanda et al. 2002; Volff and Schartl 2002; Peichel et al. 2004; Schultheis et al. 2006; Ross and Peichel 2008; Shapiro et al. 2009; Tripathi et al. 2009a). In line with sticklebacks and medaka (Kondo et al. 2001; Peichel et al. 2004; Shapiro et al. 2009), the sex linkage group in N. furzeri does show major differences in recombination rates when computed independently for males and females, and N. furzeri males consistently share a sex haplotype. Synteny analysis between N. furzeri sex-linked markers and the known medaka and stickleback sex chromosomes revealed that the sex-linked LG in N. furzeri (LG XIII) is syntenic with the O. javanicus sex chromosome (chromosome 16), raising the interesting question of whether the sex-determining regions in these two different species were derived from the same ancestral chromosome or arose independently from one another.
Color and sex determination in N. furzeri:
Genes underlying sexually attractive traits, such as bright coloration, are often located on sex chromosomes (Lindholm and Breden 2002). In guppies, many of the male color traits (dorsal fin black, central blue white spot, anterior orange spot, etc.) map to the sex chromosome, although other color traits map to autosomes (Lindholm and Breden 2002; Tripathi et al. 2008, 2009b). Similarly, in platyfish (Xiphophorus maculatus), a number of color traits (iris color, body colors, fin color) map to sex chromosomes, while others (black comets on the fin) map to autosomes (Basolo 2006). Having color and sex linked may help maintain sexual dimorphism and provide an evolutionary advantage. In N. furzeri, male color does not map to the LG containing the sex-determining region, at least in our first-generation linkage map. This could be due to the incompleteness of the map: it is conceivable that LG V and LG XIII would merge if additional markers/specimens are used. Alternatively, color and sex-determining regions may segregate independently in N. furzeri, perhaps because the evolutionary advantage of linking male tail color and sex has not emerged yet in this species.
Life span, color, and sex determination in N. furzeri:
A major advantage of developing N. furzeri as a model system is its short life span and the presence of natural populations with differences in mean and maximal life span. Our cross did not allow us to map quantitative trait loci (QTL) for longevity because a large number of F2 fish died prematurely because of the unexpected presence of the parasite Glugea sp. in the fish housing system (data not shown). Nevertheless, preliminary data suggest that there is no simple link between the yellow/red color and longevity or between life span and sex (data not shown). Additional crosses performed in controlled environmental conditions will be needed for the mapping of QTL for longevity in N. furzeri.
Concluding remarks:
Our study reports the generation of the first linkage microsatellite-based map for the short-lived fish N. furzeri. This genetic tool allowed us to identify a single locus linked with color determination in males, as well as to reveal the sex-determination system of this species. This map will also be of key importance in determining the genetic architecture of other traits that characterize this unique group of organisms, particularly differences in life span.
Acknowledgments
We thank David Kingsley and Greg Barsh for intellectual input on the project. We thank Steve Arnott from the Kingsley lab for guidance for linkage mapping analysis. We thank Craig Miller, Katie Peichel, and Michael Shapiro for helpful suggestions throughout this project. We thank Sabrina Fullhart for her help with fish husbandry and fish pictures. We thank Tom Hofmann for expert technical assistance. We are grateful to the Kingsley lab for the use of the ABI sequencer. We thank members of the Brunet lab, as well as Steve Arnott, Greg Barsh, David Kingsley, Craig Miller, Katie Peichel, Dmitri Petrov, and Michael Shapiro for critically reading the manuscript. We thank Martin Reichard for communication of his findings on N. furzeri color in the wild. This work was supported by an R21 grant from National Institutes of Health/National Institute on Aging (AG030464) (A.B.), a fellowship from the Stanford Center on Longevity (D.R.V.), and by a pact for research and innovation of the Joint Science Conference of the federal and Länder governments from the Leibniz-Gemeinschaft, Bonn/Berlin, Germany (C.E. and M.P.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.108670/DC1.
References
- Alizadeh, A., L. Z. Hong, C. B. Kaelin, T. Raudsepp, H. Manuel et al., 2009. Genetics of sex-linked yellow in the Syrian hamster. Genetics 181 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, L., 2003. Melanocortin receptor variants with phenotypic effects in horse, pig, and chicken. Ann. NY Acad. Sci. 994 313–318. [DOI] [PubMed] [Google Scholar]
- Basolo, A. L., 2006. Genetic linkage and color polymorphism in the southern platyfish (Xiphophorus maculatus): a model system for studies of color pattern evolution. Zebrafish 3 65–83. [DOI] [PubMed] [Google Scholar]
- Bonilla, C., L. A. Boxill, S. A. Donald, T. Williams, N. Sylvester et al., 2005. The 8818G allele of the agouti signaling protein (ASIP) gene is ancestral and is associated with darker skin color in African Americans. Hum. Genet. 116 402–406. [DOI] [PubMed] [Google Scholar]
- Braasch, I., M. Schartl and J. N. Volff, 2007. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 7 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, R., and J. A. Endler, 2001. Direct and indirect sexual selection and quantitative genetics of male traits in guppies (Poecilia reticulata). Evolution 55 1002–1015. [DOI] [PubMed] [Google Scholar]
- Candille, S. I., C. B. Kaelin, B. M. Cattanach, B. Yu, D. A. Thompson et al., 2007. A defensin mutation causes black coat color in domestic dogs. Science 318 1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 2004. Sex determination: primitive Y chromosomes in fish. Curr. Biol. 14 R745–R747. [DOI] [PubMed] [Google Scholar]
- Geissler, E. N., M. A. Ryan and D. E. Housman, 1988. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 55 185–192. [DOI] [PubMed] [Google Scholar]
- Genade, T., M. Benedetti, E. Terzibasi, P. Roncaglia, D. R. Valenzano et al., 2005. Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell 4 223–233. [DOI] [PubMed] [Google Scholar]
- Gross, J. B., M. Protas, M. Conrad, P. E. Scheid, O. Vidal et al., 2008. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc. Natl. Acad. Sci. USA 105 20106–20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, J. B., R. Borowsky and C. J. Tabin, 2009. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 5 e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, R., 1976. Sexual selection in Nothobranchius guentheri (Pisces: cyprinodontidae). Evolution 30 614–622. [DOI] [PubMed] [Google Scholar]
- Han, J., P. Kraft, H. Nan, Q. Guo, C. Chen et al., 2008. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4 e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, N., K. Reichwald, A. Lechel, M. Graf, J. Kirschner et al., 2009. Telomeres shorten while Tert expression increases during ageing of the short-lived fish Nothobranchius furzeri. Mech Ageing Dev 130 290–296. [DOI] [PubMed] [Google Scholar]
- Hoekstra, H. E., 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97 222–234. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., L. Du, F. H. Rodd and D. N. Reznick, 1999. Familiarity leads to female mate preference for novel males in the guppy, Poecilia reticulata. Anim. Behav. 58 907–916. [DOI] [PubMed] [Google Scholar]
- Inglima, K., A. Perlmutter and J. Markofsky, 1981. Reversible stage-specific embryonic inhibition mediated by the presence of adults in the annual fish Nothobranchius guentheri. J. Exp. Zool. 215 23–33. [DOI] [PubMed] [Google Scholar]
- Jackson, I. J., 1993. Molecular genetics: colour-coded switches. Nature 362 587–588. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., K. Naruse, S. Sasaki, Y. Nakatani, W. Qu et al., 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447 714–719. [DOI] [PubMed] [Google Scholar]
- Khoo, G., M. H. Lim, H. Suresh, D. K. Gan, K. F. Lim et al., 2003. Genetic linkage maps of the guppy (Poecilia reticulata): assignment of RAPD markers to multipoint linkage groups. Mar. Biotechnol. 5 279–293. [DOI] [PubMed] [Google Scholar]
- Kondo, M., E. Nagao, H. Mitani and A. Shima, 2001. Differences in recombination frequencies during female and male meioses of the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 78 23–30. [DOI] [PubMed] [Google Scholar]
- Lamason, R. L., M. A. Mohideen, J. R. Mest, A. C. Wong, H. L. Norton et al., 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310 1782–1786. [DOI] [PubMed] [Google Scholar]
- Le Pape, E., T. Passeron, A. Giubellino, J. C. Valencia, R. Wolber et al., 2009. Microarray analysis sheds light on the dedifferentiating role of agouti signal protein in murine melanocytes via the Mc1r. Proc. Natl. Acad. Sci. USA 106 1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm, A., and F. Breden, 2002. Sex chromosomes and sexual selection in poeciliid fishes. Am. Nat. 160(Suppl. 6): S214–S224. [DOI] [PubMed] [Google Scholar]
- Maan, M. E., O. Seehausen, L. Soderberg, L. Johnson, E. A. Ripmeester et al., 2004. Intraspecific sexual selection on a speciation trait, male coloration, in the Lake Victoria cichlid Pundamilia nyererei. Proc. Biol. Sci. 271 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Graves, J. A., 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42 565–586. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559–563. [DOI] [PubMed] [Google Scholar]
- Miller, C. T., S. Beleza, A. A. Pollen, D. Schluter, R. A. Kittles et al., 2007. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy, N. I., 2009. Conservation and convergence of colour genetics: MC1R mutations in brown cavefish. PLoS Genet. 5 e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse, K., S. Fukamachi, H. Mitani, M. Kondo, T. Matsuoka et al., 2000. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics 154 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton, H. L., R. A. Kittles, E. Parra, P. McKeigue, X. Mao et al., 2007. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 24 710–722. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, M., S. Makino, K. Yoda, H. Wada, K. Naruse et al., 1999. Construction of a linkage map of the medaka (Oryzias latipes) and mapping of the Da mutant locus defective in dorsoventral patterning. Genome Res. 9 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy, D. M., 2003. Pigment patterns: fish in stripes and spots. Curr. Biol. 13 R947–R950. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., K. S. Nereng, K. A. Ohgi, B. L. Cole, P. F. Colosimo et al., 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414 901–905. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14 1416–1424. [DOI] [PubMed] [Google Scholar]
- Price, A. C., C. J. Weadick, J. Shim and F. H. Rodd, 2008. Pigments, patterns, and fish behavior. Zebrafish 5 297–307. [DOI] [PubMed] [Google Scholar]
- Protas, M. E., and N. H. Patel, 2008. Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24 425–446. [DOI] [PubMed] [Google Scholar]
- Rees, J. L., 2003. Genetics of hair and skin color. Annu. Rev. Genet. 37 67–90. [DOI] [PubMed] [Google Scholar]
- Reichard, M., M. Polacik and O. Sedlacek, 2009. Distribution, colour polymorphism and habitat use of the African killifish Nothobranchius furzeri, the vertebrate with the shortest lifespan. J. Fish Biol. 74 198–212. [DOI] [PubMed] [Google Scholar]
- Reichwald, K., C. Lauber, I. Nanda, J. Kirschner, N. Hartmann et al., 2009. High tandem repeat content in the genome of the short-lived annual fish Nothobranchius furzeri: a new vertebrate model for aging research. Genome Biol. 10 R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold, M., K. Lahiri, N. S. Foulkes and J. Wittbrodt, 2006. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. Nat. Protoc. 1 1133–1139. [DOI] [PubMed] [Google Scholar]
- Ross, J. A., and C. L. Peichel, 2008. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 179 2173–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke, M., 2000. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18 233–234. [DOI] [PubMed] [Google Scholar]
- Schultheis, C., Q. Zhou, A. Froschauer, I. Nanda, Y. Selz et al., 2006. Molecular analysis of the sex-determining region of the platyfish Xiphophorus maculatus. Zebrafish 3 299–309. [DOI] [PubMed] [Google Scholar]
- Selz, Y., I. Braasch, C. Hoffmann, C. Schmidt, C. Schultheis et al., 2007. Evolution of melanocortin receptors in teleost fish: the melanocortin type I receptor. Gene 401 114–122. [DOI] [PubMed] [Google Scholar]
- Shapiro, M. D., B. R. Summers, S. Balabhadra, J. T. Aldenhoven, A. L. Miller et al., 2009. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr. Biol. 19 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver, M. D., E. J. Parra, S. Dios, C. Bonilla, H. Norton et al., 2003. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum. Genet. 112 387–399. [DOI] [PubMed] [Google Scholar]
- Sulem, P., D. F. Gudbjartsson, S. N. Stacey, A. Helgason, T. Rafnar et al., 2007. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 39 1443–1452. [DOI] [PubMed] [Google Scholar]
- Sulem, P., D. F. Gudbjartsson, S. N. Stacey, A. Helgason, T. Rafnar et al., 2008. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 40 835–837. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., S. Hamaguchi and M. Sakaizumi, 2008. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi. Chromosome Res. 16 801–811. [DOI] [PubMed] [Google Scholar]
- Terzibasi, E., D. R. Valenzano, M. Benedetti, P. Roncaglia, A. Cattaneo et al., 2008. Large differences in aging phenotype between strains of the short-lived annual fish Nothobranchius furzeri. PLoS One 3 e3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzibasi, E., C. Lefrancois, P. Domenici, N. Hartmann, M. Graf et al., 2009. Effects of dietary restriction on mortality and age-related phenotypes in the short-lived fish Nothobranchius furzeri. Aging Cell 8 88–99. [DOI] [PubMed] [Google Scholar]
- Tripathi, N., M. Hoffmann and C. Dreyer, 2008. Natural variation of male ornamental traits of the guppy, Poecilia reticulata. Zebrafish 5 265–278. [DOI] [PubMed] [Google Scholar]
- Tripathi, N., M. Hoffmann, D. Weigel and C. Dreyer, 2009. a Linkage analysis reveals the independent origin of Poeciliid sex chromosomes and a case of atypical sex inheritance in the guppy (Poecilia reticulata). Genetics 182 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, N., M. Hoffmann, E. M. Willing, C. Lanz, D. Weigel et al., 2009. b Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc. Biol. Sci. 276 2195–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdesalici, S., and A. Cellerino, 2003. Extremely short lifespan in the annual fish Nothobranchius furzeri. Proc. Biol. Sci. 270(Suppl. 2): S189–S191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano, D. R., E. Terzibasi, T. Genade, A. Cattaneo, L. Domenici et al., 2006. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 16 296–300. [DOI] [PubMed] [Google Scholar]
- Valverde, P., E. Healy, I. Jackson, J. L. Rees and A. J. Thody, 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 11 328–330. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Volff, J. N., 2005. Genome evolution and biodiversity in teleost fish. Heredity 94 280–294. [DOI] [PubMed] [Google Scholar]
- Volff, J. N., and M. Schartl, 2002. Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus. Cytogenet. Genome Res. 99 170–177. [DOI] [PubMed] [Google Scholar]
- von Hofsten, J., and P. E. Olsson, 2005. Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes. Reprod. Biol. Endocrinol. 3 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, H., K. Naruse, A. Shimada and A. Shima, 1995. Genetic linkage map of a fish, the Japanese medaka Oryzias latipes. Mol. Mar. Biol. Biotechnol. 4 269–274. [PubMed] [Google Scholar]
- Walter, R. B., J. D. Rains, J. E. Russell, T. M. Guerra, C. Daniels et al., 2004. A microsatellite genetic linkage map for Xiphophorus. Genetics 168 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildekamp, R. H., 2009. A World of Killies. American Killifish Association, USA.
- Wourms, J. P., 1972. The developmental biology of annual fishes. 3. Pre-embryonic and embryonic diapause of variable duration in the eggs of annual fishes. J. Exp. Zool. 182 389–414. [DOI] [PubMed] [Google Scholar]