Abstract
Doubly uniparental inheritance (DUI) of mitochondrial DNA in marine mussels (Mytiloida), freshwater mussels (Unionoida), and marine clams (Veneroida) is the only known exception to the general rule of strict maternal transmission of mtDNA in animals. DUI is characterized by the presence of gender-associated mitochondrial DNA lineages that are inherited through males (male-transmitted or M types) or females (female-transmitted or F types), respectively. This unusual system constitutes an excellent model for studying basic aspects of mitochondrial DNA inheritance and the evolution of mtDNA genomes in general. Here we compare published mitochondrial genomes of unionoid bivalve species with DUI, with an emphasis on characterizing unassigned regions, to identify regions of the F and M mtDNA genomes that could (i) play a role in replication or transcription of the mtDNA molecule and/or (ii) determine whether a genome will be transmitted via the female or the male gamete. Our results reveal the presence of one F-specific and one M-specific open reading frames (ORFs), and we hypothesize that they play a role in the transmission and/or gender-specific adaptive functions of the M and F mtDNA genomes in unionoid bivalves. Three major unassigned regions shared among all F and M unionoid genomes have also been identified, and our results indicate that (i) two of them are potential heavy-strand control regions (OH) for regulating replication and/or transcription and that (ii) multiple and potentially bidirectional light-strand origins of replication (OL) are present in unionoid F and M mitochondrial genomes. We propose that unassigned regions are the most promising candidate sequences in which to find regulatory and/or gender-specific sequences that could determine whether a mitochondrial genome will be maternally or paternally transmitted.
MARINE mussels (Mytiloida), freshwater mussels (Unionoida), and marine clams (Veneroida) are the only known animals that do not transmit their mitochondrial DNA exclusively maternally (see White et al. 2008 for a review of exceptional cases of paternal leakage in animals). The system of mitochondrial DNA transmission in these bivalves is referred to as “doubly uniparental inheritance” (DUI) and is characterized by the presence of two gender-associated mitochondrial DNA lineages that are inherited through males (male transmitted or M types) or females (female transmitted or F types), respectively (see Breton et al. 2007 and Passamonti and Ghiselli 2009 for reviews of DUI). DUI constitutes an excellent model system for studying basic aspects of mitochondrial DNA inheritance and the evolution of mtDNA genomes in general. Because DUI is the exception to the rule, understanding how bivalves evolved distinct male and female mtDNA lineages can provide important insights into the evolutionary forces that maintain strictly maternal inheritance in most animals.
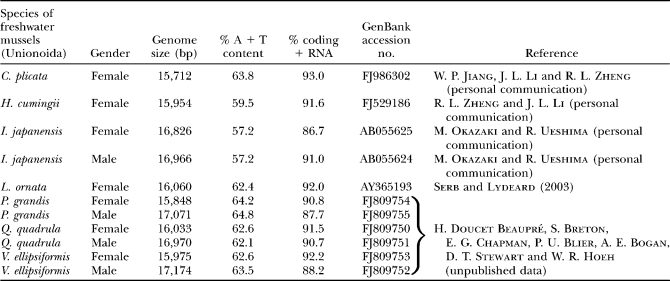
To date, complete F and M mtDNA sequences have been determined for the mytiloid mussels Mytilus edulis, M. galloprovincialis, and M. trossulus (AY484747, Boore et al. 2004; AY497292 and AY363687, Mizi et al. 2005; AY823623 and AY323624, Breton et al. 2006; and DQ198231 and DQ198225, Zbawicka et al. 2007); the veneroid clam Venerupis philippinarum (AB065374 and AB065375, M. Okazaki and R. Ueshima personal communication); and seven unionoid bivalve species [i.e., the F genome of Lampsilis ornata (Unionoida: Ambleminae: Lampsilini) (AY365193, Serb and Lydeard 2003), the F genome of Hyriopsis cumingii (Unionoida: Ambleminae: Gonideini) (FJ529186, R. L. Zheng and J. L. Li, personal communication), the F genome of Cristaria plicata (Unionoida: Anodontinae: Anodontini) (FJ986302, W. P. Jiang, R. L. Zheng and J. L. Li, personal communication), the F and M genomes of Inversidens japanensis (Unionoida: Ambleminae: Gonideini) (AB055624 and AB055625, M. Okazaki and R. Ueshima, personal communication), and recently, we have sequenced the F and M genomes of Venustaconcha ellipsiformis (Unionoida: Ambleminae: Lampsilini), Quadrula quadrula (Unionoida: Ambleminae: Quadrulini), and Pyganodon grandis (Unionoida: Anodontinae: Anodontini) (FJ809750–FJ809755, H. Doucet Beaupré, S. Breton, E. G. Chapman, P. U. Blier, A. E. Bogan, D. T. Stewart and W. R. Hoeh, unpublished data)] (Table 1). Overall, these studies have shown (i) a high level of nucleotide sequence divergence but nearly identical gene content between F and M genomes within each species, (ii) an accelerated rate of molecular evolution of both the M and the F genomes compared to other animal mitochondrial genomes, (iii) an accelerated rate of molecular evolution of M genomes compared to F genomes, (iv) an absence of atp8 in mytiloid mussels, (v) the presence of a second trnM (i.e., transfer RNA gene for methionine) in mytiloid and veneroid bivalves, (vi) recombination between M and F genomes in mytiloid mussels, (vii) periodic “role reversals” (masculinization) of female-transmitted mtDNA in mytiloid mussels that are subsequently transmitted through sperm, (viii) different gene order between F and M genomes in unionoids, and (ix) the presence of a unique extension of the cytochrome c oxidase subunit II gene in the M (but not the F) genome in unionoids (reviewed in Breton et al. 2007; see also Chapman et al. 2008). An important hypothesis that has emerged from sequencing studies of species with DUI is that gender-specific sequences and/or sequences that exhibit the highest level of nucleotide divergence between the F and M genomes (i.e., regions that are under different, potentially gender-specific selective constraints and could, therefore, have different roles in either genome) are the most likely candidates for determining whether a mitochondrial genome will be transmitted maternally or paternally (Zouros 2000; Burzyński et al. 2003; Cao et al. 2004a; Breton et al. 2006, 2007; Theologidis et al. 2007; Venetis et al. 2007; Cao et al. 2009). For example, it has been demonstrated in marine mussels that masculinized type genomes (i.e., an F genome that “masculinizes” and takes on the role of the previous M genome) are essentially recombinants composed of an F genome's coding and control regions, with an additional standard M-type control region (Burzyński et al. 2003; Breton et al. 2006; Venetis et al. 2007). This has led to the proposition that an M-type control region, particularly its most variable domain called VD1 by Cao et al. (2004a), might be necessary to confer the paternal role on genomes that are otherwise F-like (Burzyński et al. 2003; Breton et al. 2006; Venetis et al. 2007). Alternatively, the absence of masculinization or role-reversal events in freshwater bivalves coincides with the presence of a unique M genome-specific 3′ extension of the cytochrome c oxidase subunit 2 gene (Mcox2e; Curole and Kocher 2002) that could facilitate the transmission of the M genomes in freshwater bivalves (Breton et al. 2007; Chakrabarti et al. 2007; Chapman et al. 2008). To date, the control region has not been confirmed in unionoid bivalves (see Serb and Lydeard 2003). Identifying the unionoid F and M control regions is essential for a more complete understanding of DUI. Studies of mytiloid genomes that have switched from maternal to paternal transmission have provided evidence that specific sequences of the mtDNA genome that control the mode of inheritance (i.e., male or female transmission) are located in the control region (Burzyński et al. 2003; Breton et al. 2006; Venetis et al. 2007; Cao et al. 2009). It is therefore critical that the control region in unionoids be confirmed to facilitate comparative studies of the developmental control of the F and M genomes across all bivalve species that possess DUI.
TABLE 1.
Gender-associated mitochondrial genomes in unionoid bivalve species with DUI
Here we present a comparative analysis of complete F and M mitochondrial genomes of unionoid bivalve species with DUI. Our objectives are to highlight both unique features and characteristics shared among different species, with an emphasis on characterizing unassigned regions (i.e., noncoding regions that are functionally unassigned) to identify F and M sequences that could play crucial roles in replication or transcription of the mtDNA molecule and to point out particular regions of the genomes that could determine whether a genome will be transmitted by eggs or sperm. One F-specific and one M-specific open reading frames (ORFs) have been identified and, given their expression and antiquity in unionoid bivalves, we hypothesize that they are involved in the different modes of transmission and/or gender-specific adaptive functions of the M and F mtDNA genomes in unionoid bivalves. Additionally, our results reveal that unionoid mitochondrial control regions are not well defined and their locations could be variable among unionoid mitochondrial genomes. We propose that the currently unassigned regions are the most favorable candidates in which to find regulatory or gender-specific sequences that could determine whether a genome is transmitted maternally or paternally.
MATERIALS AND METHODS
Complete F and M mitochondrial sequences of unionoid bivalve species with DUI were obtained from the National Center for Biotechnology Information (NCBI) GenBank entries for the 11 genomes listed in Table 1 (it must be noted that the M genomes of C. plicata, H. cumingii, and L. ornata have not been sequenced). ClustalW (Thompson et al. 1994) was used to align sequences and MEGA 3.0 (Kumar et al. 2004) was used to calculate the proportion of nucleotide and amino acid differences (p-distances) between F and M genes and AT skew = (A − T)/(A + T) (Perna and Kocher 1995) at fourfold redundant sites for each mitochondrial protein-coding gene. Repeated elements were identified using REPFIND (Betley et al. 2002), and conserved motifs were identified using Dialign version 2.2.1 (Subramanian et al. 2008) and Jalview version 2 (Waterhouse et al. 2009). DNA secondary structures in unassigned regions were predicted using Mfold version 3.2 (Zuker 2003). Examination of ORFs was performed using the NCBI ORF Finder program (http://www.ncbi.nlm.nih.gov/projects/gorf/) with the invertebrate mitochondrial genetic code. Sequence similarity searches were performed in GenBank using BLASTN (Benson et al. 2004), BLASTX, and PSI-BLAST (Altschul et al. 1997). Gender-specific ORFs were examined using Fickett's (1982) test code algorithm. Transmembrane helices and other potential protein features of gender-specific ORFs were identified using ConPred II (Arai et al. 2004) and PredictProtein (Rost et al. 2004). Western blot analyses of eggs and testes extracts of the species V. ellipsiformis were performed as described by Chakrabarti et al. (2006).
In addition to the customary characteristics used to identify the mitochondrial control region of replication in animals (i.e., the largest noncoding region, increased AT content, and presence of repetitive elements and secondary structures) (Boore 1999; Saccone et al. 2002; Cao et al. 2004a; Saito et al. 2005; Kuhn et al. 2006; Oliveira et al. 2007; Brugler and France 2008), we also used AT-skew values of protein-coding genes at fourfold redundant sites to locate the origins of heavy (OH) and light (OL) strand replication in freshwater bivalves. In most metazoans, the mitochondrial DNA genome replicates with a strand-asynchronous, asymmetric mechanism during which the parental heavy (H) strand becomes temporarily single-stranded DNA (ssDNA) while the nascent H strand is synthesized, and when the heavy strand synthesis reaches two-thirds of the genome, it exposes the OL and initiates the synthesis of a new light (L) strand in the opposite direction (Clayton 1982; Reyes et al. 1998). Strong biases toward A + C for the L strand and G + T for the H strand are common in mitochondrial genomes and are associated with this asymmetrical replication and the extended time that the parental heavy strand spends in the mutagenically susceptible single-stranded state during the process (Reyes et al. 1998; Saccone et al. 2002; Faith and Pollock 2003). This mutational bias appears to be due to (i) spontaneous deamination of C on the single-stranded H strand that produces U, which DNA polymerase basepairs with A rather than G (consequently the percentage of C decreases and that of T increases on the H strand according to single-strand exposure), (ii) deamination of A that produces hypoxanthin, which basepairs with C rather than T (A decreases and G increases on the H strand), and (iii) oxidation of guanine on the H strand that produces 8-hydroxyguanine, which basepairs with A rather than C (G decreases and T increases on the H strand) (Reyes et al. 1998; Saccone et al. 2002; Faith and Pollock 2003; Rodakis et al. 2007). The net effect on nucleotide frequency is that cytosine and adenine may only decrease on the H strand, whereas thymine may only increase and guanine could either increase or decrease as a result of single-strand exposure (Rodakis et al. 2007). Because the increase or decrease of G on the H strand will influence the amount of C on the L strand, we consider only AT-skew values in the following section.
According to the formula AT skew = (A − T)/(A + T), skew values are distributed in the range of −1 to +1 and, thus, the compositional asymmetry increases when the absolute skew values approach one and decreases when the skew values approach zero (Saccone et al. 2002). The genes located in the vicinity of the OH remain in a single-stranded state for a relatively long time during the replication process and these regions therefore experience more mutations (Faith and Pollock 2003; Fonseca et al. 2008). Consequently, the compositional asymmetry should be greater for the genes closer to the OH (the genes closer to the OH should be less A rich and more T rich when encoded on the heavy strand and more A rich and less T rich when encoded on the light strand), and thus AT-skew values should be greater for these genes. Alternatively, the genes closer to the OL, in the direction of the L-strand synthesis, remain exposed to mutation for less time so the genes closer to the OL should present a lower compositional asymmetry with lower skew values (these genes should be more A rich and less T rich when encoded on the heavy strand and less A rich and more T rich when encoded on the light strand) (Faith and Pollock 2003; Fonseca et al. 2008). If the asymmetrical model of mtDNA replication also applies to unionoid bivalves, we can speculate that OH would be located in the vinicity of the protein-coding genes showing the greatest AT-skew value at fourfold redundant sites, while OL would be located in the vinicity of the protein-coding genes showing the lowest AT-skew values.
RESULTS AND DISCUSSION
Unassigned regions in F and M unionoid mitochondrial genomes:
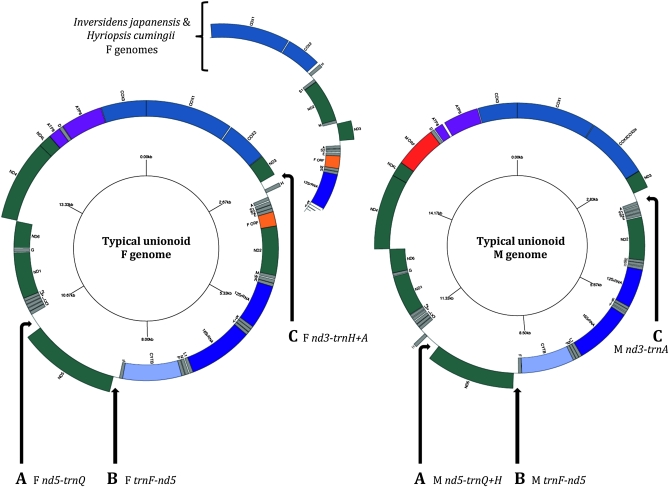
All analyzed unionoid mitochondrial genomes contain the 37 genes typically found in metazoans (Figure 1). Eleven genes (i.e., cox1, cox3, atp6, atp8, trnD, nd4l, nd4, trnH, nd5, nd3, and cox2/cox2e) are encoded on the heavy strand (H strand: G + T = ∼65 and 67% for the F and M genomes, respectively), while the remaining 26 genes are encoded on the light strand (L strand: G + T = ∼35 and 33% for the F and M genomes, respectively) (Figure 1). The only exception observed within the M lineage is the absence of a complete atp8 gene in P. grandis (H. Doucet Beaupré et al., unpublished data). In the unionoid F lineage, the mtDNAs of I. japanensis and H. cumingii share a different gene order between cox2 and 12SrRNA that could represent a derived characteristic of the F lineage in the Gonideini (Unionidae: Ambleminae). The transposition of trnH, the gene order inversion of trnD and atp8, and one F- and one M-specific open reading frames (gender-specific ORFs; see below) are responsible for the organizational differences between F and M genomes in freshwater bivalves (Figure 1). The M-specific cox2 extension also contributes to differences between F and M genomes (Figure 1).
Figure 1.—
Gene maps of the gender-associated mitochondrial genomes of unionoid mussels. Gene identities: nd1–6 and nd4l, NADH dehydrogenase subunits 1–6 and 4L (complex I in green); cytb, cytochrome b (complex III in light blue); cox1-3, cytochrome c oxidase subunits I–III (complex IV in blue); atp6 and atp8, ATP synthase subunits 6 and 8 (complex V in light purple); 12SrRNA and 16SrRNA, small and large subunits of ribosomal RNA (in purple). Transfer RNA genes (in gray) are depicted by one-letter amino acid codes; L1, L2, S1, and S2 are differentiated by their anticodon sequences CUA, UAA, AGA and UCA, respectively. F ORF, F-specific open reading frame (orange); M ORF, M-specific open reading frame (red). Genes positioned inside the white circle are encoded on the light strand and genes outside the circle are encoded on the heavy strand. Arrows A, B, and C indicate shared unassigned regions >20 bp between F and M unionoid genomes [i.e., A, nd5–trnQ (this region contains trnH in M genomes); B, trnF–nd5; and C, between nd3–trnA (this region contains trnH in P. grandis, Q. quadrula, and V. ellipsiformis F genomes)].
In total, 22–33 unassigned regions are found in F and M unionoid genomes, encompassing a total of 1148–2241 bp (F genomes) and 1526–2096 bp (M genomes), i.e., ∼10% of the total length of the mitochondrial genomes (Figure 1 and supporting information, Table S1). Similar results have been observed in mytiloid F and M genomes (i.e., ∼10% unassigned sequences) (Mizi et al. 2005; Breton et al. 2006; Zbawicka et al. 2007). In comparison, a higher proportion of unassigned sequences (i.e., >15.8% for F and >21.3% for M) is observed in the F and M genomes of the veneroid clam V. philippinarum (M. Okazaki and R. Ueshima, personal communication). Multiple intergenic regions are not unusual in mitochondrial genomes that have undergone significant rearrangements (Boore 1999). These regions often represent vestiges of pseudogenes generated by gene duplication followed by random deletions, a process that appears to be particularly important in molluscan taxa (Serb and Lydeard 2003; Akasaki et al. 2006; Boore 2006).
An examination of all complete F and M genomes shows three unassigned regions in the same relative genomic positions that are >20 nucleotides in length in all species (Figure 1, regions A, B, and C, and Table S1). We refer to these as “shared unassigned regions.” They include the regions between trnF and nd5, nd3 and trnA, and nd5 and trnQ (referred here as trnF–nd5, nd3–trnA, and nd5–trnQ). Because the nd3–trnA region contains trnH in F genomes of L. ornata, C. plicata, P. grandis, Q. quadrula, and V. ellipsiformis, we refer to it as nd3–trNa+H in F genomes. Because the nd5–trnQ region contains trnH in M genomes, we refer to it as nd5–trnQ+H in M genomes (see Figure 1, A–C, and Table S1). Interestingly, these three regions correspond to segments of the genome with a change in the direction of transcription (Figure 1). As is explained below, this arrangement marks these three regions as potential candidates for control regions (i.e., regions that contain elements involved in the regulation of replication and/or transcription of mtDNA; e.g., Boore 2006). In contrast, some unassigned regions are unique to either the F or the M unionoid genome. One relatively large unassigned region specific to the F genomes is found between trnE–nd2 (the F ORF, see below), except for the F mtDNAs of H. cumingii and I. japanensis, which possess a different gene order between cox2 and 12SrRNA and instead contain their large gender-specific unassigned regions between trnE–trnW. All unionoid M genomes possess two relatively large unassigned regions located between nd4l–trnD and atp8–atp6 [the M ORF and a noncoding region, which might serve as an origin of light strand replication (OL) in some species, see below] (Figure 1 and Table S1).
Further examination of the three shared and gender-specific “F trnE–nd2” (trnE–trnW for F I. japanensis and H. cumingii) and “M nd4l–trnD” unassigned regions reveals two categories of sequences. The first category contains sequences that possess an ORF of considerable length (i.e., the ORF makes up most of the length of the unassigned sequence) and the second category contains sequences exhibiting many characteristics typically associated with the animal mitochondrial control regions, such as presence of repeat units and sequences that can form stem–loop and hairpin structures. Because the gender-specific regions fall into the first category and the shared unassigned regions fall into the second category, we interpret the latter sequences as potential control regions. The results of the analyses of F- and M-specific ORFs and shared unassigned regions are presented in two different subsections below.
Gender-specific ORFs in F and M unionoid genomes:
The identification of gender-specific ORFs of significant length in unassigned regions [F trnE–nd2 (trnE–trnW for F I. japanensis and H. cumingii) and M trnD–nd4l; Figures 1 and 2] is particularly interesting because they could represent regulatory sequences or proteins that could be responsible for the different mode of transmission of the mtDNAs and/or gender-specific adaptive functions of the M and F mtDNA genomes in unionoid bivalves. A role in the differential segregation patterns of sperm mitochondria in mussel embryos has already been proposed for the M-specific cox2 extension that is present only in unionoid bivalve M genomes (Breton et al. 2007; Chakrabarti et al. 2007; Chapman et al. 2008). The unionoid MCOX2 protein, which is extremely variable in both its amino acid sequence and number of near C-terminal transmembrane helices among different species (Curole and Kocher 2002, 2005; Chapman et al. 2008), has been localized to both inner and outer mitochondrial membranes in sperm (Chakrabarti et al. 2007) and appears to function in reproduction (Chakrabarti et al. 2006, 2007; Chapman et al. 2008). It has been proposed that the localization of MCOX2 to the outer mitochondrial membranes likely “tags” the outer surface of unionoid M genome-bearing mitochondria and facilitates the distinct movements of the M genome-containing mitochondria, derived from the fertilizing sperm, in male and female embryos [as observed in Mytilus (Cao et al. 2004b; Cogswell et al. 2006)]. As is the case for MCOX2 (Chapman et al. 2008), no significant amino acid sequence similarity is detected with known proteins for the two new gender-specific ORFs using BLASTX and PSI-BLAST, and currently their identities/functions remain unclear. The only exception is the M ORF sequence of V. ellipsiformis, in which a putative conserved seryl-tRNA synthetase domain has been detected. Specifically, the M ORF of V. ellipsiformis exhibits a moderate degree of sequence similarity (E-value of 0.003) with the N-terminal nucleotide-binding domain of the seryl-tRNA synthetase, indicating that it could potentially be a DNA- or RNA-binding protein. Interestingly, many of the “extra” ORFs discovered in invertebrate mitochondrial genomes contain amino acid patterns characteristic of interaction with DNA (e.g., Pont-Kingdon et al. 1995, 1998; Shao et al. 2006; Gissi et al. 2008). Sequence comparisons among M ORFs reveal high variability in length (Figure 2) and a low extent of amino acid sequence similarity (∼20% similarity between each species pair comparison). Notably, a single transmembrane helix (TMH) is predicted in the 5′ half of each M ORF, and several positively charged amino acids are observed in the region following the predicted TMH (Figure 2). Amino acid sequence comparisons among F ORFs reveal that a greater degree of similarity exists, compared with M ORFs. The highest identity (∼60% amino acid identity) is observed between F ORFs from two of the most closely related species (L. ornata and V. ellipsiformis; for comparison, 72% amino acid identity is found in the highly variable atp8 gene for these two species). Between these two species, the divergence at the nucleotide level follows the pattern expected for a protein-coding gene evolving under purifying selection, i.e., where third and second codon positions exhibit the largest and smallest numbers of substitutions, respectively. Specifically, 25 (0.082%), 18 (0.059%), and 31 (0.102%) substitutions are identified in the first, second, and third codon positions, respectively. Analyses of F ORF protein sequences indicate variability in length (Figure 2), yet a single predicted transmembrane helix is present in a conserved position (i.e., in the 5′ half of the F ORFs; Figure 2) that is typically followed by a casein kinase II phosphorylation motif (data not shown).
Figure 2.—
Amino acid sequences of the potential peptides encoded by gender-specific open reading frames (F and M ORFs). The amino acids that constitute the putative transmembrane helix are indicated in boldface type and bigger characters. Positively charged amino acids are in red. The synthesized antigenic peptides used to generate antibodies for the Western blots are underlined.
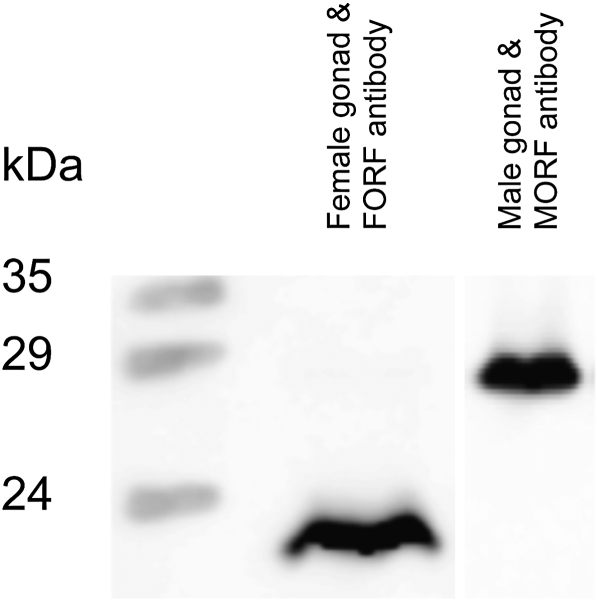
There are multiple lines of evidence indicating functionality of both novel gender-specific ORFs, likely as expressed proteins. The first evidence comes from the testcode analysis (Fickett 1982), which is used to identify coding regions on the basis of nonrandom nucleotide distributions at the third codon positions within a reading frame. Except for the F ORFs of I. japanensis and P. grandis (both ORFs having 40% probability of being protein-coding sequences), all of the unionoid F and M ORFs have been classified as protein coding or as having >77% probability of being protein-coding sequences. More importantly, Western blot analyses, using antibodies generated against peptides synthesized from the predicted F and M ORF amino acid sequences of V. ellipsiformis, indicated that the F-specific protein is effectively expressed in the female gonad and the M-specific protein is expressed in the male gonad (Figure 3).
Figure 3.—
Expression of the F and M ORFs in female and male gonads, respectively, from V. ellipsiformis. Western blots are revealed with anti-F ORF and anti-M ORF antibodies. The origins of the tissue samples for each lane are indicated. The positions of marker proteins (kDa) are shown.
To date, all analyzed F and M unionoid genomes were found to harbor an F ORF and an M ORF, respectively. The hypothesized universal presence of the two ORFs in unionoid F and M mitochondrial genomes, when viewed in the light of the unionoid fossil record (Watters 2001), indicates an origin for the ORFs >100 million years ago (MYA). Again, the retention of these ORFs in the conserved position in multiple distantly related genomes for >100 MY, as well as evidence for evolutionary relatedness of two F ORFs from two most closely related species (i.e., 60% identity at the protein level) (Chung and Subbiah 1996; Pearson 1996), argue in favor of their functionality. However, as is the case for the M-specific cox2 extension (Chapman et al. 2008), the original sources of the DNA comprising the F and M ORF regions may remain unidentified given the extremely high substitution rate in these regions and the relatively large amount of time over which these regions have accumulated substitutions.
Further, while comparisons of ORFs from distantly related taxa yield percentages of identity values in the so-called “twilight zone” of 20–25%, it is possible that they retain similar three-dimensional folds (Chung and Subbiah 1996). Indeed, the prediction of one transmembrane helix in similar positions in all F and M ORFs (i.e., in the 5′ half of the ORFs), which is also a significant support for identifying these ORFs as protein-coding genes, suggests a higher conservation of the secondary structure compared to the primary sequence. Interestingly, the mitochondrially encoded ATP8 protein is also characterized by extremely variable length and by a higher conservation of the secondary structure compared to the primary sequence (Gissi et al. 2008). Specifically, typical animal ATP8 proteins are characterized by a hydrophobic N-terminal domain and a positively charged C-terminal domain (Gray 1999; Gissi et al. 2008). Because both F and M ORFs possess one TMH followed by positively charged amino acids (Figure 2), it is tempting to speculate that they originated from within the mitochondrial genome and that the atp8 gene could have been the original copy, but no significant sequence similarity has been found between the ORFs and metazoan atp8 sequences. Presently, it is possible to suggest only that selection might maintain the general characteristics (i.e., one TMH followed by positively charged amino acids) of two functional F and M ORFs in the face of an extremely high overall amino acid substitution rate. This was also the suggestion for the M-specific cox2 extension, which, as mentioned above, appears to have a reproductive function (Chakrabarti et al. 2006, 2007; Chapman et al. 2008). Given that relatively rapid rates of evolution are frequently observed for proteins involved in reproduction (e.g., Metz et al. 1998; Swanson and Vacquier 2002; Clark et al. 2006), it is possible that the F and M ORFs might also function in reproduction. Further analyses of the F trnE–nd2 and M trnD–nd4l gender-specific unassigned regions in additional species of freshwater bivalves and further protein-based analyses will be necessary to characterize the biological significance of these ORFs.
Identifying the F and M control regions and origins of replication in unionoid bivalves:
An attempt has been made to identify the F and M control regions and/or potential signaling elements that are conserved (i) among F, (ii) among M, and/or (iii) among F and M unassigned regions in unionoid bivalves. As mentioned above, there are three shared unassigned regions >20 nucleotides in length in all species that could play crucial roles in replication or transcription of the mtDNA molecule. These include the regions trnF–nd5, nd3–trnA (this region contains trnH in L. ornata, C. plicata, P. grandis, Q. quadrula, and V. ellipsiformis F genomes), and nd5–trnQ (note that because this region contains trnH in M genomes, we refer to it as nd5–trnQ+H in M genomes) (see Figure 1, regions B, C, and A).
Typically, the main control region of animal mitochondrial genomes corresponds to (i) the longest noncoding region, (ii) a region in which repetitive elements and secondary structures frequently occur, (iii) a region containing relatively high A + T content, and/or (iv) a region associated with abrupt changes in base composition bias (Lewis et al. 1994; Boore 1999; Saccone et al. 2002; Serb and Lydeard 2003; Cao et al. 2004a; Saito et al. 2005; Kuhn et al. 2006; Oliveira et al. 2007; Brugler and France 2008). In the mytiloid mussel Mytilus, for example, the putative control region has been identified as such because it is the largest noncoding region, and it is capable of producing characteristic secondary structures (Cao et al. 2004a). In addition, it contains conserved motifs thought to play a crucial role in replication and transcription, and it is broadly similar (with specific nucleotidic regions exhibiting 60–90% identity) to the mammalian control region (Cao et al. 2004a). To our knowledge, the main control region of the veneroid clam V. philippinarum has not yet been localized or described.
The length, repeats/secondary structure, and A + T content criteria:
Based on the length criterion alone, F nd5–trnQ is the most likely candidate for the control region in all F genomes while M nd5–trnQ+H is the most likely candidate for the control region in all M genomes (except possibly for P. grandis, where the largest noncoding region is found in the nd3–trnA region) (Figure 1, region A, and Table 2). The lengths observed for F nd5–trnQ and M nd5–trnQ+H (i.e., from 81 bp for M P. grandis to 1196 bp for F I. japanensis) are comparable to the size of known metazoan control regions [e.g., 109 bp in deep-sea Bamboo corals (Brugler and France 2008) and ∼1160 bp for the F mtDNA of M. edulis (Cao et al. 2004a)]. The repeats/secondary structures criterion also suggests that the F nd5–trnQ and M nd5–trnQ+H sequences constitute the main control regions in F and M unionoid mtDNAs. The predicted location of the control region in the large unassigned F nd5–trnQ and M nd5–trnQ+H regions is particularly well supported by the noncoding features of I. japanensis (Table 3). In the F genome of this species, F nd5–trnQ is 1196 bp long (i.e., 910 bp longer than the second largest unassigned region in trnE–trnW) and it has a higher A + T content (67.5%) compared to the other parts of the genome (Tables 1 and 3; A + T for trnE–trnW = 51%). High A + T content is also a characteristic typically used to identify origins of replication (e.g., Lewis et al. 1994; Serb and Lydeard 2003; Kuhn et al. 2006). There are eight consecutive repeats of a 106-bp element with the potential to form stem-loop structures, with an additional incomplete repeat on each side of the eight-repeat cluster. Similarly, the I. japanensis M nd5–trnQ+H is the largest region (698 bp; i.e., 334 bp longer than the second largest unassigned region between nd4l–trnD) and also is A + T rich (63%) compared to the other parts of the genome (Tables 1 and 3; A + T for nd4l–trnD = 58%). It also harbors five consecutive repeats of a 107-bp element with the potential to form stem-loop structures, with an additional incomplete repeat at the 3′ end of the five-repeat cluster. Likewise, F nd5–trnQ and M nd5–trnQ+H are the longest unassigned regions of the F and M genomes of Q. quadrula (346 and 555 bp long, respectively) and both regions contain stem-loop structures and are A + T rich compared to the other parts of the genomes (65 and 70% for F nd5–trnQ and M nd5–trnQ+H, respectively). Two consecutive repeats of a 98-nucleotide element are found in Q. quadrula M nd5–trnQ+H, while short repetitive sequences (<8 bp) are observed in the F genome of the same species (Table 3). Short repetitive identical fragments are also found in all other F nd5–trnQ and M nd5–trnQ+H regions of unionoid genomes (Table 3). Overall, the extent of nucleotide divergence observed between both regions within each species (43–50%) is slightly higher than that observed for the other parts of the genomes (Tables 1 and 3). Interspecifically, the lowest amount of nucleotide sequence divergence (31–35%) was detected between the two most closely related species (i.e., between the F unassigned regions nd3–trnH, trnF–nd5, and nd5–trnQ of L. ornata and V. ellipsiformis). For the more distantly related species, pairwise comparisons within each gender revealed, as expected, higher levels of nucleotide sequence divergence (>45%).
TABLE 2.
Major unassigned regions in F and M mitochondrial genomes of freshwater mussels (Bivalvia: Unionoida)
| Species of freshwater mussels (Unionoida) | Gender | F nd5–trnQ or M nd5–trnQ+H (bp) | F nd3–trnA+Ha or M nd3–trnA (bp) | F trnF–nd5 or M trnF–nd5 (bp) | F trnE–nd2 (F ORF) (bp) | F trnE–trnWb (F ORF) (bp) | M nd4l–trnD (M ORF) (bp) | M atp8–atp6 (bp) |
|---|---|---|---|---|---|---|---|---|
| C. plicata | F | 289 | 77 | 42 | 329 | — | — | — |
| H. cumingii | F | 202 | — | 44 | — | 423 | — | — |
| I. japanensis | F | 1196 | — | 74 | — | 286 | — | — |
| I. japanensis | M | 698 | 91 | 34 | — | — | 364 | 112 |
| L. ornata | F | 247 | 130 | 135 | 282 | — | — | — |
| P. grandis | F | 450 | 71 | 56 | 305 | — | — | — |
| P. grandis | M | 81 | 248 | 36 | — | — | 1103 | 58 |
| Q. quadrula | F | 346 | 79 | 51 | 287 | — | — | — |
| Q. quadrula | M | 555 | 62 | 58 | — | — | 325 | 65 |
| V. ellipsiformis | F | 308 | 88 | 68 | 283 | — | — | — |
|
V. ellipsiformis |
M |
120 |
93 |
33 |
— |
— |
833 |
54 |
Shared unassigned regions are in boldface type.
The F genomes of I. japanensis and H. cumingii possess a different gene order in this region and are not included. Moreover, this region contains trnH in L. ornata, C. plicata, P. grandis, Q. quadrula, and V. ellipsiformis F genomes, and the length has been calculated for the segment trnH–trnA in these genomes.
I. japanensis and H. cumingii F genomes have a different gene order between cox2 and trnW and are thus the only genomes to possess a major unassigned region between trnE–trnW.
TABLE 3.
General characteristics of F nd5-trnQ and M nd5-trnQ+H
| F nd5–trnQ and M nd5–trQ+H |
|||||||
|---|---|---|---|---|---|---|---|
| Species/taxon | Length (bp) | % A + T content | No. of repetitive elements >5 bp | Longest repetitive element (bp) | Copy no. of repetitive elements | Other characteristics | % F/M divergence |
| C. plicata F | 289 | 71.1 | 8 | 8 | 2–8 | Stem-loop and hairpin structures, (A)n, (G)n, (T)n | — |
| H. cumingii F | 202 | 69.0 | 5 | 10 | 2–3 | Stem-loop structures | — |
| I. japanensis F | 1196 | 67.5 | 1 | 106 | 9 | Stem-loop structures | 43 |
| I. japanensis M | 698 | 63.0 | 1 | 107 | 5 | Stem-loop structures, (G)n | |
| L. ornata F | 247 | 64.4 | 6 | 7 | 2–3 | Hairpin structure, (A)n | — |
| P. grandis F | 450 | 70.9 | 10 | 23 | 2–3 | Stem-loop and hairpin structures, (A)n, (G)n | 43 |
| P. grandis M | 81 | 76.5 | 2 | 7 | 2 | Stem-loop structures, (A)n | |
| Q. quadrula F | 346 | 65.0 | 17 | 8 | 2–5 | Stem-loop and hairpin structures | 49 |
| Q. quadrula M | 555 | 70.3 | 33 | 98 | 2–7 | Stem-loop and hairpin structures, (A)n, (T)n | |
| V. ellipsiformis F | 308 | 67.9 | 5 | 7 | 2–3 | Hairpin structure, (A)n | 50 |
|
V. ellipsiformis M |
120 |
70.0 |
4 |
10 |
2–4 |
Hairpin structure, (G)n, (T)n |
|
Sequence divergences are given in percentages for the total number of aligned nucleotides. Stretches of nucleotides are considered when >6 bp.
Identification of conserved motifs or regions with high similarity to the other DUI-species control regions:
Previous analyses of the mytiloid mussel Mytilus have shown that the F and M main control regions can be divided into three domains on the basis of indels and patterns of nucleotide variation (Cao et al. 2004a, 2009). The middle domain of the control region encodes a hairpin structure and is the most slowly evolving part of the mitochondrial genome. In contrast, the first and last domains are among the most divergent parts of the M and F genomes. It has been suggested that this tripartite structure, which is also a characteristic of the mammalian control region, demonstrates that different parts of the control region evolve under different selective constraints (Cao et al. 2004a, 2009). In freshwater mussels (Unionoida), the high degree of sequence divergence observed among species and also between intraspecific F and M genomes presents a challenge for characterizing the structure of any of the shared unassigned regions. Alignments indicate that the portion of the shared F nd5–trnQ and M nd5–trnQ+H unassigned region closest to the trnQ gene is the most conserved part of the sequence. Interestingly, this portion consistently contains hairpin or stem-loop structures in all F and M genomes examined here, but neither the flanking regions nor the morphology of the structures appear to be conserved (Figure S1). The identification of motifs of significant sequence similarity (>60%) with elements known to have specific functions in the sea urchin and the mammalian control region was possible in the mytiloid mussel Mytilus (Cao et al. 2004a). However, this approach has not been successful in defining potential regulatory elements in unionoid genomes. We performed a search for possible signaling elements by comparing F nd5–trnQ and M nd5–trnQ+H sequences for any block of ≥10 nucleotides with nucleotide identity of at least 65% (e.g., Boore 2006). Apart from the A + T-rich region capable of forming stem-loop structures and several (A)n, (G)n, and (T)n homopolymer tracts, we found no conserved motif within each gender or between F nd5–trnQ and M nd5–trnQ+H. Similarly, the search for possible signaling elements in other long, shared regions [i.e., between nd3–trnA (nd3–trnH and trnH–trnA in F genomes) and trnF–nd5] did not reveal any conserved motif. These results indicate that F and M unionoid genomes might share more similar organization in this region at the level of secondary structures despite a low extent of similarity identifiable at the nucleotide level. In other words, a high level of DNA sequence variability might be compensated for by specific secondary structure configurations, with homopolymer tracts and stem-loop structures, which might be responsible for the regulation of mtDNA replication.
Using mitochondrial AT skews to identify the F and M origins of replication in unionoid bivalves:
As previously mentioned, a consequence of the asymmetrical model of mtDNA replication, which is thought to occur in most metazoans, is that during replication of the molecule different genes will be exposed to the single-strand state (more susceptible to mutation) for different lengths of time, depending on their position in the genome (Clayton 1982). Recently, Rodakis et al. (2007) observed a positive correlation between single-strand state duration and nucleotide composition bias (AT skews) at the less constrained protein-coding gene positions (fourfold degenerate sites) in the F and M mitochondrial genomes of the mytiloid mussel Mytilus, indicating that the replication proceeds via the asymmetric strand-displacement model, with the origins of heavy (OH) and light (OL) strand replication separated by approximately two-thirds of the genome.
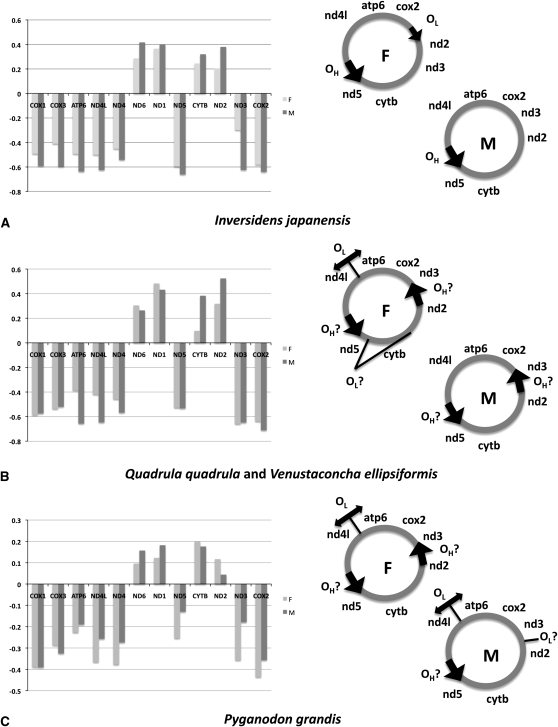
Herein, we use mitochondrial AT skews to help identify the F and M heavy and light strand replication origins in unionoid bivalves. If the asymmetrical model of mtDNA replication also applies to unionoid bivalves and if their mtDNAs are also exposed to the same type of mutations as are vertebrate mtDNAs, then we hypothesize that OH would be located in the vicinity of the protein-coding genes showing the greatest AT-skew values at fourfold redundant sites, while OL would be located in the vicinity of the protein-coding genes showing the lowest AT-skew values. Figure 3 displays AT-skew values at fourfould redundant sites for 12 F and M mitochondrial genes (we excluded atp8) within each species whose F and M genomes have been completely sequenced and the deduced locations of the putative OH and OL in each genome. According to our combined skew analyses, mitochondrial control regions are not well defined and their locations could be variable among unionoid bivalve mitochondrial genomes. In I. japanensis, OH would be located between F nd5–trnQ in the F genome and M nd5–trnQ+H in the M genome, a result that corresponds to our predictions based on the length, A + T content, and repeat/secondary structures criteria. Indeed, the heavy strand encoded nd5 and the light strand encoded nd1 and nd6 show the greatest AT-skew values (negative or positive) in both F and M I. japanensis genomes, indicating that this region would be exposed to mutagenic pressures for the longest time, and thus we can infer that it is located near the OH (Figure 4A). In the F genome of this species, a marked difference is observed between the skew values of the heavy strand encoded nd3 and all other heavy strand encoded genes. The low AT-skew value in nd3, together with the low AT-skew value of the light strand encoded nd2 gene, suggests that OL would be located between nd3 and cox2 with the direction of the L strand synthesis toward nd2 in I. japanensis F mtDNA. It should be noted that a similar pattern is observed for the F genome of H. cumingii (data not shown). Interestingly, this region (i.e., between trnH and trnS2) contains a noncoding region on the heavy strand capable of forming a stem-loop structure similar to that of other metazoan OL's (e.g., Rodakis et al. 2007; Seligmann 2008) (Figure S2, A and B). No clear skew pattern is observed for the M genome of I. japanensis; therefore, the location of its OL remains unclear.
Figure 4.—
AT-skew values in 12 mitochondrial genes (excluding atp8) of unionoid mussels and the deduced locations of the putative OH and OL in each genome. Because they share a similar pattern, V. ellipsiformis and Q. quadrula have been analyzed together. The big black arrow indicates the unassigned region located between “F nd5–trnQ” and “M nd5–trnQ+H” and the potential direction of DNA synthesis, whereas the small black arrow indicates the unassigned region located between “M nd3–trnA” and “F trnH–trnA” and the potential direction of DNA synthesis. (A) Inversidens japanensis, (B) Quadrula quadrula and Venustaconcha ellipsiformis, and (C) Pyganodon grandis.
The position of OL between nd3 and cox2 in the F genomes of I. japanensis and H. cumingii marks the location of the sole observed gene rearrangement among F unionoid genomes (I. japanensis and H. cumingii have a different gene order in this segment compared to other F mtDNAs; see Figures 1 and 4). It has previously been demonstrated that novel gene orders occur more frequently with movement of OL than would be expected by chance (Macey et al. 1997). Apparently, the region encompassing cox2–nd3–nd2 is not associated with OL in the F genomes of Q. quadrula, V. ellipsiformis, and P. grandis [Figure 4, B and C; a similar pattern is observed for the F genome of L. ornata and C. plicata (data not shown)]. The rather high skew values observed for nd3 and cox2 suggest that this region would remain exposed to mutagenic pressures for a long time and could be associated with the OH in these genomes. The same pattern is also observed for the M genomes of q. quadrula and V. ellipsiformis, but not for P. grandis (Figure 4, B and C). Although shorter in length as compared with F nd5–trnQ and M nd5–trnQ+H, the unassigned regions located between nd3–trnA in M genomes and trnH–trnA in F genomes are also favorable candidates that could be involved in replication initiation. Indeed, they possess characteristics typically used to identify origins of replication; i.e., they are A + T rich, they are capable of forming stem-loop and hairpin structures, and they contain several (A)n, (G)n, and (T)n homopolymer tracts (Table 4).
TABLE 4.
General characteristics of F trnH–trnA and M nd3–trnA
| F trnH–trnA and M nd3–trnA |
|||||||
|---|---|---|---|---|---|---|---|
| Species/taxon | Length (bp) | % A + T content | No. of repetitive elements >5 bp | Longest repetitive element (bp) | Copy no. of repetitive elements | Other characteristics | F/M divergence (%) |
| C. plicata F | 77 | 80.8 | 1 | 5 | 3–6 | Hairpin and stem-loop structures, (A)n, G)n | — |
| I. japanensis M | 91 | 62.6 | 6 | 6 | 2–3 | Hairpin and stem-loop structures | — |
| L. ornata F | 130 | 66.7 | 3 | 7 | 2–4 | Hairpin and stem-loop structures, (G)n | — |
| P. grandis F | 71 | 93.0 | 1 | 8 | 2 | Hairpin structure, (A)n, (T)n | 39.6 |
| P.grandis M | 248 | 75.4 | 4 | 10 | 2–3 | Hairpin and stem-loop structures, (A)n, (G)n, (T)n | |
| Q. quadrula F | 79 | 68.4 | 3 | 8 | 2 | Hairpin and stem-loop structures, (G)n | 53.7 |
| Q. quadrula M | 62 | 85.5 | 1 | 5 | 2 | Hairpin and stem-loop structures | |
| V. ellipsiformis F | 88 | 69.3 | 4 | 7 | 2–4 | Hairpin and stem-loop structures | 52.8 |
|
V. ellipsiformis M |
93 |
67.7 |
3 |
6 |
2 |
Hairpin and stem-loop structures |
|
Sequence divergences are given in percentages for the total number of aligned nucleotides. Stretches of nucleotides are considered when >6 bp. The F genomes of I. japanensis and H. cumingii possess a different gene order in this region and are not included.
Interestingly, two putative OL's can be detected in the F genomes of Q. quadrula and V. ellipsiformis (Figure 4B). According to its very low AT-skew value, one OL would be located in the vicinity of the cytb gene. If we assume that the H strand synthesis is unidirectional and similar to that of the I. japanensis F genome, a “unidirectional” OL could be located only within the mitochondrial tRNA gene cluster trnL1–trnN-trnP or in the 16SrRNA and the direction of the L strand synthesis is toward cytb (see Figures 1 and 4B). Seligmann et al. (2006) reported that heavy strand encoded tRNA genes can sometimes function as an OL by forming alternative secondary structures other than their classical cloverleaf structures. The possibility therefore exists that the heavy strand “mirror” sequences of trnL1–trnN–trnP could serve as origins of light strand replication in the F genomes of Q. quadrula and V. ellipsiformis (Seligmann et al. 2006; Seligmann 2008). It is of interest to note that the prominent difference in AT-skew value for the cytb gene has not been observed in the M genomes of Q. quadrula and V. ellipsiformis, in the F genome of C. plicata (data not shown), or in the F or the M genome of P. grandis (Figure 4). This is consistent with the previous suggestion that an alternative OL can frequently evolve and disappear in mtDNA sequences (Seligmann et al. 2006). Alternatively, our results can be explained by (i) a putative bidirectional OL in the unassigned region trnF–nd5 [Figures 1 and 4B; bidirectional replication has previously been demonstrated in mitochondrial genomes of vertebrates (Reyes et al. 2005; Seligmann et al. 2006)] and/or (ii) differences in transcription processes and/or selective constraints experienced by the F genomes of V. ellipsiformis and Q. quadrula.
According to our results, a second putative OL in the F genomes of Q. quadrula and V. ellipsiformis would be located in the vicinity of the atp6 gene, a region that corresponds to the gene order inversion of trnD and atp8 and that represents one of the two organizational differences between F and M genomes in unionoid bivalves (the other difference is the transposition of the trnH gene; see Figures 1 and 4B). The M genomes of the same species do not share a similar pattern (i.e., the OL is not clearly indicated in these M genomes), but the results obtained for the F genome of C. plicata (data not shown) and the F and M genomes of P. grandis also suggest the presence of a putative OL in the vinicity of atp6 (Figure 4, B and C). Again, our results are in line with and confirm previous studies showing that mitochondrial regions near origins of replication are hot spots for rearrangement and that gene rearrangements occur more frequently with displacement of OL than expected by chance (e.g., Macey et al. 1997; Mueller and Boore 2005; Brugler and France 2008). We also note that the AT-skew values suggest that replication could proceed in both directions from this OL (Figure 4, B and C). Because no unassigned region is found between atp6 and nd4l in the F genome of V. ellipsiformis, one likely candidate for an OL in this species would be the heavy strand encoded trnD gene. Consistent with the observations of Seligmann et al. (2006), the F-type trnD (but not the M-type) of V. ellipsiformis is able to form an OL-like structure, thereby suggesting it could have an alternative OL function (Figure S2, C and D, respectively). A similar result is also observed for the trnD of the F genomes of Q. quadrula and P. grandis, but not for the M genome of P. grandis (data not shown). In the P. grandis M genome, an OL-like structure is found in the unassigned region between trnD and the truncated atp8. Moreover, this genome could possess a second putative OL in the vicinity of the nd2 gene (Figure 4). Once more, this suggests that an alternative OL can frequently evolve and disappear in mtDNA sequences (Seligmann et al. 2006).
Overall, our AT-skew values suggest that the classical asymmetrical model of mtDNA replication, with one OH and one OL, might not apply to all unionoid bivalves. Our data suggest that the locations of the origins of replication in these species are variable, there are potentially multiple locations, and locations could be uni- or bidirectional. Other studies of distantly related invertebrate taxa also reported that control region locations can be highly variable [e.g., insect species (Saito et al. 2005) and corals (Brugler and France 2008; Chen et al. 2008)]. Furthermore, multiple and bidirectional mitochondrial origins of replication have been previously suggested for vertebrates (Reyes et al. 2005). However, our results could also be associated with differences in transcription processes and/or selective constraints experienced by the F and M genomes, leading to potential misinterpretation of the localizations of the OH and OL. According to the length, A + T content, and repeat/secondary structures criteria, F nd5–trnQ and M nd5–trnQ+H remain the most likely candidate OH control regions for regulating replication and/or transcription (i.e., origin of replication, initiation, or termination sites for transcription) in unionoid bivalves. Additional studies, both functional and comparative, will be required to determine the precise position of the mitochondrial origins of replication in unionoid bivalves.
Conclusion:
The description and comprehensive analysis of complete F and M mitochondrial genomes of bivalve species with DUI, more specifically the unassigned regions of unionoid species, have led to new insights into the mitogenomics of species with DUI. Our results reveal that at least three shared regions in F and M unionoid genomes could contain regulatory signals involved in the replication and/or transcription of mtDNA. According to the length, A + T content, and repeat/secondary structures criteria, likely candidate regions for the OH origin of replication would be F nd5–trnQ and M nd5–trnQ+H. AT-skew values of protein-coding genes at fourfold degenerate sites have also been used to identify the location of OH and OL control regions. Our results reveal that (i) two regions (i.e., F nd5–trnQ and M nd5–trnQ+H and M nd3–trnA and F trnH–trnA) are potential OH control regions for regulating replication and/or transcription and (ii) multiple and potentially bidirectional OL origins of replication are present in unionoid F and M mitochondrial genomes. In other words, unionoid mitochondrial control regions are seldom well defined using skew values and their locations could be variable among genomes and species.
Finally, although uncharacterized mitochondrial ORFs are essentially absent in vertebrates, their presence in other animal groups is not unprecedented (Burger et al. 2003a; Gissi et al. 2008). For example, ORFs that exhibit no significant sequence similarity to known proteins have been recently discovered in the Cnidaria and Porifera (e.g., Shao et al. 2006; Flot and Tillier 2007; Wang and Lavrov 2008). Such metazoan ORFs could be (i) homologous to ancestral bacterial protein-coding genes, (ii) the product of mitochondrial gene duplication events, or (iii) the result of DNA transferred from nuclear genomes. However, testing the above hypotheses will likely be difficult due to the ORFs' highly divergent sequences (Burger et al. 2003b). Analyses of complete mitochondrial genomes from additional bivalve species, particularly basal taxa, and further protein-based studies are needed to elucidate the number, taxonomic distribution, evolution, and function of uncharacterized ORFs in this group as well as the molecular features that are associated with the developmental regulation and transmission genetics of DUI.
Acknowledgments
The authors thank R. Shandilya and S. Vijayaraghavan for their assistance in the laboratory. This study was supported by research grants from the National Science Foundation (to W.R.H.), the National Sciences and Engineering Research Council (NSERC) (to P.U.B. and D.T.S.), and by research funds from the Friends of the North Carolina State Museum of Natural Sciences (to A.E.B.). S.B. and H.D.P. were respectively financially supported by a NSERC fellowship and a Fonds Québécois de la Recherche sur la Nature et les Technologies scholarship.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.110700/DC1.
References
- Akasaki, T., M. Nikaido, K. Tsuchiya, S. Segawa, M. Hasegawa et al., 2006. Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Mol. Phylogenet. Evol. 38 648–658. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Scäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai, M., H. Mitsuke, M. Ikeda, J. X. Xia, T. Kikuchi et al., 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32 W390–W393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D., I. Karsch-Mizrachi, D. Lipman, J. Ostell and D. Wheeler, 2004. GenBank: update. Nucleic Acids Res. 32 D23–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley, J. N., M. C. Frith, J. H. Graber, S. Choo and J. O. Deshler, 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 12 1756–1761. [DOI] [PubMed] [Google Scholar]
- Boore, J. L., 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L., 2006. The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda). BMC Genomics 7 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L., M. Medina and L. A. Rosenberg, 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 21 1492–1503. [DOI] [PubMed] [Google Scholar]
- Breton, S., G. Burger, D. T. Stewart and P. U. Blier, 2006. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.). Genetics 172 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, S., H. D. Beaupré, D. T. Stewart, W. R. Hoeh and P. U. Blier, 2007. The unusual system of doubly uniparental inheritance of mtDNA: Isn't one enough? Trends Genet. 23 465–474. [DOI] [PubMed] [Google Scholar]
- Brugler, M. R., and S. C. France, 2008. The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): genome structure and putative origins of replication are not conserved among octocorals. J. Mol. Evol. 67 125–136. [DOI] [PubMed] [Google Scholar]
- Burger, G., M. W. Gray and F. B. Lang, 2003. a Mitochondrial genomes: anything goes. Trends Genet. 19 709–716. [DOI] [PubMed] [Google Scholar]
- Burger, G., F. B. Lang, H. P. Braun and S. Marx, 2003. b The enigmatic mitochondrial ORF ymf39 codes for ATP synthase chain b. Nucleic Acids Res. 31 2353–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyński, A., M. Zbawizka, D. O. Skibinski and R. Wenne, 2003. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 20 388–392. [DOI] [PubMed] [Google Scholar]
- Cao, L., E. Kenchington, E. Zouros and G. C. Rodakis, 2004. a Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.). Genetics 167 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., E. Kenchington and E. Zouros, 2004. b Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis). Genetics 166 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., B. S. Ort, A. Mizi, G. Pogson, E. Kenchington et al., 2009. The control region of maternally and paternally inherited mitochondrial genomes of three species of the sea mussel genus Mytilus. Genetics 181 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, R., J. M. Walker, D. T. Stewart, R. J. Trdan, S. Vijayaraghavan et al., 2006. Presence of a unique male-specific extension of C-terminus to the cytochrome c oxidase subunit II protein coded by the male-transmitted mitochondrial genome of Venustaconcha ellipsiformis (Bivalvia: Unionoidea). FEBS Lett. 580 862–866. [DOI] [PubMed] [Google Scholar]
- Chakrabarti, R., J. M. Walker, E. G. Chapman, S. P. Shepardson, R. J. Trdan et al., 2007. Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Lett. 581 5213–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E. G., H. Piontkivska, J. M. Walker, D. T. Stewart, J. P. Curole et al., 2008. Extreme primary and secondary protein structure variability in the chimeric male-transmitted cytochrome c oxidase subunit II protein in freshwater mussels: evidence for an elevated amino acid substitution rate in the face of domain-specific purifying selection. BMC Evol. Biol. 8 165–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., C. F. Dai, S. Plathong, C. Y. Chiou and C. A. Chen, 2008. The complete mitochondrial genomes of needle corals, Seriatopora spp. (Scleractinia: Pocilloporidae): an idiosyncratic atp8, duplicated trnW gene, and hypervariable regions used to determine species phylogenies and recently diverged populations. Mol. Phylogenet. Evol. 46 19–33. [DOI] [PubMed] [Google Scholar]
- Chung, S. Y., and S. Subbiah, 1996. A structural explanation for the twilight zone of protein sequence homology. Structure 4 1123–1127. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., J. E. Aagaard and W. J. Swanson, 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131 11–22. [DOI] [PubMed] [Google Scholar]
- Clayton, D. A., 1982. Replication of animal mitochondrial DNA. Cell 28 693–705. [DOI] [PubMed] [Google Scholar]
- Cogswell, A. T., E. L. Kenchington and E. Zouros, 2006. Segregation of sperm mitochondria in two- and four-cell embryos of the blue mussel Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA. Genome 49 799–807. [DOI] [PubMed] [Google Scholar]
- Curole, J. P., and T. D. Kocher, 2002. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol. Biol. Evol. 19 1323–1328. [DOI] [PubMed] [Google Scholar]
- Curole, J. P., and T. D. Kocher, 2005. Evolution of a unique mitotype-specific protein-coding extension of the cytochrome c oxidase II gene in freshwater mussels (Bivalvia: Unionoida). J. Mol. Evol. 61 381–389. [DOI] [PubMed] [Google Scholar]
- Faith, J. J., and D. D. Pollock, 2003. Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics 165 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett, J. W., 1982. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 10 5303–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot, J.-F., and A. Tillier, 2007. The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: the putative D-loop and a novel ORF of unknown function. Gene 401 80–87. [DOI] [PubMed] [Google Scholar]
- Fonseca, M. M., D. Posada and J. D. Harris, 2008. Inverted replication of vertebrate mitochondria. Mol. Biol. Evol. 25 805–808. [DOI] [PubMed] [Google Scholar]
- Gissi, C., F. Iannelli and G. Pesole, 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101 301–320. [DOI] [PubMed] [Google Scholar]
- Gray, M. W., 1999. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 9 678–687. [DOI] [PubMed] [Google Scholar]
- Kuhn, K., B. Streit and K. Schwenk, 2006. Conservation of structural elements in the mitochondrial control region of Daphnia. Gene 420 101–112. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lewis, D. L., C. L. Farr, A. L. Farquhar and L. S. Kaguni, 1994. Sequence, organization, and evolution of the A+T region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 11 523–538. [DOI] [PubMed] [Google Scholar]
- Macey, J. R., A. Larson, N. B. Ananjeva, Z. Fang and T. J. Papenfuss, 1997. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14 91–104. [DOI] [PubMed] [Google Scholar]
- Metz, E. C., R. Robles-Sikisaka and V. D. Vacquier, 1998. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc. Natl. Acad. Sci. USA 95 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizi, A., E. Zouros, N. Moschonas and G. C. Rodakis, 2005. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritances mode of mtDNA. Mol. Biol. Evol. 22 952–967. [DOI] [PubMed] [Google Scholar]
- Mueller, R. L., and J. L. Boore, 2005. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol. Biol. Evol. 22 2104–2112. [DOI] [PubMed] [Google Scholar]
- Oliveira, M. T., A. M. L. Azeredo-Espin and A. C. Lessinger, 2007. The mitochondrial DNA control region of Muscidae flies: evolution and structural conservation in a Dipteran context. J. Mol. Evol. 64 519–527. [DOI] [PubMed] [Google Scholar]
- Passamonti, M., and F. Ghiselli, 2009. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 28 1–12. [DOI] [PubMed] [Google Scholar]
- Pearson, W. R., 1996. Effective protein sequence comparison. Methods Enzymol. 266 227–258. [DOI] [PubMed] [Google Scholar]
- Perna, N. T., and D. T. Kocher, 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41 353–358. [DOI] [PubMed] [Google Scholar]
- Pont-Kingdon, G. A., N. A. Okada, J. L. Macfarlane, C. T. Beagley, D. R. Wolstenholme et al., 1995. A coral mitochondrial MutS gene. Nature 375 109–111. [DOI] [PubMed] [Google Scholar]
- Pont-Kingdon, G. A., N. A. Okada, J. L. Macfarlane, C. T. Beagley, C. D. Watkins-Sims et al., 1998. Mitochondrial DNA of the coral Sarcophyton glaucum contains a gene for a homologue bacterial MutS: a possible case of gene transfer from the nucleus to the mitochondrion. J. Mol. Evol. 46 419–431. [DOI] [PubMed] [Google Scholar]
- Reyes, A., C. Gissi, G. Pesole and C. Saccone, 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 15 957–966. [DOI] [PubMed] [Google Scholar]
- Reyes, A., M. Y. Yang, M. Bowmaker and I. J. Holt, 2005. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 280 3242–3250. [DOI] [PubMed] [Google Scholar]
- Rodakis, G. C., L. Cao, A. Mizi, E. L. Kenchington and E. Zouros, 2007. Nucleotide content gradients in maternally and paternally inherited mitochondrial genomes of the mussel Mytilus. J. Mol. Evol. 65 124–136. [DOI] [PubMed] [Google Scholar]
- Rost, B., G. Yachdav and J. Liu, 2004. The PredictProtein Server. Nucleic Acids Res. 32 W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, C., C. Gissi, A. Reyes, A. Larizza, E. Sbisà et al., 2002. Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene 286 3–12. [DOI] [PubMed] [Google Scholar]
- Saito, S., K. Tamura and T. Aotsuka, 2005. Replication origin of mitochondrial DNA in insects. Genetics 171 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligmann, H., 2008. Hybridization between mitochondrial heavy strand tDNA and expressed light strand tRNA modulates the function of heavy strand tDNA as light strand replication origin. J. Mol. Biol. 379 188–199. [DOI] [PubMed] [Google Scholar]
- Seligmann, H., N. M. Krishnan and B. J. Rao, 2006. Possible multiple origins of replication in primate mitochondria: alternative role of tRNA sequences. J. Theor. Biol. 241 321–332. [DOI] [PubMed] [Google Scholar]
- Serb, J. M., and C. Lydeard, 2003. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca). Mol. Biol. Evol. 20 1854–1866. [DOI] [PubMed] [Google Scholar]
- Shao, Z., G. Shannon, O. Y. Chaga and D. V. Lavrov, 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependant DNA polymerase. Gene 381 92–101. [DOI] [PubMed] [Google Scholar]
- Subramanian, A. R., M. Kaufmann and B. Morgenstern, 2008. DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol. Biol. 3 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Theologidis, I., C. Saavedra and E. Zouros, 2007. No evidence for absence of paternal mtDNA in male progeny from pair matings of the mussel Mytilus galloprovincialis. Genetics 176 1367–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetis, C., I. Theologidis, E. Zouros and G. C. Rodakis, 2007. A mitochondrial genome with a reversed transmission route in the Mediterranean mussel Mytilus galloprovincialis. Gene 406 79–90. [DOI] [PubMed] [Google Scholar]
- Wang, X., and D. V. Lavrov, 2008. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Desmonspongiae. PLoS ONE 3 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A. M., J. B. Procter, D. M. A. Martin, M. Clamp and G. J. Barton, 2009. Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters, G. T., 2001. The evolution of the Unionacea in North America, and its implications for the worldwide fauna, pp. 281–307 in Ecology and Evolution of the Freshwater Mussels Unionoida, edited by G. Bauer and K. Wächtler. Springer-Verlag, Berlin.
- White, D. H., J. N. Wolff, M. Pierson and N. J. Gemmell, 2008. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 17 4925–4942. [DOI] [PubMed] [Google Scholar]
- Zbawicka, M., A. Burzyński and R. Wenne, 2007. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene 406 191–198. [DOI] [PubMed] [Google Scholar]
- Zouros, E., 2000. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet. Syst. 75 313–318. [DOI] [PubMed] [Google Scholar]
- Zuker, M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]