Abstract
The correlation coefficient is commonly used as a measure of the divergence of gene expression profiles between different species. Here we point out a potential problem with this statistic: if measurement error is large relative to the differences in expression, the correlation coefficient will tend to show high divergence for genes that have relatively uniform levels of expression across tissues or time points. We show that genes with a conserved uniform pattern of expression have significantly higher levels of expression divergence, when measured using the correlation coefficient, than other genes, in a data set from mouse, rat, and human. We also show that the Euclidean distance yields low estimates of expression divergence for genes with a conserved uniform pattern of expression.
IT is now possible to measure the expression levels of thousands of genes in multiple tissues at multiple times. This has led to investigations into the evolution of gene expression and how the pattern of expression changes on a genomic scale. In some analyses, the evolution of expression is considered only within one tissue, but in many studies the evolution across multiple tissues is investigated. In this latter case, the evolution of an expression profile—a vector of expression levels of a gene across several tissues—is considered.
Several different statistics have been proposed to measure the divergence between gene expression profiles. The two most popular measures are the Euclidean distance (Jordan et al. 2005; Kim et al. 2006; Yanai et al. 2006; Urrutia et al. 2008) and Pearson's correlation coefficient (Makova and Li 2003; Huminiecki and Wolfe 2004; Yang et al. 2005; Kim et al. 2006; Liao and Zhang 2006a,b; Xing et al. 2007; Urrutia et al. 2008). The correlation coefficient is often subtracted from one, so that the statistic varies from zero, when there has been no expression divergence, to a maximum of two; we refer to this statistic as the Pearson distance. Here we describe a significant shortcoming of the Pearson distance that is not shared by the Euclidean distance.
To investigate properties of these two measures of expression divergence, we compiled a data set of 2859 orthologous genes from human, mouse, and rat for which we had microarray expression data from nine homologous tissues: bone marrow, heart, kidney, large intestine, pituitary, skeletal muscle, small intestine, spleen, and thymus). The expression data for rat came from Walker et al. (2004), the mouse data from Su et al. (2004), and the human data from Ge et al. (2005). Each tissue experiment had two replicates in mouse, a varying number of replicates in rat, and one in humans; some genes were also matched by multiple probe sets. To obtain an average across experiments and probe sets we processed the data as follows:
Raw CEL files of gene expression levels were obtained from the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo/).
The results from the mouse, rat, and human arrays were normalized separately using both the MAS5 (Affymetrix 2001) and the RMA algorithms (Irizarry et al. 2003) as implemented in Bioconductor (Gentleman et al. 2004). The results are qualitatively similar for the two normalization procedures, although recent analyses suggest that MAS5 normalization is generally better (Ploner et al. 2005; Lim et al. 2007).
The expression of each gene within a tissue was averaged across experiments and probe sets.
We computed expression distances (ED) between orthologous gene expression profiles, for each of the three species comparisons, rat–mouse, rat–human, and mouse–human, according to the two different distance metrics, the Euclidean distance and the Pearson distance:
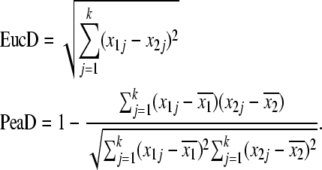 |
(1) |
Here xij is the expression level of the gene under consideration in species i in tissue j, and 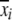 is the average expression level of the gene in species i across tissues. Expression levels are known in a total of k tissues.
is the average expression level of the gene in species i across tissues. Expression levels are known in a total of k tissues.
Because expression levels are measured on different microarray platforms in the three species, we compute relative abundance (RA) values, before calculating the Euclidean distance (Liao and Zhang 2006a). The RA is the expression of a gene in a particular tissue divided by the sum of the expression values of that gene across all tissues. We calculated RA values to remove “probe” effects (the tendency for a gene to bind its probe set on one platform more efficiently than on another platform). Because of probe effects it is not easy to distinguish absolute changes in expression and differences in binding efficiency. Calculating RA values removes this problem from the Euclidean distance. Pearson's distance does not change under such a rescaling and so this is unnecessary.
In some analyses the logarithm of the expression or RA values are used (e.g., Makova and Li 2003; Kim et al. 2006; Xing et al. 2007), and in others the expression values are used without this transformation (e.g., Huminiecki and Wolfe 2004; Jordan et al. 2005; Yang et al. 2005; Liao and Zhang 2006a,b; Yanai et al. 2006; Urrutia et al. 2008). We calculated both the Pearson and the Euclidean distances on log-transformed and untransformed expression values. The results are qualitatively similar so here we present only the results obtained using the logarithm of the expression or RA values.
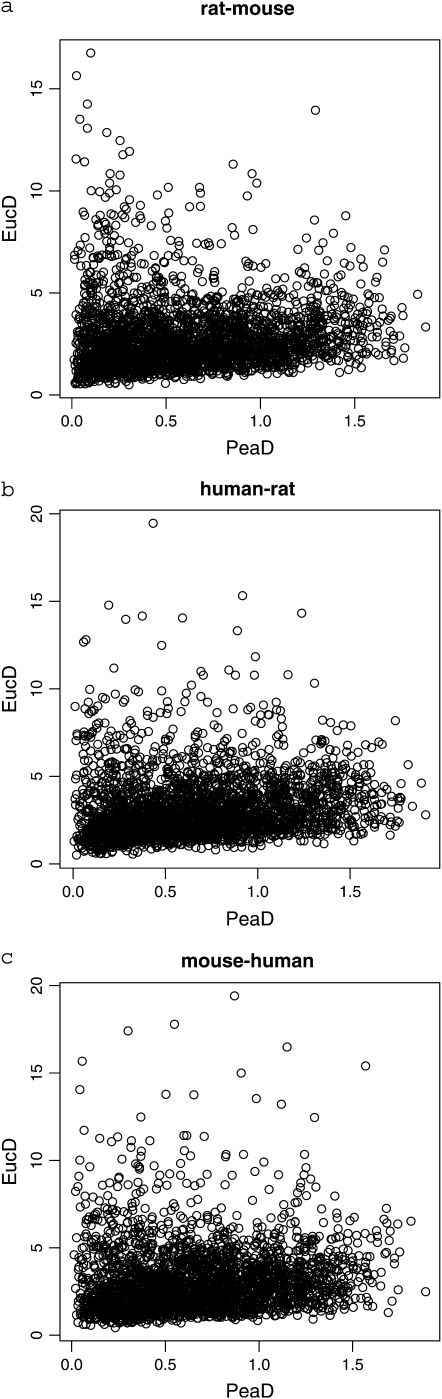
It is natural to expect the two measures of expression divergence to be positively correlated with one another; however, the Euclidean and Pearson distances are almost completely uncorrelated (MAS5 normalization, mouse–rat correlation coefficient = 0.06, human–rat r = 0.13, human–mouse r = 0.10; RMA normalization, mouse–rat correlation coefficient = −0.12, human–rat r = −0.00, human–mouse r = −0.08; Figure 1). This could, plausibly, be because the two statistics measure different aspects of divergence. However, irrespective of this, there is a potential problem associated with the Pearson distance. Imagine that we have a gene that is expressed at identical levels in all tissues in two species (i.e., expression levels are uniform between tissues and also between species). We quite reasonably assume that measured expression levels contain noise. Thus each measured expression level (xij) is the sum of the (assumed) uniform expression level and an independent random number representing noise. In this case there is no real divergence in the expression profile between the species. However, the two measures of divergence may differ greatly in this case. The Euclidean distance reflects only the noise present in the data and hence will be small if the noise is small. By contrast, the Pearson distance will have a value close to 1 since the second term in PeaD in Equation 1 will be close to zero, reflecting the fact that the noise components of different expression levels are independent. Thus the Pearson distance will give the impression that expression divergence is great, but all this apparent divergence is noise. This will be a problem with Pearson's distance whenever measurement error is of the same magnitude as the differences in expression between tissues. This will therefore tend to be a problem for lowly expressed genes, where measurement error can be large relative to the true value.
Figure 1.—
The correlation between the Euclidean and Pearson distances for (a) mouse–rat, (b) human–rat, and (c) human–mouse. Only the results from MAS5 normalization are shown; qualitatively similar results were obtained with RMA.
The above example is unrealistic because real gene expression profiles are rarely perfectly uniform. To investigate whether this shortcoming of the Pearson distance is a problem in real data sets, we determined genes with a relatively uniform pattern of expression in all three species considered above. To do this we computed the entropy of a gene's expression, which is a measure of uniformity in expression across tissues (Schug et al. 2005): the higher the value of the entropy, the more uniform is the expression. We calculated the entropy for each gene in each of the three species, averaged these across species, and then took those genes in the upper quartile of mean entropy values as a data set of genes with a relatively conserved pattern of uniform expression.
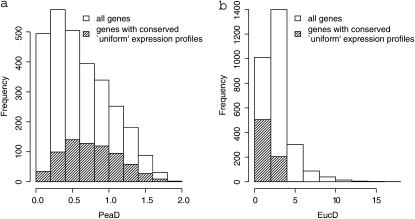
It is natural to expect those genes with a conserved uniform pattern of expression to have relatively low expression divergence; however, on average these genes have significantly higher Pearson distances than other genes (Table 1; Figure 2; supporting information, Figure S1 and Figure S2). By contrast, the Euclidean distance shows the pattern one would anticipate; all of the conserved uniform genes have low expression divergence. It therefore seems likely that the Pearson distance is sensitive to measurement error and hence may not be a good measure of expression divergence.
TABLE 1.
The median expression divergence for genes that have a conserved uniform pattern of expression (upper quartile of mean entropy values) vs. all other genes
| Data set | Statistic | Conserved uniform genes | Other genes | Wilcoxon test P-value |
|---|---|---|---|---|
| MAS5 normalization | ||||
| Mouse–rat | Euclidean | 1.66 | 2.79 | <10−15 |
| Pearson | 0.70 | 0.47 | <10−15 | |
| Human–mouse | Euclidean | 1.67 | 3.13 | <10−15 |
| Pearson | 0.78 | 0.58 | <10−15 | |
| Human–rat | Euclidean | 1.83 | 3.21 | <10−15 |
| Pearson | 0.78 | 0.58 | <10−15 | |
| RMA normalization | ||||
| Mouse–rat | Euclidean | 0.59 | 1.40 | <10−15 |
| Pearson | 0.82 | 0.38 | <10−15 | |
| Human–mouse | Euclidean | 0.59 | 1.58 | <10−15 |
| Pearson | 0.81 | 0.48 | <10−15 | |
| Human–rat | Euclidean | 0.58 | 1.55 | <10−15 |
| Pearson |
0.73 |
0.50 |
<10−15 |
|
Figure 2.—
The distribution of expression divergence values for those genes with a uniform pattern of expression that is conserved across species vs. the distribution for all genes for (a) Pearson and (b) Euclidean distances for mouse–rat. We present similar values for human–mouse and human–rat in Figure S1 and Figure S2. Only the results from MAS5 normalization are shown; qualitatively similar results were obtained with RMA.
We note that there are two additional advantages of the Euclidean distance. First, it can take into account differences in the absolute level of expression if those data are available, either because the method of assay allows this, for example, if ESTs, SAGE, sequencing, or RNA-Seq data are used, or because expression in the two species has been assessed on the same platform using probes that are conserved between the two species. Second, the square of the Euclidean distance is expected to increase linearly with time. Khaitovich et al. (2004) have previously shown that the squared difference in log expression level increases linearly with time under a Brownian motion model of gene expression evolution. It is therefore expected that the squared Euclidean distance will increase with time since the squared Euclidean distance is the sum of the squared differences across tissues. We prove this in File S1; we also show that this linearity holds, approximately, when relative abundance values are used (see also Pereira et al. 2009).
Acknowledgments
We are grateful to a referee for helpful comments. V.P. and A.E.W. were supported by the Biotechnology and Biological Sciences Research Council.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.110247/DC1.
References
- Affymetrix, 2001. Statistical Algorithms Reference Guide. Affymetrix, Santa Clara, CA.
- Ge, X., S. Yamamoto, S. Tsutsumi, Y. Midorikawa, S. Ihara et al., 2005. Interpreting expression profiles of cancers by genome wide survey of breadth of expression in normal tissues. Genomics 86 127–141. [DOI] [PubMed] [Google Scholar]
- Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling et al., 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki, L., and K. H. Wolfe, 2004. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 14 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis et al., 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Jordan, I. K., L. Marino-Ramirez and E. V. Koonin, 2005. Evolutionary significance of gene expression divergence. Gene 345 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., G. Weiss, M. Lachmann, I. Hellmann, W. Enard et al., 2004. A neutral model of transcriptome evolution. PLoS Biol. 2 E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, R. S., H. Ji and W. H. Wong, 2006. An improved distance measure between the expression profiles linking co-expression and co-regulation in mouse. BMC Bioinformatics 7 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, B.-Y., and J. Zhang, 2006. a Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol. Biol. Evol. 23 530–540. [DOI] [PubMed] [Google Scholar]
- Liao, B. Y., and J. Zhang, 2006. b Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol. Biol. Evol. 23 1119–1128. [DOI] [PubMed] [Google Scholar]
- Lim, W. K., K. Wang, C. Lefebvre and A. Califano, 2007. Comparative analysis of microarray normalization procedures: effects on reverse engineering gene networks. Bioinformatics 23 i282–i288. [DOI] [PubMed] [Google Scholar]
- Makova, K. D., and W. H. Li, 2003. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, V., D. Enard and A. Eyre-Walker, 2009. The effect of transposable element insertions on gene expression evolution in rodents. PLoS ONE 4 e4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner, A., L. D. Miller, P. Hall, J. Bergh and Y. Pawitan, 2005. Correlation test to assess low-level processing of high-density oligonucleotide microarray data. BMC Bioinformatics 6 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, J., W. P. Schuller, C. Kappen, J. M. Salbaum, M. Bucan et al., 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6 R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching et al., 2004. A gene atlas of the mouse and human protein coding transcriptomes. Proc. Natl. Acad. Sci. USA 101 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, A. O., L. B. Ocana and L. D. Hurst, 2008. Do Alu repeats drive the evolution of the primate transcriptome? Genome Biol. 9 R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J. R., A. I. Su, D. W. Self, J. B. Hogenesch, H. Lapp et al., 2004. Applications of a rat multiple tissue gene expression data set. Genome Res. 14 742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Z. Ouyang, K. Kapur, M. P. Scott and W. H. Wong, 2007. Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol. Biol. Evol. 24 1283–1285. [DOI] [PubMed] [Google Scholar]
- Yanai, I., J. O. Korbel, S. Boue, S. K. McWeeney, P. Bork et al., 2006. Similar gene expression profiles do not imply similar tissue functions. Trends Genet. 22 132–138. [DOI] [PubMed] [Google Scholar]
- Yang, J., A. I. Su and W. H. Li, 2005. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol. Biol. Evol. 22 2113–2118. [DOI] [PubMed] [Google Scholar]