Abstract
Purpose
FAS is a cell surface receptor involved in apoptotic signal transmission. Deregulation of this pathway results in down regulation of apoptosis and subsequent persistence of a malignant clone. A single nucleotide polymorphism resulting in guanine-to-adenine (G→A) transition in the FAS promoter region (position –1377) is thought to reduce stimulatory protein 1 (SP1) transcription factor binding and decrease FAS expression. Previous work has shown increased risk of developing acute myeloid leukemia (AML) in adult patients with a variant allele at this site. The same authors have shown that the presence of an adenine residue rather than a guanine residue at –1377 bp significantly attenuates transcription factor SP1 binding and may contribute to a reduction in FAS expression and ultimately to the enrichment of apoptosis-resistant clones in AML. We hypothesized that FAS genotype by altering susceptibility to apoptosis might impact outcome of childhood AML therapy.
Experimental Design
440 children treated for de novo AML on a uniform protocol were genotyped for FAS 1377.
Results
There were no significant differences in overall survival (OS), event-free survival (EFS), treatment-related mortality (TRM), or relapse rate between patients with FAS 1377GG genotype vs. 1377GA/1377AA genotypes.
Conclusion
FAS1377 genotype does not alter outcome of de novo AML in children.
Introduction
Human FAS (TNFRSF6/CD95/APO-1) protein is a cell surface receptor, belonging to the family of tumor necrosis factor receptors, and is involved in apoptotic signal transmission (1,2). It is a 48-kDa Type I membrane glycoprotein composed of 3 domains; an extracellular domain comprising of 3 cysteine-rich motif subdomains characteristic of the superfamily, a transmembrane domain, and a highly conserved intracellular domain known as a death domain (3). Binding to the receptor by the FAS ligand (CD95L) triggers receptor trimerization and subsequent assembly of the death-inducing signaling complex (4,5). Germline mutations or deletions within FAS, resulting in a loss or a reduction in receptor function, have been shown to cause autoimmune lymphoproliferative syndrome as well as an overall increased risk of hematological malignancies (6). Dysregulation of this pathway is believed to result in down regulation of apoptosis, allowing subsequent persistence of a malignant clone.
FAS expression levels may also be affected by mutations or polymorphisms in the promoter region of FAS, particularly when they affect the transcription binding sites. A single nucleotide polymorphism resulting in guanine-to-adenine (G→A) transition in the FAS promoter region occurs at position -1377, affecting a SP1 transcription factor binding site. An adenine residue at this position significantly reduces SP1 binding compared to guanine residue, causing a decrease in FAS expression. Functional germline and somatic mutations in the FAS gene and perhaps also in the FASL gene that lead to decreased expression of FAS and/or increased expression of FAS Ligand (FASL) favors malignant transformation and progression (4) by impairing apoptotic signal transduction and are associated with an increased risk of cancer (7,8,9,10). A recent case-control study in adults indicated increased risk of developing AML in patients with variant allele (A) at this site (11). In addition, reduced expression of Fas-associated protein with death domain (FADD), the main adaptor for transmission of FAS signaling, is associated with decreased response to chemotherapy in AML cells (12). We hypothesized that FAS -1377 polymorphism by altering susceptibility to apoptosis might impact outcome of AML therapy.
Patients and Methods
Patients
The study population included 440 children with de novo AML treated on Children’s Cancer Group (CCG) therapeutic studies CCG-2941(n=36) and CCG-2961(n=404) between 1995 and 2002. Clinical data, including age, sex, white blood cell (WBC) count at diagnosis, race, presence of chloroma, presence of CNS disease, and immunophenotype were collected prospectively (Table 1). Cases were classified on the basis of criteria established and revised by the French-American-British (FAB) Cooperative Study Group by central pathology review. All FAB categories except acute promyelocytic leukemia (APL – AML M3) were eligible for enrollment and were treated with the same chemotherapy regimens.
Table 1.
Demographics of patients and distribution of FAS genotypes
| Characteristic | GG (N=347) | GA (N=81) | AA (N=12) | GG vs GA |
GA vs AA |
GG vs AA |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | P | P | P | ||
| Age: median & range (yrs) | 9.4 | (0.01 – 20.9) | 11.5 | (0.47 – 19.0) | 10.1 | (1.64 – 16.2) | 0.12 | 0.99 | 0.50 | |
| WBC: median & range | 20600 | (1000 –860000) | 18400 | (300 – 373300) | 17700 | (1500 –848000) | 0.67 | 0.52 | 0.38 | |
| Study | ||||||||||
| CCG-2941 | 30 | 9% | 6 | 7% | 0 | 0% | 0.89 | 1.00 | 0.61 | |
| CCG-2961 | 317 | 91% | 75 | 93% | 12 | 100% | ||||
| Gender | ||||||||||
| Male | 189 | 55% | 49 | 61% | 6 | 50% | 0.39 | 0.54 | 0.99 | |
| Female | 158 | 45% | 32 | 39% | 6 | 50% | ||||
| Race | ||||||||||
| White | 246 | 71% | 54 | 67% | 6 | 50% | 0.52 | 0.34 | 0.19 | |
| Black | 31 | 9% | 4 | 5% | 1 | 8% | 0.34 | 0.51 | 1.00 | |
| Hispanic | 49 | 14% | 18 | 22% | 1 | 8% | 0.10 | 0.45 | 1.00 | |
| Asian | 5 | 1% | 4 | 5% | 4 | 33% | 0.07 | 0.01 | <0.01 | |
| Other | 15 | 4% | 1 | 1% | 0 | 0% | 0.33 | 1.00 | 1.00 | |
| Unknown | 1 | 0 | 0 | |||||||
| Response at end of first course |
||||||||||
| Remission | 290 | 87% | 71 | 90% | 12 | 100% | 0.55 | 0.59 | 0.38 | |
| PD | 23 | 7% | 7 | 9% | 0 | 0% | 0.71 | 0.59 | 1.00 | |
| Die | 22 | 7% | 1 | 1% | 0 | 0% | 0.10 | 1.00 | 1.00 | |
| W/D or unevaluable | 12 | 2 | 0 | |||||||
Chemotherapy Treatment Regimen
CCG-2961 study was a randomized phase III trial of intensively timed induction, consolidation, and intensification therapy for pediatric patients with previously untreated AML or MDS (13). The study was conducted between August 1996 and December 2002. CCG 2941 was a feasibility pilot of the same chemotherapy regimen that preceded the randomized study. Induction included 5 drugs: idarubicin, etoposide, dexamethasone, cytarabine, and 6-thioguanine (IDA DCTER) given on days 0-3 followed by 5 drugs (daunorubicin, etoposide, dexamethasone, Ara-C and 6-thioguanine) (DCTER) given on days 10–13 (14). Upon recovery of white blood cell and platelet counts, patients were randomly assigned to consolidation therapy consisting of the same sequence of drugs or to fludarabine/ cytarabine /idarubicin. Intrathecal cytarabine was used for CNS prophylaxis. Patients with matched-related donors were assigned to allogeneic marrow transplant intensification. Pre-transplant cytoreduction was busulfan and cyclophosphamide. Patients without a related donor received high-dose cytarabine/L-asparaginase (Capizzi II), and additional intrathecal cytarabine. After recovery from chemotherapy, patients were randomized again to either receive Interleukin-2 or standard follow-up care. Transplanted patients were not eligible for randomization to interleukin-2.
FAS genotyping
DNA extracted from diagnostic marrow samples using standard methods was normalized to 10 ng/µl. Primers were synthesized based on the sequence of the Apo-1/FAS gene reported in Gene Bank (X87625).
For each sample testing, two PCR reactions in different wells were performed; one reaction detected the wild-type allele and the other detected the mutant allele using mismatch amplification assay coupled with real time PCR (MAMA assay). This assay is based on the concept that two mismatched nucleotides instead of a single mismatch at the 3’ end of the primer effectively abrogates PCR amplification efficiency.
For each reaction, a 20 ng DNA template was added to the reaction mixture, containing a final concentration of 0.2 µM of each primer -specific forward primer (F1 or F2) and the common reverse primer and sybergreen mastermix (Applied biosystems, CA, USA). RT-PCR was performed using an ABI 7700 Thermocycler for 40 cycles, with annealing temperature of 62°C and a total of 50 µl reaction volume.
DNA from normal controls was extracted using standard techniques and genotyped as described for cases. Genotyping results were duplicated in 10% of samples; concordance between repeats was 100%. Furthermore, 10% of the samples were also genotyped using direct sequencing; concordance with MAMA genotyping was 100%.
Statistical Analysis
Data were analyzed from CCG-2941 and CCG-2961 through April 2005 and October 2006, respectively. The significance of observed differences in proportions was tested using the Chi-squared test and Fisher’s exact test when data were sparse. The Mann-Whitney test was used to determine the significance between differences in medians (15). The Kaplan-Meier method was used to calculate estimates of overall survival (OS), event-free survival (EFS) and disease-free survival (DFS) (16). Estimates are reported with their Greenwood standard errors (17). Differences in these estimates were tested for significance using the log-rank statistic (18). OS is defined as time from study entry to death from any cause. EFS is defined as time from study entry to failure at the end of two courses, relapse or death from any cause. DFS is defined as time from the end of one course of therapy to failure at the end of two courses, relapse or death from any cause. Cumulative incidence estimates were used to determine relapse rate (RR) and treatment-related mortality (TRM). RR is defined as time from the end of one course of therapy to failure at the end of two courses, relapse or death from progressive disease where deaths from non-progressive disease were competing events. TRM is defined as time from study entry to death from non-progressive disease where failures at the end of two courses, relapses and deaths from progressive disease were competing events. Differences between RR or TRM estimates were tested for significance using Gray’s test (19). Children lost to follow-up were censored at their date of last known contact or at a cutoff 6 months prior to April 2005 (CCG-2941) or October 2006 (CCG-2961). Cox regression was used for multivariate models that looked at differences between groups adjusting for study assignment, age, gender, race, and WBC count. Reported p-values represent comparisons between patients with FAS 1377GG genotype vs. those with FAS 1377GA or 1377AA genotypes.
Results
FAS Genotype and Outcome
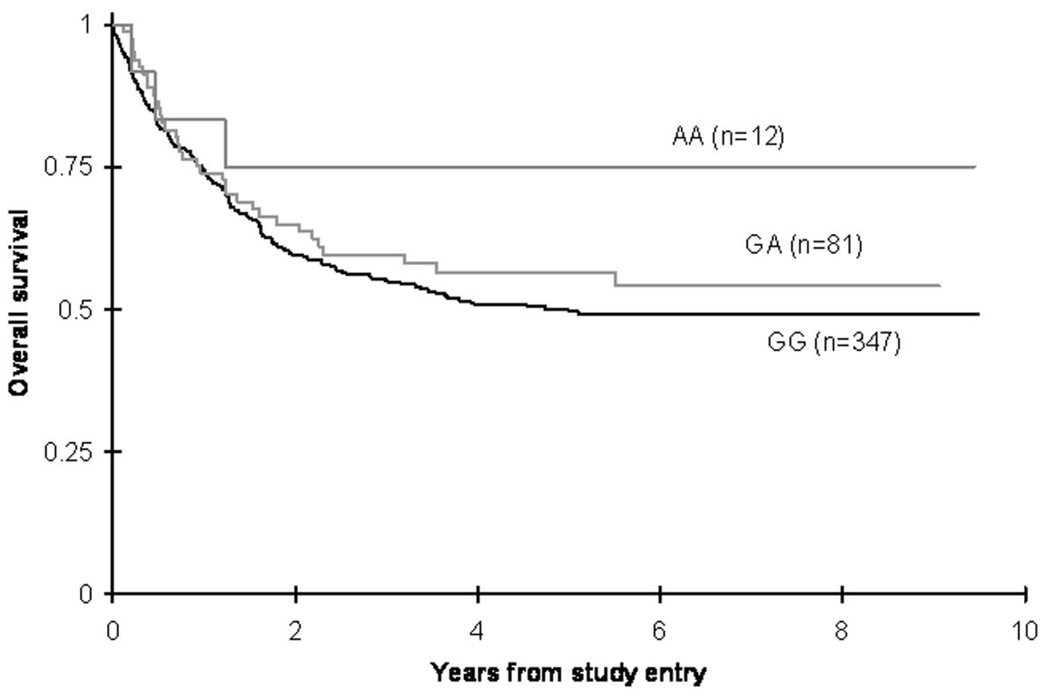
There were no significant differences in OS from study entry between patients with FAS1377GG, 1377GA or 1377AA genotypes (50 ± 6% vs. 57 ± 11% vs. 75 ± 25% respectively at 5 years; log-rank p = 0.21). Although point estimate for OS for AA genotype was 75% compared to 50% for GG genotype, the AA group included only small number of patients (n=12) (Figure 1, Table2). Analysis of EFS from study entry in different genotypes also showed similar estimates, 40 ± 5% in FAS 1377GG vs. 44 ± 11% in 1377GA vs. 67 ± 27% in 1377AA patients at 5 years (log rank p= 0.36).
Figure 1. Overall survival of pediatric AML patients according to FAS 1377 polymorphism status.
Difference in OS from study entry between patients with FAS1377GG vs. 1377GA vs. 1377AA genotypes (p=0.21).
Table 2.
Treatment outcomes according to FAS genotype.
| FAS 1377 GG | FAS 1377 GA | FAS 1377 AA | p-value | |
|---|---|---|---|---|
| 5 year OS | 50 ± 6% | 57 ± 11% | 75 ± 25% | 0.21 |
| 5 year EFS | 40 ± 5% | 44 ± 11% | 67 ± 27% | 0.36 |
| 5 year DFS | 46 ± 6% | 50 ± 12% | 67 ± 27% | 0.63 |
| 5 year TRM | 18 ± 4% | 14 ± 8% | 17 ± 22% | 0.42 |
| 5 year RR | 41 ± 6% | 38 ± 12% | 19 ± 24% | 0.47 |
There was no difference in treatment related mortality (TRM) from study entry between different genotypes (18 ± 4% in GG vs. 14 ± 8% in GA vs. 17 ± 22% in AA patients) at 5 years (p=0.42). Relapse rates (RR) from end of one course of therapy for patients in remission were also similar; 41 ± 6% in FAS 1377GG vs. 38 ± 12% in 1377GA vs. 19 ± 24% in 1377AA patients at 5 years (p=0.47).
Multivariate analyses that adjusted for study assignment, age, gender, race, and WBC also suggested that FAS genotype did not modify OS or EFS. Thus, FAS genotype was not significantly associated with either resistant disease or treatment-related toxicity.
Discussion
In this pediatric study we found that FAS1377 genotype does not alter outcome of de novo AML in children. Role of FAS variants in leukemogenesis is supported by studies that report increased risk of lymphoproliferative disease and certain hematologic malignancies associated with defects in the FAS pathway. Down-regulation of apoptosis prolongs the normal cellular life span, allows cells to acquire mutations and facilitates tumor progression, for example, BCL2 transgenic mice with targeted expression in myeloid cells develop myeloproliferation similar to human chronic myelomonocytic leukemia; however, they rarely go on to develop acute leukemia. Interestingly, when mice constitutively expressing BCL2 are crossed onto a FAS−/− background, approximately 15% develop AML, implicating FAS-mediated apoptosis in the pathogenesis of AML (20).
In humans, germline mutations or deletions within FAS have been shown to cause autoimmune lymphoproliferative syndrome, a condition associated with generalized lymphoproliferation and systemic autoimmunity, as well as an overall increased risk of hematological malignancies (6). Also, aberrant apoptosis is known to be important in the pathogenesis of AML (6) because up-regulation of the antiapoptotic protein BCL2 is a common feature of leukemic blasts and may be a critical event in myeloid transformation (21). Myeloblasts are known to express high levels of FAS, and functional deficiencies of FAS signaling have been shown to be important in several subtypes of AML, providing further evidence for FAS-mediated apoptosis in the etiology of AML (22).
These same mechanisms potentially make these AML cells more resistant to therapy. As shown by Sibley et al (11) the presence of an adenine residue rather than a guanine residue at −1377 bp significantly attenuates transcription factor SP1 binding and may contribute to a reduction in FAS expression and ultimately to the enrichment of apoptosis-resistant clones in AML. However, our study did not show significant impact of FAS genotype on outcome of childhood AML. It is possible that this FAS variant may be of functional importance in children with AML when combined with polymorphic alleles of other genes in the apoptosis pathway. Mechanistic studies further defining the functionality of FAS polymorphisms will help clarify the importance of variants at this site.
Statement of Clinical Relevance
Available response of children to chemotherapy for acute myeloid leukemia (AML) remains a challenge to the cure of this difficult disease. The development of personalized medicine allows access to potential avenues to the development of the individualized therapy and optimize outcomes for each individual child. In this study we looked at a polymorphic locus and it’s influence on outcome, and describe the relevance of variation at the FAS locus to chemotherapy response for childhood AML. In the future we anticipate that chemotherapy regimens will be individualized to the patient’s genotype. We anticipate that further candidate studies such as the one presented here will help individualize therapy for children with AML.
Acknowledgments
Supported by Gregory H. Reaman U10 CA098543- 06 Children’s Oncology Group Chair’s Grant Stella MARGARET. Davies R01 CA093552- 01 Host factors and etiology of leukemia
References
- 1.Itoh N, Yonehara S, Ishii A, et al. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 2.Oehm A, Behrmann I, Falk W, et al. PH Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor family. J Biol Chem. 1992;267:10709–10715. [PubMed] [Google Scholar]
- 3.Itoh N, Nagata S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 4.Muschen M, Warskulat U, Beckmann MW. Defining CD95 as a tumor suppressor gene. J Mol Med. 2000;78:312–325. doi: 10.1007/s001090000112. [DOI] [PubMed] [Google Scholar]
- 5.Yonehara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieux-Laucat F, Le Deist F, Hivroz C, et al. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 7.Davidson WF, Giese T, Fredrickson TN. Spontaneous development of plasmacytoid tumors in mice with defective FAS-FAS ligand interactions. J Exp Med. 1998;187:1825–1828. doi: 10.1084/jem.187.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters AM, Kohfink B, Martin H, et al. Defective apoptosis due to a point mutation in the death domain of CD95 associated with autoimmune lymphoproliferative syndrome, T-cell lymphoma, and Hodgkin’s disease. Exp Hematol. 1999;27:868–874. doi: 10.1016/s0301-472x(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee SH, Shin MS, Park WS, et al. Alterations of FAS (Apo-1/CD95) gene in transitional cell carcinomas of urinary bladder. Cancer Res. 1999;59:3068–3072. [PubMed] [Google Scholar]
- 10.Takahashi T, Tanaka M, Brannan CI, et al. Generalized lymphoproliferative disease in mice, caused by a point mutation in the FAS ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 11.Sibley K, Rollinson S, Allan JM, et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63:4327–4330. [PubMed] [Google Scholar]
- 12.Tourneur L, Delluc S, Levy V, et al. Absence or low expression of fas-associated protein with death domain in acute myeloid leukemia cells predicts resistance to chemotherapy and poor outcome. Cancer Res. 2004;64:8101–8108. doi: 10.1158/0008-5472.CAN-04-2361. [DOI] [PubMed] [Google Scholar]
- 13.Lange BJ, Gerbing RB, Feusner J, et al. Mortality in overweight and underweight children with acute myeloid leukemia. JAMA. 2005;293:203–211. doi: 10.1001/jama.293.2.203. [DOI] [PubMed] [Google Scholar]
- 14.Lange BJ, Dinndorf P, Smith FO, et al. Pilot study of idarubicin-based intensive timing induction therapy for children with previously untreated acute myeloid leukemia: Children’s Cancer Group (CCG) Study 2941. J Clin Oncol. 2004;22:150–156. doi: 10.1200/JCO.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Statistics. 1947;18:50–60. [Google Scholar]
- 16.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 17.Greenwood M. The natural duration of cancer: Reports on Public Health and Medical Subjects. London, United Kingdom: Her Majesty’s Stationery Office; 1926. pp. 1–33. [Google Scholar]
- 18.Peto R, Peto J. Asymptotically efficient rank in variant text procedures. J R Stat Soc. 1972;135:185–206. [Google Scholar]
- 19.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. The Annals of Statistics. 1988;16:1141–1154. [Google Scholar]
- 20.Traver D, Akashi K, Weissman IL, Lagasse E. Mice defective in two apoptosis pathways in the myeloid lineage develop acute myeloblastic leukemia. Immunity. 1998;9:47–57. doi: 10.1016/s1074-7613(00)80587-7. [DOI] [PubMed] [Google Scholar]
- 21.Delia D, Aiello A, Soligo D, et al. bcl-2 proto-oncogene expression in normal and neoplastic human myeloid cells. Blood. 1992;79:1291–1298. [PubMed] [Google Scholar]
- 22.Komada Y, Zhou YW, Zhang XL, et al. FAS receptor (CD95)-mediated apoptosis is induced in leukemic cells entering G1B compartment of the cell cycle. Blood. 1995;86:3848–3860. [PubMed] [Google Scholar]