Summary
Complex signaling events control tumor invasion in three-dimensional (3D) extracellular matrices. Recent evidence suggests that cells utilize both matrix metalloproteinase (MMP)-dependent and MMP-independent means to traverse 3D matrices. Herein, we demonstrate that lysophosphatidic-acid-induced HT1080 cell invasion requires membrane-type-1 (MT1)-MMP-mediated collagenolysis to generate matrix conduits the width of a cellular nucleus. We define these spaces as single-cell invasion tunnels (SCITs). Once established, cells can migrate within SCITs in an MMP-independent manner. Endothelial cells, smooth muscle cells and fibroblasts also generate SCITs during invasive events, suggesting that SCIT formation represents a fundamental mechanism of cellular motility within 3D matrices. Coordinated cellular signaling events are required during SCIT formation. MT1-MMP, Cdc42 and its associated downstream effectors such as MRCK (myotonic dystrophy kinase-related Cdc42-binding kinase) and Pak4 (p21 protein-activated kinase 4), protein kinase Cα and the Rho-associated coiled-coil-containing protein kinases (ROCK-1 and ROCK-2) coordinate signaling necessary for SCIT formation. Finally, we show that MT1-MMP and Cdc42 are fundamental components of a co-associated invasion-signaling complex that controls directed single-cell invasion of 3D collagen matrices.
Keywords: MT1-MMP, Invasion, Cdc42, HT1080, LPA, Three-dimensional, Nucleus, Metastasis, Extracellular matrix
Introduction
Tumor cell metastasis and invasion predict poor survival outcomes in cancer patients (Sporn, 1996). In order for malignant cells to disseminate from primary growths, neoplastic cells must navigate a dense, cross-linked extracellular matrix (ECM) composed mostly of type I collagen (Chambers et al., 2002; Hanahan and Weinberg, 2000). Recently, contrasting theories about dispersion of cancer cells through the ECM have emerged. One hypothesis proposes a requisite role for the membrane-tethered collagenase membrane-type-1 matrix metalloproteinase (MT1-MMP) (also known as MMP14) via its ability to proteolytically degrade matrix constituents such as type I collagen (Fisher et al., 2006; Li et al., 2008; Sabeh et al., 2004; Seiki, 2003). These reports suggest that MT1-MMP-expressing tumor cells harbor an intrinsic capability to navigate the ECM due to the collagenolytic activity of this enzyme. Despite numerous studies implicating MMPs (MT1-MMP in particular) in tumor invasion (Cruz-Munoz et al., 2006; Hojilla et al., 2003; Itoh and Nagase, 2002; Stetler-Stevenson and Yu, 2001), clinical trials targeting these enzymes yielded poor results (Coussens et al., 2002). However, most drugs in clinical trials targeted a broad range of MMPs, and recent work demonstrates that certain MMPs might directly (via modulation of the extracellular environment) or indirectly (through initiation of vascular regression) affect disease progression (Berrier and Yamada, 2007; Egeblad and Werb, 2002; Saunders et al., 2005). Nonetheless, these results, combined with additional data, spawned an alternative hypothesis for tumor invasion: tumor cells adapt an `amoeboid' phenotype akin to the amoeba Dictyostelium discoideum or to leukocytes that facilitates navigation of ECM barriers by mechanical means without proteolysis (Friedl et al., 2001; Wolf et al., 2003; Wolf et al., 2007).
Although MT1-MMP expression correlates with metastatic ability, not all metastatic tumors express this enzyme (Sato et al., 2005). Additionally, when tumor cells that express MT1-MMP (such as HT1080s) are embedded in dense physiologically cross-linked type I collagen (i.e. 2.0-5.0 mg/ml type I collagen), and broad-scale MMP inhibitors are added, MMP-independent invasion is not observed (Fisher et al., 2006; Sabeh et al., 2004). These discrepancies suggest cooperation between both collagenolytic MMP-dependent invasion and MMP-independent invasion, which might relate to previously described `amoeboid' motility events.
Here, we demonstrate that invading tumor cells (HT1080s) generate single-cell invasion tunnels (SCITs) via MT1-MMP that are the width of cellular nuclei. Once SCITs form, HT1080s can move within a 3D matrix in an MMP-independent manner. The formation of SCITs requires unique molecular signaling events involving MT1-MMP, Cdc42, Cdc42 effectors, protein kinase Cα (PKCα) and Rho-associated coiled-coil-containing protein kinases 1 and 2 (ROCK-1 and ROCK-2; abbreviated here to ROCK-1/2). These signaling events are stimulated by addition of lysophosphatidic acid (LPA). We demonstrate that non-tumor cells such as endothelial cells, fibroblasts and coronary artery smooth muscle cells also create SCITs during invasion of 3D matrices. Overall, these results suggest a mechanism for tumor dispersion whereby cells utilize MT1-MMP-mediated proteolysis to generate SCITs (which are the diameter of the nucleus). These provide a temporal framework for proteolytic, followed by non-proteolytic, means of cell motility in 3D matrices. The results also reveal distinct signaling mechanisms vital for cell motility in a 3D matrix environment.
Results
Tumor cells require MT1-MMP to initiate but not to sustain motility in 3D collagen matrices
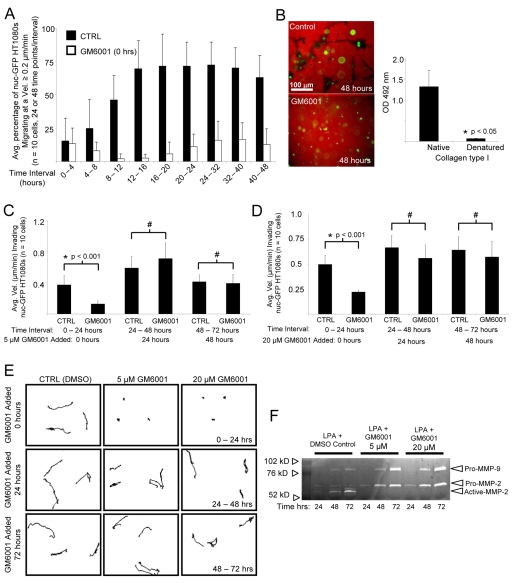
LPA induces HT1080 invasion of 3D collagen matrices in an MT1-MMP-dependent manner and the distances and velocities of invading cells can be quantitated over time (Fig. 1A) (Fisher et al., 2006). When GM6001 (a broad-scale MMP inhibitor) is added at 5 or 20 μM doses to invading 3D cultures at the initiation of invasion (time 0), motility is markedly reduced and these doses block pro-MMP-2 activation over a 72-hour period (Fig. 1F). However, migration velocity remains constant when GM6001 is added after 24 or 48 hours (Fig. 1C-E). Movies demonstrating the motility of nuclear GFP (nuc-GFP)-labeled HT1080s in 3D are presented under these conditions (supplementary material Movies 1-4). The data demonstrate that MMPs are required to initiate motility in 3D collagen matrices (Fig. 1, supplementary material Movie 2), but are not required to maintain motility once sufficient time has elapsed (supplementary material Movie 4). By contrast, cell movement is observed when GM6001 is added after 48 hours of culture (supplementary material Movie 4), which is directly comparable to control cell movement in 3D collagen matrices (Fig. 1, supplementary material Movie 2).
Fig. 1.
MMP-dependent HT1080 cell invasion in 3D collagen matrices precedes the appearance of MMP-independent invasive behavior. (A) nuc-GFP HT1080s were seeded in 3D collagen matrices with LPA and stimulated to invade for 48 hours in the presence or absence of 5 μM GM6001. Data are expressed as mean velocity of invading cells ± s.d. (error bars) for n=10 cells per condition over the indicated time intervals. (B) Assays set up as above were stained for tunnels using an anti-type-I-collagen antibody that only recognizes native collagen in ELISA assays, as shown in the right panel. (C-E) Assays were quantitated for invasion velocities (n=10 cells) using real-time analysis in the absence or presence of GM6001 at 5 μM (C) or 20 μM (D). Statistical significance *P<0.001; #, not statistically significant.. (E) Representative tracings of HT1080 motility are shown under the indicated conditions and time intervals. (F) Conditioned medium obtained from the indicated cultures and time points were analyzed using gelatin zymography.
For the first 16 hours, the percentage of a specified time interval that cells require to achieve velocities ≥0.2 μm/min steadily increases. After 16 hours, cells sustain velocities ≥0.2 μm/min for more than 75% of the designated interval, suggesting that cells require ∼16-20 hours to achieve maximum velocity (Fig. 1A). We hypothesized that matrix proteolysis is the rate-limiting step for cellular velocity in a 3D matrix. Thus, we reasoned that after ∼24 hours, the matrix is sufficiently degraded to sustain maximum velocity and to allow MMP-independent cellular motility, and this result is demonstrated in Fig. 1C-E.
Invading cells generate SCITs
Cellular velocity is markedly reduced when cells are embedded in 3D collagen compared with collagen-coated 2D substrates (Fisher et al., 2006). We hypothesized that the increase in cellular velocity in the first 16 hours was attributable to matrix degradation and the generation of SCITs. At later time points, cells do not require MMPs for motility because once SCITs form, matrix degradation is no longer required (i.e. the matrix contains spaces for cells to move within).
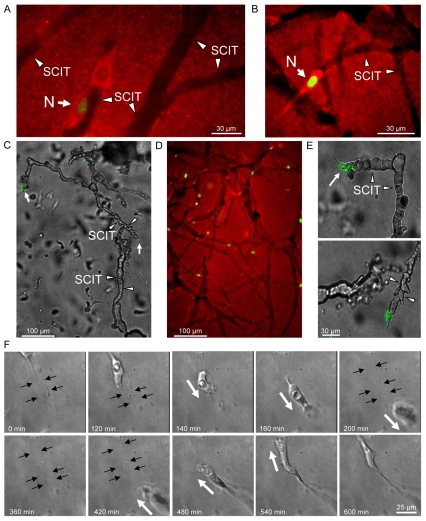
To demonstrate the existence of SCITs in our assays, nuc-GFP HT1080s were embedded in 2.5 mg/ml collagen gels containing 1 μM LPA and allowed to invade. SCITs were observed with fluorescence microscopy using an anti-collagen I antibody that recognizes only native collagen and not denatured collagen (Fig. 1B, Fig. 2A,B,D). The non-proteolyzed collagen matrix stains red, whereas the areas of collagen degradation (SCITs) do not stain. Fig. 2A demonstrates a cell migrating along the edge of a SCIT (arrowheads denote SCIT borders). These images demonstrate the presence of SCITs and provide a permanent record of the migration paths of cells during cellular invasion. A previous elegant assay used gold particles to monitor cell motility and migration paths on 2D surfaces (Albrecht-Buehler, 1977). In many respects, the detection of SCITs is an analogous approach to the detection of cell motility in 3D matrices.
Fig. 2.
HT1080s generate SCITs during invasion of 3D collagen matrices. (A,B) nuc-GFP HT1080s were allowed to invade, and representative SCITs were detected by immunofluorescence using an anti-collagen I antibody. Arrowheads denote the SCIT border. Arrows denote the nucleus (N). (C) SCITs were injected at a single site with silicone oil. (D) Comparison of an oil-injected tunnel and tunnels detected by immunofluorescence. (E) Higher-power image of invading cell (arrow) at the terminal end of a tunnel (arrowheads). (F) Representative images from a time-lapse movie of a nuc-GFP HT1080 traveling within a SCIT. Black arrows denote margins of SCITs. White arrows denote direction of cell motility.
SCITs are hollow continuous spaces in a 3D matrix
The observation that cells use proteolysis in order to make paths to invade 3D collagen matrices has been previously described (Bayless and Davis, 2003; Gaggioli et al., 2007; Sabeh et al., 2004; Wolf et al., 2003; Wolf et al., 2007). However, there exists little evidence that single cells generate these paths, that tunnels persist and are patent, or that cells can traverse tunnels once they have formed. For SCITs to function as matrix conduits for cellular migration, we hypothesized that they must be hollow, continuous, and patent (similar to an underground tunnel). To demonstrate this, micropipette-guided silicone-oil-perfusion of the SCITs was performed. Remarkably, extensive SCIT networks were observed as a result of a single injection (Fig. 2C). In GM6001-treated cultures, no tunnels were perfused with 20 injections (data not shown), showing that MMP-dependent proteolysis was necessary to create these SCIT physical spaces. A low power fluorescent image corroborated the numerous SCITs present in a single field (Fig. 2D). These data indicate that multiple SCITs can intersect to create vast, patent and continuous tunnel networks. The presence of HT1080s (GFP-tagged) at the terminal end of the tunnels (Fig. 2C-E) suggests that proteolysis occurs at the leading edge of the cell and that tunnels form behind invading cells. Using time-lapse microscopy, we identified numerous instances where cells traversed a pre-existing SCIT. Representative time-lapse images of a cell traveling through a SCIT and then subsequently returning within the same structure are shown in Fig. 2F. The black arrows denote the SCIT borders and the white arrows denote the direction of motility. A movie displaying the entire 10-hour transit is provided (supplementary material Movie 5). Together, these data support the hypothesis that SCITs are continuous, hollow and patent structures in a 3D matrix that can function as conduits for cellular migration.
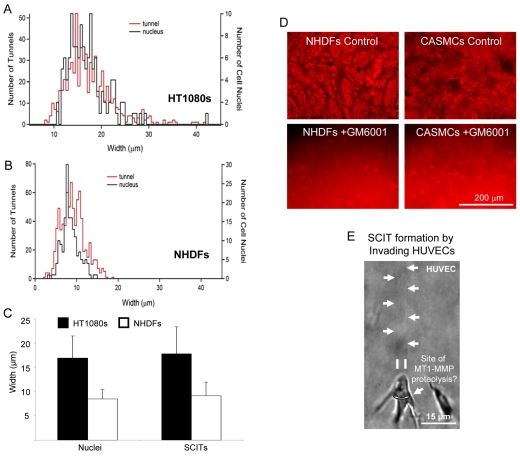
SCIT width is equivalent to the diameter of nuclei in invading cells
The images in Fig. 2 suggest that the width of SCITs is nearly identical to the diameter of nuclei within invading cells. To quantitate this observation, a histogram of nuclear width and SCIT width from collagen-stained cultures was generated by measuring SCIT width and nuclear diameter from fluorescently labeled images (Fig. 3A-C). Both the histogram measurements and the image measurements revealed nuclear and SCIT mean widths of ∼18 μm for HT1080 fibrosarcoma cells and ∼9 μm for normal human dermal fibroblasts (NHDFs). These data demonstrate that SCIT width is dictated by the width of the nucleus of the invading cell forming the SCIT. The data suggest that MMP-dependent mediated proteolysis must in some manner be linked to a cell membrane location in close proximity to the position of nuclei during invasive events. LPA-induced invasion leads to SCIT formation behind the cell, and with a width identical to that of the nucleus of the cell invading.
Fig. 3.
Invading cells generate SCITs the width of cellular nuclei. (A) Histogram demonstrating the number of nuclei (black) or tunnels (SCITs) (red) is displayed for a given width. (B) An overlay of the histograms with the y-axes rescaled (tunnel on left, nuclei on right). (C) Comparison of the average width of nuclei and SCITs from the histograms in A and B. Data are expressed as mean width in μm ± s.d. (n=172 for nuclei; 790 for SCITs). (D) CaSMCs or NHDFs invaded 2.5 mg/ml collagen matrices containing 200 ng/μl PDGF-BB in the presence or absence of 5 μM GM6001. (E) HUVECs were induced to invade 3D collagen matrices with 1 μM sphingosine-1-phosphate and 200 ng/ml of stromal-derived factor-1α for 24 hours.
MMP-generated SCITs are fundamental regulators of motility in 3D collagen matrices
The density and composition of a 3D matrix can influence cellular motility within it (Yamada and Cukierman, 2007; Zaman et al., 2006). Recent reports suggest that cells acquire a non-proteolytic amoeboid phenotype in response to MMP inhibition (Wolf et al., 2003; Wolf et al., 2007). These data argue that cells might not generate SCITs at lower matrix concentrations because escape strategies exist or because lattice porosity is such that cells can navigate the matrix independently of remodeling. However, our data demonstrate that invading cells produce SCITs at low concentrations of collagen (1.0, 2.0 and 3.0 mg/ml) (supplementary material Fig. S1A). Additionally, GM6001 abolishes SCIT formation and cell invasion even at a concentration of 1.0 mg/ml collagen. These data show that when cells navigate physiologic cross-linked collagen even at low densities, MT1-MMP-dependent SCIT formation occurs.
To demonstrate that other non-tumor cell lines generate SCITs during invasion, NHDFs or coronary artery smooth muscle cells (CaSMCs) were embedded within collagen matrices containing platelet-derived growth factor (PDGF)-BB and allowed to invade for 48 hours. Images were acquired as described in the Materials and Methods section. SCITs were present in all cases and SCIT formation was abolished in the presence of GM6001 for all cell types tested (Fig. 3D). Single ECs are also capable of SCIT formation during invasive events (Fig. 3E, supplementary material Fig. S2) (Bayless and Davis, 2003). ECs, NHDFs and vascular smooth muscle cells have been previously reported to invade collagen in an MT1-MMP-dependent manner (Filippov et al., 2005; Sabeh et al., 2004; Saunders et al., 2006; Stratman et al., 2009). Collectively, these data demonstrate that SCIT formation is a fundamental required step for cellular motility and invasive behavior in 3D collagen matrices. SCITs could also provide a mechanism for the observed phenomenon of MMP-independent motility in such matrices.
MT1-MMP is required for SCIT formation
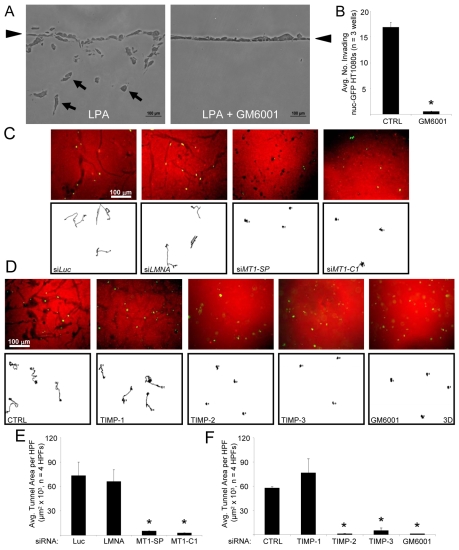
MMPs play a seminal role in HT1080 invasion from a monolayer in our assays. This is demonstrated in previous work (Fisher et al., 2006) and also in a cross-section of invading HT1080 cells from a monolayer surface (arrowheads) (Fig. 4A). The cross-section demonstrates complete blockade of LPA-induced HT1080 invasion in the presence of GM6001 (Fig. 4A,B). MT1-MMP is a membrane-tethered collagenase that acts as a principle modulator of the pericellular environment (Hotary et al., 2000). Due to the prominent role of MT1-MMP in collagenolysis (Holmbeck et al., 2004) and its documented role in tumor invasion (Fisher et al., 2006; Sato et al., 2005), we designed experiments to determine the role of MT1-MMP in SCIT formation.
Fig. 4.
MT1-MMP is required to form SCITs during cellular invasion. (A) Cross-sections of invading HT1080 cultures demonstrating the effects of 1 μM LPA and GM6001 on invasion of 2.5 mg/ml collagen matrices. Arrows denote invading cells. Arrowheads denote the monolayer. (B) Quantitation of A. Data are expressed as mean number of invading cells per well (n=3 wells ± s.d., 10 fields per well). (C) The indicated siRNA-treated HT1080s were assayed for invasion, and representative cell motility tracings were obtained. Gels were stained with an anti-collagen I antibody to evaluate SCIT formation after 48 hours. (D) Untreated HT1080 cells were suspended in 3D collagen matrices and cultures were either left untreated, treated with the indicated recombinant TIMPs at 5 μg/ml, or GM6001 was added at 5 μM. Cell motility tracings and stains for SCITs were performed. (E,F) Quantitation of C or D. SCIT areas were measured using MetaMorph in each field. Data are expressed as the mean area in μm2 ± s.d. (n=4). Statistical significance *P<0.05.
HT1080s treated with siRNAs targeting MT1-MMP yielded markedly fewer SCITs than HT1080s treated with control siRNAs [Luciferase (Luc), and Lamin A/C (LMNA)] (Fig. 4C-E). The average total area of multiple SCITs was tabulated using MetaMorph software. siRNAs targeting MT1-MMP [with both a Smartpool (SP) and a single siRNA (C1)] have marked blocking effects on SCIT area compared to controls (Fig. 4C-E). Specificity and efficacy of MT1-MMP knockdown was confirmed with western blot analysis and inhibition of pro-MMP-2 activation using gelatin zymography over a 72-hour period of culture (supplementary material Fig. S3). SCIT formation is markedly blocked by the addition of TIMP-2 or TIMP-3 (which block MT1-MMP) but not by the addition of TIMP-1 (which does not block MT1-MMP activity) (Fig. 4D,F). Collectively, these data demonstrate a required role for MT1-MMP in SCIT formation during HT1080 cell invasion of 3D collagen matrices.
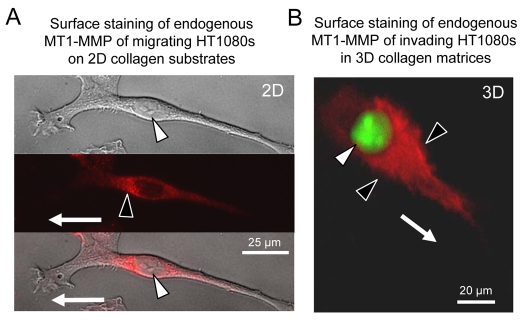
In support of these results, immunostaining (in the absence of cell permeabilization) of migrating and invading HT1080 cells reveals cell-surface-expressed MT1-MMP in a proximal cell surface and perinuclear region in the direction of motility (Fig. 5A,B). Our findings show that cells with differing nuclear widths create SCIT diameters that are the widths of their nuclei (Fig. 3). Thus, we propose that SCIT formation occurs to create sufficient physical space within the ECM to allow movement of cells with their nuclei through 3D matrices.
Fig. 5.
Cell-surface MT1-MMP in HT1080 cells is localized to a plasma membrane region in a perinuclear location and is concentrated proximal to the nucleus in the direction of migration and invasion. (A) HT1080 cells on collagen substrates were probed with anti-MT1-MMP in the absence of detergent so that only cell-surface MT1-MMP was detected. White arrowhead indicates the nucleus; black arrowhead indicates cell-surface MT1-MMP staining. Arrow indicates migration direction. (B) Fluorescent overlay image of LPA-induced invading nuc-GFP HT1080 in 3D collagen probed with anti-MT1-MMP in the absence of detergent. White arrowhead indicates nucleus; black arrowhead indicates MT1-MMP localization on the cell surface in the front of the nucleus and in the direction of invasion (white arrow).
SCIT formation requires MT1-MMP, Cdc42, PKCα and ROCK-1/2 signaling
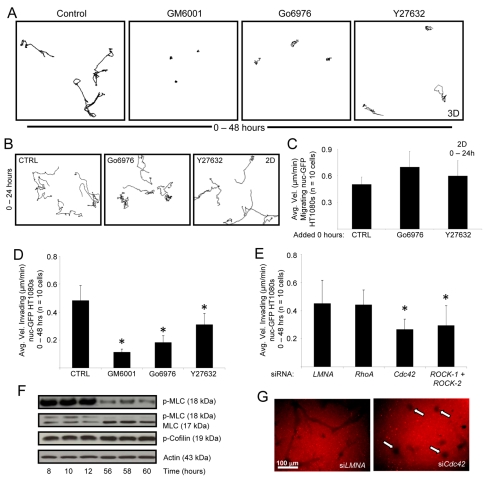
To address the signaling mechanisms involved in SCIT formation, we designed a pharmacological screen to identify inhibitors of SCIT formation. Kinase inhibitors were added to cultures at time 0 and the velocity of invasion of single cells within gels was assessed (supplementary material Fig. S1B). As a result of this screen, the effects of Y27632 or hydroxyfasudil (HF) (inhibitors of ROCK-1/2) and Go6976 (an inhibitor of PKCα and PKCβ) identified two molecular signaling pathways for further investigation, ROCK-1/2 and PKCα, respectively.
The results of the screen are summarized in Fig. 6A,B. GM6001, Y27632 and Go6976 all inhibit SCIT formation. Cells treated with Y27632 do not travel a substantial distance from their origin, but instead, exhibit a `carousel' phenotype whereby cells spin in place (supplementary material Movie 6). Thus, velocity (Δx/Δt) is artificially increased if cells travel many pixel lengths in a non-directional manner (cells do not travel far from their origin). Therefore, the tracings are provided to demonstrate distance traveled from origin (Fig. 6A,D). Of great interest is that the blocking effect of these inhibitors is selective to HT1080 cells in 3D collagen matrices in that they have no blocking influence on motility of HT1080 cells on 2D collagen-coated substrates (Fig. 6B,C). Interestingly, we find that Go6976 can block HT1080 motility within pre-existing SCITs whereas Y27632 does not (supplementary material Fig. S1C), which raises the interesting strategy of identifying new molecular requirements controlling cell motility within SCITs.
Fig. 6.
SCIT formation requires Cdc42, PKCα and ROCK-1/2 signaling. (A) 48-hour cell tracings from invading HT1080 3D cultures are shown after the addition of the specified inhibitors at 0 hours. (B) 24-hour tracings of migrating HT1080 cells on 2D collagen substrates. (C,D) Quantitation of A and B, respectively. (E) nuc-GFP HT1080s were transfected with the indicated siRNA and stimulated to invade. Data are expressed as mean velocity of invading cells ± s.d. for n=10 cells per condition. (F) Invading HT1080 cultures were lysed at the designated time points for western blot analysis. (G) nuc-GFP HT1080s were transfected with the indicated siRNA, stimulated to invade, then fixed and stained with an anti-collagen-I antibody. Statistical significance *P<0.05.
The roles for ROCK-1 and ROCK-2 in SCIT formation are corroborated by both the pharmacological effects of Y27632 and HF and the siRNA suppression of ROCK-1/2 (Fig. 6E, supplementary material Fig. S1B). Also, nuc-GFP HT1080s treated with siRNAs against Cdc42 and ROCK-1/2, but not against RhoA or control LMNA, were inhibited in 3D invasion assays. siRNAs targeting ROCK-1 or ROCK-2 individually had no effect (data not shown). Interestingly, nuc-GFP HT1080s treated with Cdc42 siRNA exhibit a similar `carousel' phenotype to Y27632-treated cells compared to control cells, which productively invade and show directional motility (supplementary material Movies 6-9). Marked blockade of invasion and motility are observed following suppression of Cdc42 and ROCK-1/2 siRNA (Fig. 6E, Fig. 7A,B), as evidenced by analysis of motility and SCIT formation (Fig. 6G). In addition, real-time video analysis reveals defective motility (i.e. lack of sustained directional motility) following Cdc42 knockdown compared to control-siRNA-treated HT1080 cells (supplementary material Movies 8, 9). The inhibitory influence of Cdc42 siRNA is specific in this case to 3D collagen matrices because this same treatment has no inhibitory effect on HT1080 cell motility on 2D collagen-coated surfaces (Fig. 7C,D). Previous studies show that blockade of MT1-MMP function or expression also selectively blocks motility of HT1080 cells in 3D collagen matrices but not on 2D collagen substrates (Fisher et al., 2006).
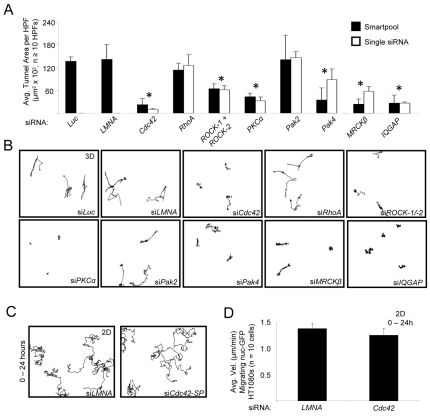
Fig. 7.
Essential functional role for Cdc42, Cdc42 effectors, PKCα and ROCK-1/2 in LPA-induced HT1080 cell invasion and SCIT formation in 3D collagen matrices. (A) siRNA suppression of nuc-GFP HT1080 cells was performed using either Smartpools or single siRNAs for each of the indicated genes. After 48 hours, cultures were stained using anti-collagen antibodies and the SCIT area determined. Statistical significance *P<0.05. (B) Representative HT1080 motility tracings are shown with the indicated siRNA treatments over 48 hours. (C,D) Cells treated with control siRNA (LMNA) or Cdc42 siRNA were examined for motility on 2D collagen substrates over 24 hours. Representative tracings are shown in C and quantitation of the results is shown in D.
To determine whether relevant kinase signaling congruent with our pharmacologic inhibition and siRNA data was observed during SCIT formation (8-12 hours), western blot detection of phospho-myosin light chain (MLC-P) at designated time points was performed. MLC-P is markedly induced in the early phases of invasion but interestingly, not at later stages (Fig. 6F), which is consistent with a role for ROCKs (or MRCK, myotonic dystrophy kinase-related Cdc42-binding kinase) in SCIT formation. Interestingly, phospho-cofilin is not regulated even though it is a downstream target of ROCK (Worthylake and Burridge, 2003).
The Cdc42 effectors MRCKβ, Pak4 and IQGAP-1 coordinate MT1-MMP-dependent HT1080 cell invasion of 3D collagen matrices
Cdc42-mediated signaling catalyzes cell behaviors through downstream effectors with affinity for Cdc42-GTP (Hall, 2005). A unique feature of Cdc42 is its ability to control directional motility and cell polarization through its functional role in centrosome reorientation on 2D matrix substrates (Etienne-Manneville, 2004; Etienne-Manneville and Hall, 2003). The influence of Cdc42 on cell motility and invasive functions in 3D matrix environments has not been addressed. Our work on endothelial cell lumen and tube formation in 3D collagen matrices (Bayless and Davis, 2002; Davis et al., 2007; Iruela-Arispe and Davis, 2009; Koh et al., 2008; Koh et al., 2009) shows a requirement for Cdc42 along with various downstream effectors including Pak2, Pak4 and Par3 (Koh et al., 2008; Koh et al., 2009). To address the potential importance of known Cdc42 effectors during HT1080 invasion of 3D collagen matrices, we assessed the functional role of Pak2, Pak4, MRCKβ and IQGAP-1 using siRNA suppression experiments (Fig. 7A,B). siRNA suppression of Pak4, a Cdc42-specific Pak (Abo et al., 1998), MRCKβ and IQGAP-1 inhibit LPA-induced HT1080 invasion and SCIT formation (invasion distance from origin and tunnel area, respectively) (Fig. 7A,B). By contrast, Pak2 siRNA suppression had no effect on this process. We also showed that siRNAs to PKCα blocked motility, whereas siRNAs to RhoA had no effect (Fig. 7A,B). In each case, we utilized a Smartpool and a separate individual siRNA for each molecule to confirm their biological effects. Western blots were used to confirm protein knockdown for each siRNA (supplementary material Fig. S4).
In our system, LPA stimulates directional motility and invasion in 3D collagen matrices and an intriguing study demonstrated that Cdc42 controls rearward movement of the nucleus (Gomes et al., 2005) during centrosome repositioning in response to LPA in a manner that depends on the Cdc42 effector MRCK (Leung et al., 1998) as well as on myosin (Gomes et al., 2005). These data appear highly related to our findings here. We hypothesized that MT1-MMP and Cdc42 form a molecular complex that regulates SCIT formation and that these complexes assemble to a greater extent in response to LPA treatment.
Invasion-signaling complexes containing both Cdc42 and MT1-MMP control HT1080 cell motility and SCIT formation in 3D collagen matrices
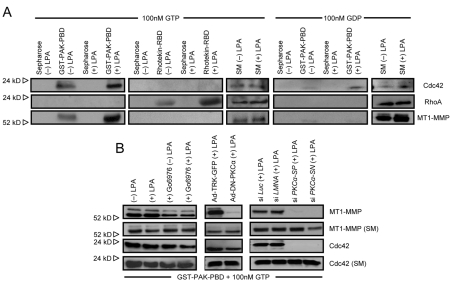
When HT1080s are treated with LPA and the lysates incubated with PAK-GST-PBD beads that only bind activated Cdc42, endogenous MT1-MMP is strongly co-precipitated compared with controls (Fig. 8A). The LPA-induced binding of MT1-MMP to activated Cdc42 is specific because no MT1-MMP is present when lysates are incubated with Rhotekin-RBD beads that bind activated RhoA (Fig. 8A). LPA does induce RhoA activation (Fig. 8A), consistent with previous reports (Kranenburg and Moolenaar, 2001). The binding of Cdc42 depends on the presence of GTP and does not occur in the presence of GDP. Importantly, co-association of endogenous Cdc42 with MT1-MMP only occurs in the presence of GTP and not with GDP and is only detected using Pak beads and not Rhotekin or control beads (Fig. 8A). These results demonstrate the specificity of these binding reactions. Equal loading of starting material was confirmed using western blots for Cdc42, RhoA and MT1-MMP (Fig. 8B). These data are consistent with the siRNA data in Fig. 7A and demonstrate a role for Cdc42 but not RhoA in 3D HT1080 cell invasion. We also show that blockade of PKCα leads to decreased Cdc42 activation and markedly decreased association of MT1-MMP with Cdc42-containing invasion signaling complexes (Fig. 8B). This result was demonstrated using Go6976, an inhibitor of PKCα, use of a dominant negative PKCα adenoviral vector and siRNA suppression of PKCα in comparison to controls. In each case, blockade of PKCα leads to marked decreases in co-association of endogenous MT1-MMP with Cdc42-GTP (Fig. 8B), further confirming that PKCα plays a crucial role in Cdc42- and MT1-MMP-dependent HT1080 cell invasion events.
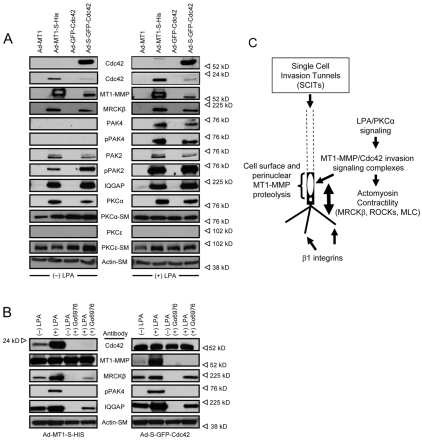
Fig. 8.
Invasion signaling complexes containing MT1-MMP and Cdc42, but not RhoA, assemble in a PKCα-dependent manner in the presence of LPA. (A) HT1080s were treated with or without 1 μM LPA and lysates were collected. Lysates were incubated with Sepharose, GST-PAK-PBD beads or Rhotekin-RBD beads in the presence of 100 nM GTP-γS or GDP. Bound proteins were probed for the indicated protein using western blots. SM, starting material. (B) HT1080 cells were either left untreated or pretreated with Go6976 for 2 hours prior to being treated with or without LPA at 1 μM for 30 minutes. Lysates were incubated with PAK beads as in A in the presence of GTP. Separately, HT1080 cells were transfected using adenoviral vectors carrying dominant-negative PKCα or control GFP or were treated with siRNAs to PKCα versus control Luc and LMNA siRNAs. Lysates were processed as mentioned above to assess the extent of endogenous Cdc42 or MT1-MMP binding to PAK beads.
Fig. 8 demonstrates that endogenous Cdc42 and MT1-MMP proteins are both present together in an invasion-signaling complex that shows increased associations in the presence of LPA. To further confirm these data and address the specific role of Cdc42 and associated proteins in this process, we transfected HT1080 cells using adenoviral vectors carrying control or S-epitope-tagged MT1-MMP or GFP-Cdc42 constructs. We have previously demonstrated that both of these constructs are biologically active and perform like their endogenous protein counterparts in functional assays using endothelial cells (Koh et al., 2008; Koh et al., 2009; Stratman et al., 2009). HT1080 cells were treated with or without LPA for 30 minutes and lysates were prepared and incubated with S-protein Sepharose to capture the expressed S-tagged Cdc42 or MT1-MMP and their associated proteins. S-GFP-Cdc42 captures MT1-MMP and MT1-MMP-S captures Cdc42 and this is stimulated by LPA (Fig. 9). This binding is specific because no binding is detected in the absence of an S-tag (control GFP-Cdc42 or MT1-MMP) (Fig. 9A) and no binding occurs using S-tagged constructs when lysates are incubated with control Sepharose beads (not shown). The data confirm that Cdc42 and MT1-MMP are present together in a signaling complex and that their association is enhanced by LPA treatment of HT1080 cells. The data using both endogenous (Fig. 8) and epitope-tagged Cdc42 and MT1-MMP (Fig. 9), demonstrate that they co-associate during cell invasion of 3D collagen matrices.
Fig. 9.
Invasion signaling complexes containing MT1-MMP and Cdc42 show co-association with known Cdc42 effectors as well as PKCα in a manner that is stimulated by LPA treatment. (A) Adenoviral vectors carrying MT1-MMP-S and S-GFP-Cdc42 with or without an S-tag were used to infect HT1080 cells overnight. Cells were then incubated with or without 1 μM LPA for 30 minutes, followed by detergent lysis and incubation with S-protein beads. Bound proteins were analyzed on western blots using the indicated antibodies. SM, starting material (B) The same protocol as in A was performed, except that cells were pretreated with or without 10 μM Go6976 for 2 hours prior to treatment with LPA. (C) Scheme detailing the role of MT1-MMP and Cdc42 invasion-signaling complexes and SCITs in single-cell invasion of 3D matrices. Proteolysis occurs directly adjacent to the nucleus on the cell membrane and SCITs are created. Coupling of the actomyosin contractility with matrix attachment via β1 integrins on invadopodia propels the cell forward until it approaches a physical collagen barrier. Perinuclear cell surface MT1-MMP then locally degrades this matrix, allowing the cell to move with its rigid nucleus. This orients the cell in 3D matrices and ensures directional proteolysis and subsequent motility, a process that also depends on Cdc42 and its effectors.
To further investigate relevant Cdc42 effectors controlling these events, strong associations of MRCKβ, Pak4, Pak2 and IQGAP are also observed using either S-GFP-Cdc42 or MT1-MMP-S (and not with the control constructs) and, in most cases, these interactions are enhanced by LPA addition (Fig. 9). These data indicate that Cdc42 and Cdc42 effectors control both HT1080 cell invasion and SCIT formation in 3D collagen matrices and are present in a multiprotein invasion-signaling complex (along with MT1-MMP) whose association is enhanced by LPA. Thus, these molecular interactions directly correlate with HT1080 cell invasive behavior and SCIT formation.
To address the potential influence of known signaling pathways on the association of this multicomponent invasion-regulatory complex, we added an inhibitor (Go6976) with specificity for the conventional PKC isoforms α and β. The addition of Go6976 inhibits HT1080 cell invasion and SCIT formation (Fig. 6), and interestingly causes dissociation of the Cdc42 and MT1-MMP complex. Neither S-GFP-Cdc42 nor MT1-MMP-S constructs are able to co-precipitate MT1-MMP or Cdc42 in the presence of Go6976, respectively. This lack of association indicates further the specificity of these experiments but also directly correlates with the ability of Go6976 to block HT1080 cell invasion (Fig. 6). In addition, associations of MRCKβ, Pak4 and IQGAP-1 with Cdc42 or MT1-MMP were also attenuated. Further support for this conclusion is that PKCα, but not PKCε, is co-captured using either S-GFP-Cdc42 or MT1-MMP-S expression in a manner stimulated by LPA treatment (Fig. 9A). Both PKCα and PKCε are equally expressed in the starting material lysates. These data confirm our siRNA data demonstrating the roles of select Cdc42 effectors and PKCα during HT1080 invasion and SCIT formation in 3D collagen matrices. Collectively, the data presented support the conclusion that HT1080 invasion and SCIT formation require MT1-MMP, Cdc42, various Cdc42 effectors (including IQGAP-1, MRCKβ and Pak4), PKCα and ROCK-1/2.
Discussion
Contrasting mechanisms for dispersion of tumor cells in 3D matrix environments have been described in recent years (Fisher et al., 2006; Gaggioli et al., 2007; Maniotis et al., 1999; Sabeh et al., 2004; Sabeh et al., 2009; Wilkinson et al., 2005; Wolf et al., 2003; Wolf et al., 2007; Wyckoff et al., 2006) and substantiate a role for MT1-MMP in tumor invasion and metastasis as well as in tumor formation and progression (Ueno et al., 1997; Yana and Seiki, 2002). Yet, the modalities of proteinase-mediated and proteinase-independent invasion remain mutually exclusive. The data presented here propose a unifying mechanism for MT1-MMP-dependent matrix proteolysis and MMP-independent invasion of 3D matrices. Invasion generates SCITs that are the width of a cellular nucleus and that cells can later traverse in a manner that does not depend on MMPs. This work demonstrates a temporal mechanism for cell motility in 3D matrices that requires preceding proteolysis; elucidates a signaling cascade for SCIT formation involving MT1-MMP, Cdc42 and the downstream effectors IQGAP-1, MRCKβ and Pak4, as well as PKCα and ROCK1/2; and provides new insights into the molecular control of cellular motility in 3D matrix environments.
SCITs function as conduits for MMP-independent migration in 3D collagen matrices
Previous work indicates that invading cells generate matrix tunnels (Bayless and Davis, 2003; Gaggioli et al., 2007; Sabeh et al., 2004; Stratman et al., 2009). HT1080s do not invade, form SCITs, nor achieve a maximum velocity if broad-spectrum MMP inhibitors GM6001, TIMP-2 or TIMP-3 are added prior to the onset of invasion (Figs 1, 4). HT1080s transfected with siRNAs targeting MT1-MMP do not form SCITs (Fig. 4) and express a phenotype identical to HT1080s treated with GM6001 or with TIMP-2 and TIMP-3, indicating that MT1-MMP is essential for SCIT formation in 3D collagen matrices. However, when GM6001 is added to invading cultures after 24 or 48 hours, maximum cellular velocity continues (Fig. 1C-E) and no blocking effect is observed. Most cells achieve maximum velocity at ∼24 hours (Fig. 1A), suggesting that SCIT formation is the rate-limiting step for motility in the 3D matrix. After 24 hours, cells maintain maximum velocity due to their ability to navigate within SCITs (Fig. 2F; supplementary material Movie 5). Our results suggest that highly invasive tumor cells such as HT1080 cells can migrate within 3D collagen matrices in an MMP-independent manner that is related to previously described descriptions of amoeboid motility (Wolf et al., 2003; Wolf et al., 2007). However, our results differ from those presented previously in that we do not observe this phenomenon if we add MMP inhibitors at the beginning of the 3D culture (Figs 1, 4). By contrast, when HT1080 cells create SCITs through MT1-MMP-dependent proteolysis, they are able to migrate through these physical spaces (see Figs 1, 2) as early as after 24 hours of culture (Fig. 1A). Possible differences in our models compared to previous publications on these questions are that we use unmodified HT1080 cells, serum-free conditions, LPA addition to the collagen matrices, and cross-linked rat tail collagen type I matrices (Fisher et al., 2006).
Although MT1-MMP facilitates invasion of some cells, others do not express this enzyme (Sato et al., 2005) and numerous tumor lines do not invade in our assays (Fisher et al., 2006). During tumorigenesis, a subpopulation of cells might acquire invasive capacity. Other `non-invasive' cells could exploit the SCIT of the `leading cell' and disseminate from the tumor nidus by simple migration. Alternatively, other cell types (e.g. fibroblasts, ECs and smooth muscle cells) might generate SCITs that could be utilized by malignant tumor cells (Fig. 3) (Gaggioli et al., 2007). Tumor entry into pre-existing SCITs generated by non-tumor cell types could occur by movement or growth into such spaces, thus gaining access to lymphatic or blood vascular networks without the necessity for MT1-MMP-directed matrix degradation.
HT1080s require LPA, MT1-MMP, Cdc42, PKCα and ROCK-1/2 to generate SCITs
In this report, we have identified molecules and signaling pathways fundamental to SCIT formation: LPA, MT1-MMP, Cdc42, various Cdc42 effectors, PKCα and ROCK-1/2. These proteins appear necessary for tumor cells to sustain directed motility over significant distances in a 3D matrix environment. Disruption of Cdc42 and ROCK-1/2 causes cells to rotate or oscillate in a small circular area with no directional component (Figs 6, 7; supplementary material Movie 6). Both MT1-MMP and Cdc42 co-associate in HT1080 cells when stimulated by LPA (Figs 8, 9). This co-association can be disrupted by the PKCα inhibitor Go6976 as well as by the use of siRNAs targeting PKCα or a dominant-negative mutant (Figs 8, 9).
The ROCK inhibitors Y27632 and HF also inhibit invasion in our assays (Fig. 6). ROCKs inhibit myosin phosphatase leading to increasing MLC-P (Kimura et al., 1996), and western blot analysis reveals strong phosphorylation of MLC at the onset of invasion and subsequent SCIT formation (Fig. 6F). HT1080s treated with siRNAs targeting Cdc42 exhibit a `carousel' phenotype similar to ROCK-1/2-inhibited HT1080s. Although many reports suggest that ROCK-1 and ROCK-2 are downstream effectors of RhoA and RhoC (Croft and Olson, 2006; Maekawa et al., 1999), siRNA targeting of either Rho isoform (Fig. 7 and data not shown) has no influence on SCIT formation or invasion. Because neither RhoA nor RhoC appear to regulate invasion in our system, our findings that MT1-MMP does not co-precipitate with activated RhoA after LPA treatment is consistent with these findings (Fig. 8A).
Our results differ from previous reports describing a Rho-ROCK-MLC-P axis in non-proteolytic invasion via matrix deformation (Sahai and Marshall, 2003; Wyckoff et al., 2006). In the work presented here, no MMP-independent invasion or SCIT formation occurred for any collagen concentration tested. We observed no MMP-independent invasive behavior for any cell type in our models (tumor cells, ECs, pericytes, NHDFs and CaSMCs) when MMP inhibitors were added from the beginning of the cultures. Invasion distances approach 0.5-1.0 mm over 48-72 hours and these distances are completely blocked by MMP inhibitors. We observed no productive MMP-independent movement of cell types (i.e. sustained directional motility away from the origin) unless cells first created tunnel spaces via MT1-MMP-dependent proteolysis.
MT1-MMP-dependent SCIT formation allows movement of cells and their respective nuclei through 3D matrices
SCIT width is nearly identical to nuclear width. On 2D collagen substrates, MT1-MMP localizes preferentially in a perinuclear distribution in the direction of cell motility (Fig. 5). Given that cell motility and the accompanying movement of the cellular nucleus require MMPs, we hypothesize that in our assays (where invasion requires MMPs) the nucleus is unable to deform sufficiently due to the restrictive nature of the cross-linked collagen matrix. Interestingly, numerous invadopodial processes extend from HT1080 cells and other cell types during the invasion process when GM6001 is added, suggesting that these processes extend into collagen matrices in an MMP-independent manner (not shown). These processes, through β1-integrin-based signaling (data not shown), probably generate the tractional forces needed to propel the cell and nucleus forward during invasion and thus allow MT1-MMP, which is presented on the cell membrane concentrated in a proximal perinuclear region (Fig. 5), to digest matrix during this process.
We favor a model where LPA signaling facilitates HT1080 invasion of 3D matrices by coordinating the invasion machinery: MT1-MMP, Cdc42, actomyosin contractility (ROCK-1/2–MRCKβ–MLC-P) and integrin-mediated adhesion signaling. When a cell orients in a 3D matrix, possibly via activation by LPA and PKCα, MT1-MMP distributes in a perinuclear cell-membrane location in the direction of invasion to degrade the matrix and generate a path for the restricting point of the cell, the nucleus. Interestingly, the crucial functional role for Cdc42 in controlling cell polarity and its direct link to MT1-MMP-mediated invasive behavior suggests a fundamental mechanism whereby polarized cell motility in 3D matrices can be regulated. Importantly, blockade of Cdc42 failed to block motility on 2D collagen substrates, suggesting that its influence shows specificity for 3D matrix environments. Thus, it is of great significance that Cdc42 and MT1-MMP co-associate within invasion-signaling complexes to selectively control cellular invasive behavior in 3D matrices (Fig. 9C).
Materials and Methods
Reagents
LPA (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate sodium salt) was from Avanti Polar Lipids. Rat tail collagen type I was prepared as described (Bornstein, 1958). Antibodies: human MT1-MMP (RP1–MMP-14, Abcam or 2010-1, Epitomics); RhoA (ARH01, Cytoskeleton); Cdc42, ROCK-1 and ROCK-2 (610929, 611136 and 610623 BD Transduction Laboratories); Lamin A/C (MAB3211, Chemicon); Actin (JLA-20, Calbiochem); MLC-2, phospho-cofilin and MLC-P (3672, 3311, and 3671, Cell Signaling Technology) were used for western blot analysis (Salazar et al., 1999). Other reagents were from Sigma-Aldrich.
Cell culture
All cell lines were purchased from American Type Culture Collection and were grown in DMEM (Invitrogen) containing 10% fetal bovine serum (FBS) in T-75 flasks. Human umbilical vein ECs (HUVECs) were purchased from Lonza and used from passages 2-6. Lentiviral-selected cell lines were supplemented with Blasticidin (Invitrogen) at 5-15 μg/ml. nuc-GFP-labeled HT1080s were prepared as described previously (Fisher et al., 2006).
Microscopy and imaging
An inverted fluorescence microscope (Eclipse TE2000-U; Nikon) was used to visualize nuc-GFP HT1080 3D invasion. Time-lapse imaging of living cells used the Eclipse microscope with a CoolSNAP HQ camera (Photometrics) and MetaMorph software (Molecular Devices) and was performed in a temperature-controlled chamber set to 37°C with continuous flow of 5% CO2. For fluorescent time-lapse microscopy, a CFI plan-Fluor 10× with an NA of 0.30A lens was used. Time-lapse images and Z-stacks were converted in MetaMorph to QuickTime format.
To image non-fluorescent SCITs, collagen gels were polymerized in glass-bottom 384-well tissue culture plates (VWR Scientific Products) supplemented with clear DMEM (Invitrogen). A LEICA DMI 6000B microscope captured images with an Orca 1394 camera (Hamamatsu) under transmitted light (brightfield) with a 40× dry objective.
Tunnel microperfusion with silicone oil
Micropipettes were pulled from borosilicate glass capillary tubing (1.0 mm O.D.; 0.5 mm I.D.; Frederick Haer and Co.) and sharpened to a tip diameter of 2 μm. Pipette positioning was controlled using a Leitz micromanipulator under brightfield illumination on the stage of a Zeiss ACM upright microscope. The pipette tip was positioned near visible HT1080s. When the pipette tip penetrated a tunnel, the tunnel filled directly after oil ejection from the pipette. Tunnel perfusion was recorded using a digital video recorder (Panasonic) through a CCTV system (Toshiba IK-C30 color CCD camera).
Assessment of HT1080 motility in 3D collagen matrices
HT1080s (1.0×105) stably expressing nuc-GFP were cultured in 2.5 mg/ml collagen matrices containing 1 μM LPA in 384- or 96-well tissue culture plates (VWR Scientific) supplemented with DMEM and a 1:250 dilution of reduced serum II supplement (RSII+) as described previously (Fisher et al., 2006). Cells were allowed to invade for up to 72 hours, fluorescent images were captured using MetaMorph, and cell tracings were generated by MetaMorph as described (Fisher et al., 2006).
siRNA transfection
siGENOME SMARTpool human MT1-MMP, two 21-nucleotide single MT1-MMP siRNAs (custom and scrambled) described previously (Sabeh et al., 2004), RhoA, Cdc42, ROCK-1, ROCK-2, Pak2, Pak4, MRCK-β and IQGAP were all purchased from Dharmacon. Lamin A/C and a luciferase GL2 duplex siRNA oligo (Dharmacon) were utilized as controls. Single siRNA duplexes were purchased from Invitrogen. The sequences are as follows: Cdc42, 5′-CCGGUGGAGAAGCUGAGGUCAUCAU-3′ and 5′-AUGAUGACCUCAGCUUCUCCACCGG-3′; MT1-MMP, 5′-CAGGCCGACAUCAUGAUCUUCUUUG-3′ and 5′-CAAAGAAGAUCAUGAUGUCGGCCUG-3′; IQGAP, 5′-GCCUCCACUUUAGACACACUGAUAA-3′ and 5′-UUAUCAGUGUGUCUAAAGUGGAGGC-3′; MRCKα, 5′-UAAAGCAUUUCAUACAUACAGACCC-3′ and 5′-GGGUCUGUAUGUAUGAAAUGCUUUA-3′; MRCKβ, 5′-UAAAUCACCACCCACAUAGUAAUCC-3′ and 5′-GGAUUACUAUGUGGGUGGUGAUUUA-3′; Pak2, 5′-CCUGUGCCUCUAACAAGCGAUUCUA-3′ and 5′-UAGAAUCGCUUGUUAGAGGCACAGG-3′; Pak4, 5′-UGCUUGCGCAGGUCCAUCUUCUUGA-3′ and 5′-UCAAGAAGAUGGACCUGCGCAAGCA-3′; PKCα, 5′-CCUAAAGGCUGAGGUUGCUGAUGAA-3′ and 5′-UUCAUCAGCAACCUCAGCCUUUAGG-3′; Rock-1, 5′-CCGAGACACUGUAGCACCAGUUGUA-3′ and 5′-UACAACUGGUGCUACAGUGUCUCGG-3′; Rock-2, 5′-UGGAAGAAAUCAGACAGCAUCCUUU-3′ and 5′-AAAGGAUGCUGUCUGAUUUCUUCCA-3′; RhoA, 5′-GAAAGACAUGCUUGCUCAUAGUCUU-3 and 5′-AAGACUAUGAGCAAGCAUGUCUUUC-3′. Primers were resuspended in DEPC-H2O at 20 nM and used at 20 pM final concentration. All siRNA transfections were performed in 20% FBS as described previously (Fisher et al., 2006). Cells were harvested for assays 18 hours after the second siRNA transfection.
Immunostaining of invading cultures
Cultures were fixed in 2% paraformaldehyde and then either permeabilized with Triton X-100 or not. Primary antibodies (anti-collagen I or anti-MT1-MMP) were added with the appropriate blocking serum overnight. Cultures were washed in phosphate-buffered saline and incubated with the appropriate secondary antibody (Alexa Fluor 594; Invitrogen). Fluorescent images were taken with a CFI plan-Fluor 10× with a lens of 0.30 NA.
Nuclear diameter measurements
Diameter measurements for invasion tunnels and HT1080 or NHDF cell nuclei were binned according to their respective widths and plotted as histograms using Igor Pro (Wavemetrics). Diameters were calculated using MetaMorph.
Gelatin zymography and western blotting
Gelatin zymography and western blot analysis were performed as described previously (Salazar et al., 1999). All phospho-antibodies were probed with ECL Plus (Amersham).
Preparation and use of S-tagged recombinant MT1-MMP
Recombinant adenoviral vectors carrying S-tag-GFP-Cdc42 were prepared as described (Koh et al., 2008). Cells were infected, and lysates prepared and incubated with S protein-Sepharose as described (Koh et al., 2008). Recombinant adenoviral MT1-MMP-S-tag was prepared by cloning full-length MT1-MMP into pIEX-5 vector (Novagen) using BglII and KpnI sites. The primers used were; (5′-AGAGATCTGCCACCATGTCTCCCGCCCCAAGACCC-3′) and (5′-AGGGTACCGACCTTGTCCAGCAGGGAACGCTG-3′). The MT1-MMP-S-tag construct was amplified using the following primers; (5′-AGGTCGACGCCACCATGTCTCCCGCCCCAAGACCC-3′) and (5′-AGTCTAGATTAGTGATGGTGATGGTGATGGTG-3′) and cloned into pAdTrack-CMV into SalI and XbaI sites.
Pull down assays with PAK-GST-PBD and Rhotekin-RBD beads
T25s of HT1080s were lysed with or without 1 μM LPA in detergent lysis buffer of 1% Triton X-100 in 1× TBS, pH 8.0, containing complete EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics), 150 μg/μl high-purity collagenase and 100 nM GTPγS or GDP (Calbiochem). Lysates were incubated at 4°C for 30 minutes and centrifuged at 16,000 g for 20 minutes at 4°C. Supernatants were incubated with GST-PAK-PBD or Rhotekin-RBD protein agarose beads (Cytoskeleton) for 45 minutes at 4°C and washed four times with buffer. Bound active Cdc42 and MT1-MMP proteins were detected by western blot analysis.
Statistical analysis
Statistical analysis was performed using Microsoft Excel. Statistical significance was set at P<0.05; t-tests assuming equal variances were used when analyzing two groups within a single experiment (test versus control).
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/24/4558/DC1
This work was supported by NIH grants HL79460 and HL87308 to G.E.D. Deposited in PMC for release after 12 months.
References
- Abo, A., Qu, J., Cammarano, M. S., Dan, C., Fritsch, A., Baud, V., Belisle, B. and Minden, A. (1998). PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17, 6527-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht-Buehler, G. (1977). The phagokinetic tracks of 3T3 cells. Cell 11, 395-404. [DOI] [PubMed] [Google Scholar]
- Bayless, K. J. and Davis, G. E. (2002). The Cdc42 and Rac1 GTPases are required for capillary lumen formation in three-dimensional extracellular matrices. J. Cell Sci. 115, 1123-1136. [DOI] [PubMed] [Google Scholar]
- Bayless, K. J. and Davis, G. E. (2003). Sphingosine 1-phosphate markedly induces matrix metalloproteinase and integrin-dependent human endothelial cell invasion and lumen formation in three-dimensional collagen and fibrin matrices. Biochem. Biophys. Res. Comm. 312, 903-913. [DOI] [PubMed] [Google Scholar]
- Berrier, A. L. and Yamada, K. M. (2007). Cell-matrix adhesion. J. Cell Physiol. 213, 565-573. [DOI] [PubMed] [Google Scholar]
- Bornstein, M. B. (1958). Reconstituted rat-tail collagen used as a substrate for tissue cultures on coverslips in Maximov slides and rollertubes. Lab Invest. 7, 134-137. [PubMed] [Google Scholar]
- Chambers, A. F., Groom, A. C. and MacDonald, I. C. (2002). Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2, 563-572. [DOI] [PubMed] [Google Scholar]
- Coussens, L. M., Fingleton, B. and Matrisian, L. M. (2002). Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295, 2387-2392. [DOI] [PubMed] [Google Scholar]
- Croft, D. R. and Olson, M. F. (2006). The Rho GTPase effector ROCK regulates cyclin A, cyclin D1, and p27Kip1 levels by distinct mechanisms. Mol. Cell. Biol. 26, 4612-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Munoz, W., Sanchez, O. H., Di Grappa, M., English, J. L., Hill, R. P. and Khokha, R. (2006). Enhanced metastatic dissemination to multiple organs by melanoma and lymphoma cells in timp-3–/– mice. Oncogene 25, 6489-6496. [DOI] [PubMed] [Google Scholar]
- Davis, G. E., Koh, W. and Stratman, A. N. (2007). Mechanisms controlling human endothelial lumen formation and tube assembly in three-dimensional extracellular matrices. Birth Defects Res. C. Embryo Today 81, 270-285. [DOI] [PubMed] [Google Scholar]
- Egeblad, M. and Werb, Z. (2002). New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161-174. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S. (2004). Cdc42-the centre of polarity. J. Cell Sci. 117, 1291-1300. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S. and Hall, A. (2003). Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature 421, 753-756. [DOI] [PubMed] [Google Scholar]
- Filippov, S., Koenig, G. C., Chun, T. H., Hotary, K. B., Ota, I., Bugge, T. H., Roberts, J. D., Fay, W. P., Birkedal-Hansen, H., Holmbeck, K. et al. (2005). MT1-matrix metalloproteinase directs arterial wall invasion and neointima formation by vascular smooth muscle cells. J. Exp. Med. 202, 663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, K. E., Pop, A., Koh, W., Anthis, N. J., Saunders, W. B. and Davis, G. E. (2006). Tumor cell invasion of collagen matrices requires coordinate lipid agonist-induced G-protein and membrane-type matrix metalloproteinase-1-dependent signaling. Mol. Cancer 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P., Borgmann, S. and Brocker, E. B. (2001). Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J. Leukoc. Biol. 70, 491-509. [PubMed] [Google Scholar]
- Gaggioli, C., Hooper, S., Hidalgo-Carcedo, C., Grosse, R., Marshall, J. F., Harrington, K. and Sahai, E. (2007). Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9, 1392-1400. [DOI] [PubMed] [Google Scholar]
- Gomes, E. R., Jani, S. and Gundersen, G. G. (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451-463. [DOI] [PubMed] [Google Scholar]
- Hall, A. (2005). Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891-895. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. and Weinberg, R. A. (2000). The hallmarks of cancer. Cell 100, 57-70. [DOI] [PubMed] [Google Scholar]
- Hojilla, C. V., Mohammed, F. F. and Khokha, R. (2003). Matrix metalloproteinases and their tissue inhibitors direct cell fate during cancer development. Br. J. Cancer 89, 1817-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck, K., Bianco, P., Yamada, S. and Birkedal-Hansen, H. (2004). MT1-MMP: a tethered collagenase. J. Cell Physiol. 200, 11-19. [DOI] [PubMed] [Google Scholar]
- Hotary, K., Allen, E., Punturieri, A., Yana, I. and Weiss, S. J. (2000). Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 149, 1309-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruela-Arispe, M. L. and Davis, G. E. (2009). Cellular and molecular mechanisms of vascular lumen formation. Dev. Cell 16, 222-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, Y. and Nagase, H. (2002). Matrix metalloproteinases in cancer. Essays Biochem. 38, 21-36. [DOI] [PubMed] [Google Scholar]
- Kimura, K., Ito, M., Amano, M., Chihara, K., Fukata, Y., Nakafuku, M., Yamamori, B., Feng, J., Nakano, T., Okawa, K. et al. (1996). Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245-248. [DOI] [PubMed] [Google Scholar]
- Koh, W., Mahan, R. D. and Davis, G. E. (2008). Cdc42- and Rac1-mediated endothelial lumen formation requires Pak2, Pak4 and Par3, and PKC-dependent signaling. J. Cell Sci. 121, 989-1001. [DOI] [PubMed] [Google Scholar]
- Koh, W., Sachidanandam, K., Stratman, A. N., Sacharidou, A., Mayo, A. M., Murphy, E. A., Cheresh, D. A. and Davis, G. E. (2009). Formation of endothelial lumens requires a coordinated PKCε-, Src-, Pak- and Raf-kinase-dependent signaling cascade downstream of Cdc42 activation. J. Cell Sci. 122, 1812-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranenburg, O. and Moolenaar, W. H. (2001). Ras-MAP kinase signaling by lysophosphatidic acid and other G protein-coupled receptor agonists. Oncogene 20, 1540-1546. [DOI] [PubMed] [Google Scholar]
- Leung, T., Chen, X. Q., Tan, I., Manser, E. and Lim, L. (1998). Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18, 130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. Y., Ota, I., Yana, I., Sabeh, F. and Weiss, S. J. (2008). Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol. Biol. Cell 19, 3221-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, M., Ishizaki, T., Boku, S., Watanabe, N., Fujita, A., Iwamatsu, A., Obinata, T., Ohashi, K., Mizuno, K. and Narumiya, S. (1999). Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285, 895-898. [DOI] [PubMed] [Google Scholar]
- Maniotis, A. J., Folberg, R., Hess, A., Seftor, E. A., Gardner, L. M., Pe'er, J., Trent, J. M., Meltzer, P. S. and Hendrix, M. J. (1999). Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am. J. Pathol. 155, 739-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh, F., Ota, I., Holmbeck, K., Birkedal-Hansen, H., Soloway, P., Balbin, M., Lopez-Otin, C., Shapiro, S., Inada, M., Krane, S. et al. (2004). Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J. Cell Biol. 167, 769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh, F., Shimizu-Hirota, R. and Weiss, S. J. (2009). Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 185, 11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E. and Marshall, C. J. (2003). Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5, 711-719. [DOI] [PubMed] [Google Scholar]
- Salazar, R., Bell, S. E. and Davis, G. E. (1999). Coordinate induction of the actin cytoskeletal regulatory proteins gelsolin, vasodilator-stimulated phosphoprotein, and profilin during capillary morphogenesis in vitro. Exp. Cell Res. 249, 22-32. [DOI] [PubMed] [Google Scholar]
- Sato, H., Takino, T. and Miyamori, H. (2005). Roles of membrane-type matrix metalloproteinase-1 in tumor invasion and metastasis. Cancer Sci. 96, 212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W. B., Bayless, K. J. and Davis, G. E. (2005). MMP-1 activation by serine proteases and MMP-10 induces human capillary tubular network collapse and regression in 3D collagen matrices. J. Cell Sci. 118, 2325-2340. [DOI] [PubMed] [Google Scholar]
- Saunders, W. B., Bohnsack, B. L., Faske, J. B., Anthis, N. J., Bayless, K. J., Hirschi, K. K. and Davis, G. E. (2006). Coregulation of vascular tube stabilization by endothelial cell TIMP-2 and pericyte TIMP-3. J. Cell Biol. 175, 179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki, M. (2003). Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 194, 1-11. [DOI] [PubMed] [Google Scholar]
- Sporn, M. B. (1996). The war on cancer. The Lancet 347, 1377-1381. [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson, W. G. and Yu, A. E. (2001). Proteases in invasion: matrix metalloproteinases. Semin. Cancer Biol. 11, 143-152. [DOI] [PubMed] [Google Scholar]
- Stratman, A. N., Saunders, W. B., Sacharidou, A., Koh, W., Fisher, K. E., Zawieja, D. C., Davis, M. J. and Davis, G. E. (2009). Endothelial cell lumen and vascular guidance tunnel formation requires MT1-MMP-dependent proteolysis in 3D collagen matrices. Blood. 114, 237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, H., Nakamura, H., Inoue, M., Imai, K., Noguchi, M., Sato, H., Seiki, M. and Okada, Y. (1997). Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res. 57, 2055-2060. [PubMed] [Google Scholar]
- Wilkinson, S., Paterson, H. F. and Marshall, C. J. (2005). Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7, 255-261. [DOI] [PubMed] [Google Scholar]
- Wolf, K., Mazo, I., Leung, H., Engelke, K., von Andrian, U. H., Deryugina, E. I., Strongin, A. Y., Brocker, E. B. and Friedl, P. (2003). Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160, 267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, K., Wu, Y. I., Liu, Y., Geiger, J., Tam, E., Overall, C., Stack, M. S. and Friedl, P. (2007). Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9, 893-904. [DOI] [PubMed] [Google Scholar]
- Worthylake, R. A. and Burridge, K. (2003). RhoA and ROCK promote migration by limiting membrane protrusions. J. Biol. Chem. 278, 13578-13584. [DOI] [PubMed] [Google Scholar]
- Wyckoff, J. B., Pinner, S. E., Gschmeissner, S., Condeelis, J. S. and Sahai, E. (2006). ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16, 1515-1523. [DOI] [PubMed] [Google Scholar]
- Yamada, K. M. and Cukierman, E. (2007). Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601-610. [DOI] [PubMed] [Google Scholar]
- Yana, I. and Seiki, M. (2002). MT-MMPs play pivotal roles in cancer dissemination. Clin. Exp. Metastasis 19, 209-215. [DOI] [PubMed] [Google Scholar]
- Zaman, M. H., Trapani, L. M., Sieminski, A. L., Mackellar, D., Gong, H., Kamm, R. D., Wells, A., Lauffenburger, D. A. and Matsudaira, P. (2006). Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 103, 10889-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.