Abstract
Purpose
In an effort to generate inducible RPE-specific Cre mice, we recently used a 3.0-kb human vitelliform macular dystrophy-2 (VMD2) promoter (PVMD2) to control the expression of a two-gene system that contained a PVMD2 directed reverse tetracycline-dependent transactivator (rtTA) gene, and the tetracycline-responsive element (TRE) directed cre gene. While most transgenic lines demonstrated the anticipated Cre activity in the RPE, we also identified a mouse line (VMD2-cre) that had unanticipated Cre activity in the neural retina, including Müller glial cells. Müller cells play important roles in the function and maintenance of the retina, and this mouse line would be potentially useful for conditional gene targeting in Müller glia. We therefore characterized the timing, inducibility, and cell specificity of Cre expression, as well as Müller cell-specific efficiency of Cre-mediated recombination in this mouse line.
Methods
Transgenic mice carrying cassettes of human PVMD2-rtTA and TRE-cre were generated. Cre expression was characterized using a Cre-activatable lacZ reporter mouse line (R26R) and a floxed interleukin 6 signal transducing receptor (gp130) mouse line.
Results
β-Galactosidase (β-gal) assay and immunohistochemical analysis of VMD2-cre/R26R double transgenic mice indicated that Cre activity was detected in cells located in the inner nuclear layer, with prominent expression of β-gal in Müller cells. Cre activity was also detected in photoreceptors in the outer nuclear layer. PCR analysis demonstrated that Cre-mediated recombination initiated by embryonic day 15. Immunohistochemical analysis indicated that Cre-mediated deletion of floxed gp130 gene occurred in 52 percent of the retinal Müller cells. Retinal function and morphology were normal in 10-month-old VMD2-cre mice.
Conclusion
We generated a transgenic cre mouse that is useful to study gene activation and inactivation in retinal Müller cells.
Keywords: transgenic mice, Müller cells, Cre/lox, VMD2
INTRODUCTION
One of the recent breakthroughs in molecular medicine is the use of gene targeting to elucidate protein function in mice. However, disruption of an essential gene expressed in multiple tissues or cell-types often causes embryonic or neonatal lethality, obscuring its role in a target cell or in the adult (Ferrara et al., 1996; Rucker et al., 2000). The Cre/lox-based conditional gene knockout strategy has become a method of choice to circumvent this problem. Cre recombinase of bacteriophage P1 catalyzes site-specific DNA recombination both intra- and intermolecularly between target 34-bp loxP sites (Sauer and Henderson, 1988). Efficient Cre-mediated excision of DNA between directly repeated loxP sites is the molecular basis for conditional gene knockout in mice (for review, see (Le and Sauer, 2000)). With a goal to obtain inducible RPE-specific Cre mice, we recently generated transgenic mice expressing Cre recombinase under the control of a 3.0-kb human VMD2 (vitelliform macular dystrophy; VMD) promoter directed reverse tetracycline-inducible gene expression system (Le et al., 2008). The human VMD2 gene is associated with Best disease and is thought to be preferentially expressed in the RPE (Marquardt et al., 1998; Petrukhin et al., 1998). To our surprise, approximately half of the Cre-expressing transgenic lines demonstrated Cre activity in the neural retina, with prominent Cre-activated β-galactosidase (β-gal) staining in Müller cells. Müller cells are the principle supporting glial cells in the neural retina and they participate in many essential functions including homeostasis, neural protection, angiogenesis, and perhaps neural regeneration (for review, see (Bringmann et al., 2006)). Therefore, this transgenic mouse line may have important utility for studies related to gene function in Müller cells. This report describes the characterization of this mouse line.
MATERIALS AND METHODS
Animals care and treatment
All animal procedures followed the guidelines of the ARVO statement for the “Use of Animals in Ophthalmic and Vision Research” and were approved by the Institutional Animal Care and Use Committees at the University of Oklahoma Health Sciences Center, the Dean A. McGee Eye Institute, and the Oklahoma Medical Research Foundation. Doxycycline was administered through the drinking water of pregnant mothers at a concentration of 0.5 mg/mL in 5% sucrose from embryonic day 15 to postnatal day 2.
Transgenic mice were generated as described previously (Le et al., 2008). Mice carrying the floxed gp130 gene (gp130f/f) were a gift from Dr. W. Müller of University of Cologne, Germany (Betz et al., 1998). PCR analysis for the cre transgene was carried out with primers a (5′AGG TGT AGA GAA GGC ACT TAG C 3′) and b (5′ CTA ATC GCC ATC TTC CAG CAG G 3′) to detect a 411-bp product. PCR analysis for the rtTA transgene was carried out using primers c (5′–CGG CCT TGA ATT GAT CAT ATG CGG –3′) and d (5′-TCA AAC TCG AAG TCG GCC ATA TCC-3) to detect a 398-bp product. PCR diagnosis for the Cre-activatable lacZ reporter gene in R26R mice was carried out with primers e (5′-GAG TTG CGT GAC TAC CTA CGG-3′) and f (5′-GGC TTC ATC CAC CAC ATA CAG G-3′) to detect a 495-bp product. PCR analysis for Cre-mediated deletion of the floxed gp130 gene was performed as described previously (Yao et al., 2005), using primers g (5′-GAT GAA GGT GGG AAA GAT GGG CCG GAA TTC-3′) and h (5′-GGA AGG ATC AGG AAC ATT AGG CCA GAT GTG-3′) to detect a 600-bp product.
β-Galactosidase assay
The β-gal assay on retinal whole mounts and retinal sections was performed as described previously (Le et al., 2006; Le et al., 2008). Liquid β-gal assay was carried out according to a previous procedure with some modification (Le et al., 2008). Eyes were enucleated and stored at −80°C immediately. On the day of assay, the frozen retina was homogenized in 200 μl of Z-buffer containing 50 mM sodium phosphate buffer (pH 7.0), 10 mM KCl, and 1 mM MgSO4. The cell debris was removed by centrifugation (10,000 rpm for 10 min on a bench top centrifuge), 106 μl of the supernatant was mixed with 21 μl of 2-nitrophenyl β-D-galactopyronoside (ONPG, 4 mg/ml in water) and incubated at 30°C for 30 min. The reaction was stopped by adding 53 μl of 1 M Na2CO3 to the reaction mixture and the optical density was measured at 420 and 550 nm. Protein concentrations were measured with a BCA protein assay kit (Pierce, Rockford, IL). β-Gal activity was calculated as 1000 × [OD420 − (1.75 · · OD550)]/time × protein (mg) (Rajala et al., 2004). Relative activity was the percentage of specific activity compared with that from VMD2-cre/R26R mice under a non-induced condition.
Immunoblotting and immunohistochemisty
Immunoblotting for Cre was performed according to a previous method (Le et al., 2006), using an anti-Cre antibody (Novagen). Immunohistochemisty (IHC) was performed as described previously (Ueki et al., 2008). Briefly, the cornea and lens were removed and the eye cups were fixed with 4% paraformaldehyde in PBS for 30 min. Eye cups were then cryoprotected with a series of sucrose solution (10% – 30%) and cross sections of the retina were obtained using a cryostat. Retinal cross sections were immunostained using anti-β-gal antibody (#7−063100, 5 Prime-3 Prime, Boulder, CO; 1:100 dilution), anti-pSTAT3 antibody (#9131, Cell Signaling Technology, Danvers, MA; 1:50 dilution) and anti-glutamine synthetase (GS) antibody (#MAB302; Chemicon, Temecula, CA; 1:1000 dilution). To detect gp130 activation, VMD2-cre/gp130f/f mice were injected intravitreally with a gp130 ligand, leukemia inhibitory factor (LIF), as described previously (Ueki et al., 2008). Eyes were collected 30 minutes after the injection. STAT3 phosphorylation was detected with IHC. Confocal imaging was performed using a FluoView FV500 confocal laser scanning microscope (Olympus). Imaging and microscope settings were identical in all samples. For quantification, sections were co-stained for pSTAT3 and GS. pSTAT3-postive and GS-positive cells were counted in a 100 μm stretch of the inner nuclear layer in images obtained with a confocal microscope (10 μm stack). The frequency of Cre-mediated recombination was calculated based on the number of pSTAT3-positive and GS-positive Müller cell nuclei relative to the total GS-positive Müller cell nuclei.
Electroretinography and retinal morphology
For scotopic and photopic electroretinography (ERG), mice were dark adapted overnight. The ERG was performed with a UTAS-E 3000 ERG system (LKC technologies, Inc., Gaithersburg, MD), according to a previously method (Le et al., 2004; Quiambao et al., 1997; Xu et al., 2000). Retinal morphology was examined in hematoxylin and eosin (H & E) stained sections, as described previously (Le et al., 2006).
RESULTS
Generation of Cre-expressing transgenic mice
In a previous study, we used a 3.0-kb promoter of the human VMD2 gene to generate tetracycline-inducible RPE-specific Cre mice (Le et al., 2008). The human VMD2 gene is thought to be expressed preferentially in the RPE (Marquardt et al., 1998; Petrukhin et al., 1998). The transgenic mice were generated using a so-called tet-on system to control Cre expression (Gossen et al., 1995). The VMD2 promoter (PVMD2) was used to direct the expression of the tetracycline inducible transactivator gene rtTA. The rtTA should then drive the expression of the tetO controlled cre gene in the presence of doxycycline (Figure 1). Our cre transgene included a translationally optimized cre, and the intron-containing mouse metallothionein (MT-I) polyadenylation signal. This expression vector has a proven record of success in transgenic studies (Le et al., 2004; Le et al., 2006). The transgenic DNA fragments, the PVMD2-rtTA and tetO-PhCMV-cre, were co-injected into zygotes, and were co-integrated into a single chromosome in all transgenic founders, as demonstrated in the PCR analysis of segregation patterns for more then ten generations (data not shown). Co-integration of both transgenes onto a single chromosome provides a distinct advantage over separate transgene integration and it will reduce the required breeding for future downstream studies in conditional gene knockout studies. All germline-transmitted mice appeared to be normal in size, morphology, and behavior. Five of the ten Cre-expressing transgenic lines demonstrated different levels of Cre activity in the retinal Müller cells. One of them showed substantial Cre-activated β-gal reporter expression in the Müller cells and was characterized further (see discussion below). This line of VMD2-cre mice were also bred with floxed gp130 (gp130ff) mice (Betz et al., 1998). Neither the VMD2-cre/R26R mice nor the VMD2-cre/gp130ff mice had a detectible retinal degeneration phenotype, and thus were suitable for downstream characterization.
Figure 1.
Schematic drawing of transgenes in recombinant plasmids pLE119 and pLE116. PVMD2, promoter of human VMD2 gene; rtTA, structure gene for reverse-tet system transactivator; SV40 A(n), simian virus 40 polyadenylation signal; PhCMV, immediate early promoter of human cytomegalovirus; cre, structure for Cre; MT-I A(n), mouse metallothionein polyadenylation signal. XhoI and HindIII are restriction enzymes that were used to release the transgenes from the vectors.
Analysis of inducible and developmental Cre activity
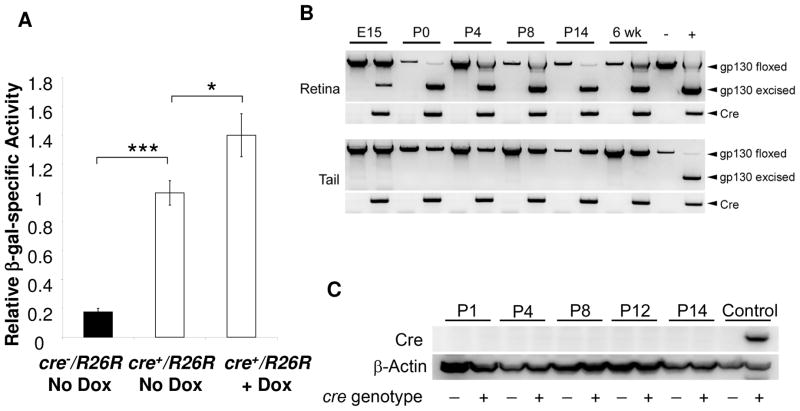
To determine the inducibility of Cre expression in the VMD2-cre mice, we utilized the Cre-activatable β-gal reporter (R26R) mice. In R26R mice β-gal can be activated after Cre-mediated excision of a floxed transcriptional “STOP” DNA segment (Soriano, 1999). Cre expression was analyzed quantitatively through indirect measurement of Cre-activated β-gal enzyme activity using retinal homogenates of F1 VMD2-cre/R26R mice. To determine the basal level of Cre expression under a non-induced condition, the pregnant mothers were placed on an autoclaved diet (LabDiet 5010; Statewide Supply, Oklahoma City, OK) that demonstrated a lower basal level of Cre expression in a previous study (Le et al., 2008), presumably due to heat-inactivation of the trace amount of tetracycline derivatives in the diet. To induce Cre expression in the pups, the pregnant females were then supplied with doxcycyline (Dox), a tetracycline derivative, in drinking water from embryonic day 15 (E15) to postnatal day 10 (P10). Protein homogenates from P15 retina were used in the β-gal assay. Compared with the β-gal-specific activity produced by non-transgenic controls, significantly more (82.5−/+2.4%) of β-gal activity was expressed in the retina of VMD2-cre/R26R mice under non-induced condition (Figure 2A), indicating that the tetracycline gene expression system was not tightly controlled. The induced VMD2-cre/R26R mice demonstrated 40.0−/+15.0% of more β-gal-specific activity, compared with that in non-induced mice (Figure 2A). This result suggests that the tetracycline gene expression system was leaky and there was limited inducibility in this mouse line.
Figure 2.
Analysis of Cre expression in the retina of VMD2-cre/R26R mice. A: In vivo induction of Cre-activated β-gal expression by measuring relatively specific activity in retinal homogenates from VMD2-cre/R26R mice. Induction was carried out from E15 to P10 and β-gal activity assay was carried out at P15. Error bars: standard errors. ***: p<0.001. *: p<0.05. Significant amount of β-gal was expressed without induction. However, the induced VMD2-cre/R26R mice demonstrated 40.0−/+15.0% of more β-gal activity. B: PCR analysis of Cre-mediated recombination in the retina of the VMD2-cre/gp130f/f mice. C: Western blot analysis of Cre expression in the postnatal retina of VMD2-cre/gp130f/f mice. Cre function was detected in the retina at embryonic day 15 (E15). However, Cre protein in the retina was undetectable postnatally with Western blot.
To determine the time-course of Cre-mediated recombination, we performed PCR analysis diagnostic for Cre-mediated excision at gp130 locus. Cre-mediated recombination in the VMD2-cre/gp130f/f mice was detected in mice as early as embryonic day 15 (E15) (Figure 2B), suggesting that Cre expression was initiated during embryonic development and before the development of the retinal Müller cells. To estimate the level of Cre expression, immunohistochemistry (IHC) and immunoblotting were performed using an anti-Cre antibody that had a proven record of success in the characterization of other retinal cell-specific Cre mice (Le et al., 2004; Le et al., 2006). However, no positively stained cells in retinal sections (data not shown) or specific immunoblotting signals from retinal homogenates were identified in mice from birth to two weeks of age (Figure 2C). These results are not unexpected, since Cre activity was detected as early as E15. In addition, a productive Cre-mediated recombination requires only eight copies of Cre recombinase in a particular cell (Guo et al., 1997), and therefore, the minimal level of protein required for a productive Cre-mediated recombination is likely much lower than the threshold of detecting Cre expression by IHC and immunoblotting.
Localization and efficiency of Cre expression
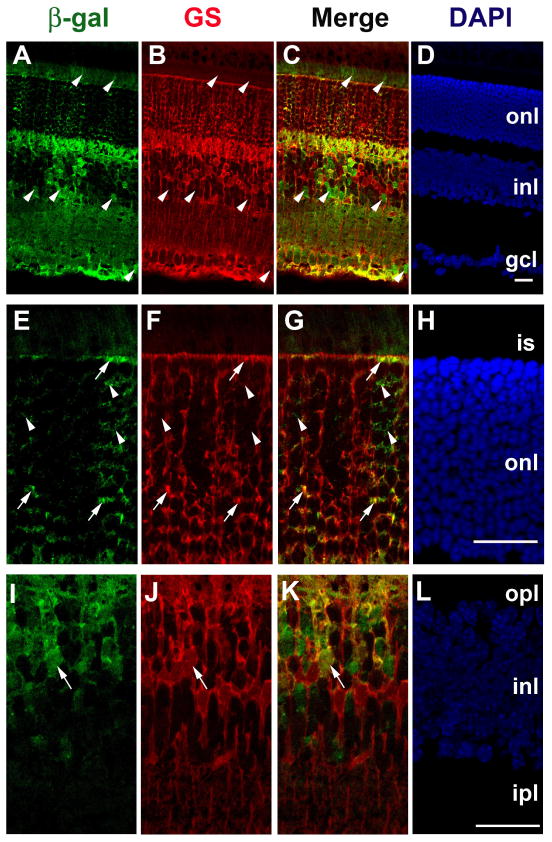
To localize Cre function in the retina, we performed a β-gal staining assay on retinal sections from VMD2-cre/R26R mice. Cre function was detected in neural retina, with clearly identifiable expression pattern in the retinal Müller cells (Figure 3). We then performed IHC on retinal sections using antibodies against β-gal and glutamate synthetase (GS), a Müller cell-specific marker. β-Gal was detected in the retinal Müller cells and unknown inner nuclear layer neurons. β-Gal was also observed in some photoreceptors (Figure 4). The activation of β-gal in VMD2-cre+/R26R mice only requires Cre-mediated excision of a single allele, and may over-represent the efficiency of Cre-mediated deletion of two alleles in conditional gene inactivation studies. To determine the efficiency of Cre-mediated gene excision in Müller cells, we used mice homozygous for the floxed gp130 gene. Activation of STAT3 by tyrosine phosphorylation is a direct signaling event downstream of gp130 in all retinal cells, including Müller cells (Ueki et al., 2008). To quantify deletion of floxed gp130 we took an advantage of STAT3 activation in the presence of leukemia inhibitory factor (LIF). Injection of LIF induced the activation of STAT3 in all Müller cells in VMD2-cre -/gp130f/f mice (Figure 5A–C). In contrast, STAT3 was not activated in 52.1−/+2.6% of Müller cells in VMD2-cre+/gp130f/f mice (Figure 5D–F). Since STAT3 activation by LIF requires gp130, this result indicates that the frequency of Cre-mediated excision of two floxed gp130 alleles occurred in 52.1% of Müller cells (Figure 5G).
Figure 3.
Localization of Cre expression in VMD2-cre/R26R mice with β-gal staining assay. A–B: Overall β-gal staining pattern in retinal flat-mounts (A) sections (B). C: β-Gal staining in a P5 retinal section. D: Localization of Cre function in Müller cells in a retina section after a brief (3 h) β-gal staining. Arrows point to distinct Cre expression pattern in Müller cells. Scale bar equals to 100 μm. Outer nuclear layer (onl) and inner nuclear layer (inl) are labeled. Cre function was localized to neural retina, including Müller cells.
Figure 4.
Localization of Cre expression in six-week-old VMD2-cre/R26R mice with IHC. A–D: Representative IHC image of Cre-activated β-gal expression in retinal section. E–H and I–L: high magnification images. Nuclei were counterstained with DAPI. Outer nuclear layer (onl), inner nuclear layer (inl), and ganglion cell layer (gcl) are labeled. Scale bars equal to 25 μm. Cre-activated β-gal was colocalized with glutamate synthetase (GS), a Müller cell marker (arrows). Some photoreceptors and unidentified INL neurons were also positive for Cre activated β-gal (arrowheads).
Figure 5.
Quantitative analysis of Cre-mediated recombination at gp130 locus in Müller cells by detecting phosphorylated STAT3 (pSTAT3), an activated downstream target of gp130, in response to leukemia inhibitory factor (LIF). A–E: Single slice (0.5 μm) confocal microscopic images of pSTAT3 in inner nuclear layer of VMD2-cre−/gp130f/f mice (A–C) and VMD2-cre+/gp130f/f mice (D–F). GS: glutamate synthetase. Scale bar: 25 μm. G: Frequency of Cre-mediated recombination in Müller cells. Error bars: standard errors. ***: p<0.001 in t-test. Activation of STAT3 (pSTAT3) was detected by IHC 30 min after LIF injection. While LIF induced activation of STAT3 in all Müller cells in VMD2-cre−/gp130f/f mice (A, C) in response to LIF, STAT3 was not activated in 52.1% of Müller cells in VMD2-cre+/gp130f/f (arrowheads in D–F), suggesting that Cre-mediated recombination occurred in 52.1% of Müller cells at gp130 locus.
Retinal integrity
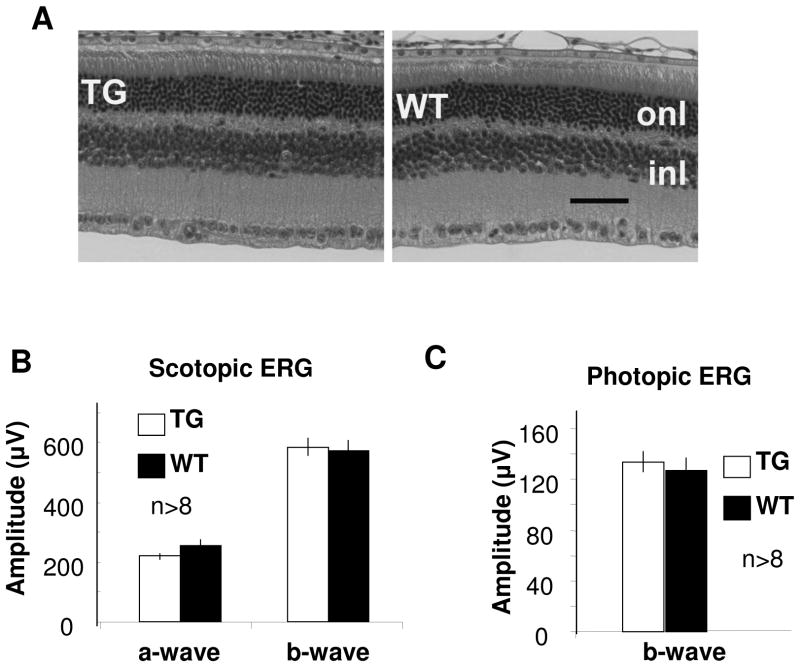
Previously studies demonstrated that mouse opsin promoter directed over-expression of Cre in rods results in photoreceptor degeneration (Jimeno et al., 2006; Le et al., 2006). Although the underline mechanism is unclear, over-expression of Cre can cause undesired chromosomal rearrangement in mammals (Loonstra et al., 2001; Schmidt et al., 2000). To determine the usefulness of our transgenic mice, we determined the effect of Cre expression in the retina over a ten-month period. Since a direct consequence of Müller cell death is photoreceptor degeneration, the retinal morphology and function was examined in the VMD2-cre mice. We did not detect any significant morphological changes in the mice (Figure 6A). In addition, the scotopic and photopic ERG analysis showed that these transgenic mice had normal photoreceptor function (Figure 6B–C). These results suggest that Cre expression did not cause any detectable change in retinal integrity in VMD2-cre mice, and thus the mice can be used for gene function studies.
Figure 6.
Retinal morphology and function of 10-month-old VMD2-cre mice. A: representative H&E-stained retinal sections of transgenic (TG) and wild-type (WT) mice. Outer nuclear layer (ONL) and inner nuclear layer (INL) are labeled. Scale bar equal to 50 μm. B–C: scotopic and photopic ERG amplitudes. Flash intensity was 138 cd × s/m2 for scotopic ERG and 79 cd × s/m2 photopic ERG. Error bars represent standard errors. Statistical analysis (t-test) showed that p-values for all waves were >0.05. At least eight mice were included in each group. No significant change in retinal morphology and function was detected in the VMD2-cre mice.
DISCUSSION
To generate inducible RPE-specific cre mice, we recently used a 3.0-kb human VMD2 promoter to control a binary system that expressed tetracycline-inducible Cre recombinase (Le et al., 2008), and identified a mouse line that had relatively strong Cre activity in Müller cells during retinal development. Although the inducible system in this VMD2-cre mouse line was not tightly controlled and the inducibility was less then onefold (Figure 2A), an inherited problem of the tetracycline-inducible gene expression technology, this mouse line offers an option for Cre/lox based conditional gene expression in retinal Müller cells. The efficiency of Cre-mediated recombination at gp130 locus in Müller cells is 52%, which is comparable to the efficiency in two of our published studies (Rajala et al., 2008; Zheng et al., 2006). This is not unexpected since conditional gene knockout studies usually have less than 100% efficiency of Cre-mediated recombination. With approximately 50% of Müller cells undergoing Cre-mediated recombination, this transgenic mouse line is ideal for generating genetic mosaics, a widely used method to investigate cell lineages, patterns of growth, and gene function, and to deal with the challenge in phenotypic analysis of mice generated with an all-or-none genetic approach (Collinson et al., 2004).
Of the ten VMD2-cre lines we have generated, five demonstrated Cre activated β-gal reporter expression in retinal Müller cells (data not shown). This result was surprising as the VMD2 promoter was expected to drive Cre expression in the RPE. Expression of our transgenes in retinal Müller cells is perhaps a characteristic of transient VMD2 promoter activity in retinal progenitor cells. This expression pattern is seemly contradictory to previous findings that VMD2 expression was localized to the RPE (Bakall et al., 2003; Esumi et al., 2004; Marquardt et al., 1998; Petrukhin et al., 1998), However, we cannot exclude transient transcription of VMD2 in non-RPE lineage since the cell-specific expression pattern of VMD2 promoter during embryonic development was not the focus of these earlier studies. To our knowledge, only one previous study dealt with embryonic expression of bestrophin, which demonstrated the expression of its mRNA in the eye at embryonic day 15 and there was no information regarding tissue-specific expression (Bakall et al., 2003). It is also important to point out two caveats of our study that prevent current mice from being sufficient to define VMD2 expression in retinal progenitor cells definitively. The first caveat is that the VMD2 promoter used in these transgenic animals may not contain all control elements necessary for identical expression patterns of the endogenous gene. The second caveat is that our expression system is a two transgene system. The unexpected expression pattern may reflect the use of this binary system, in which the transgenes are interacting to direct gene expression. Since Cre-mediated excision is not usually reversible, a transient expression of our transgenes in development will result in permanent removal of loxP flanked DNA. Therefore, regardless of the reasons behind the unexpected expression pattern, we definitely show that Cre activity is detected in the retina before E15 in the transgenic line described. Since we did not observe significant difference in Cre-activated β-gal activity between inductions from E15 to birth or P1 to P5, as judged by β-gal staining assay (data not shown), the VMD2 promoter is likely transiently active in progenitor cells, including those leading to Müller cells. However, we cannot completely rule out a possibility of very low level of Cre expression in the Müller cells of this mouse line after birth.
In this VMD2-cre mouse, we observed near absence of Cre function in the RPE (Figure 3). Since the presence of RPE-specific Cre function in another VMD2-cre mouse line derived from identical transgenes (Le et al., 2008), a lack of RPE-specific expression in this mouse line is probably due to a positional effect that is common to transgenics (Jaenisch, 1988). In this study, we also observed Cre activity in a subset of un-identified neurons in inner nuclear layer (INL) and some photoreceptor cells (Figure 4). This result may be a consequence of Cre function in their progenitor cells during embryonic development. In the VMD2-cre+/gp130f/f mice, less than 5% of rods lost STAT3 activation (data not shown), suggesting that most photoreceptors retained at least one copy of gp130. No other neural population in the retina lost a gp130 response. However, in the single hit β-gal reporter assay, we did observe β-gal expression in other neurons in the inner retina. End users of this mouse line should be aware of this extra-Müller cell recombination activity when designing their studies.
Acknowledgments
We thank W. Zheng and Y. W. Le for technical assistance, Dr. U. Hochgeschwender of Microinjection Core Facility at the Oklahoma Medical Research Foundation for the generation of the transgenic mice, Drs. N. Esumi and D. Zack for providing human VMD2 promoter DNA, and Dr. D. Sherry for helpful discussion. This study was supported by NIH grants RR17703, EY16459, and EY12190, ADA grant 1-06-RA-76, AHAF grant M2008-059, FFB grant BR-CMM-0808-0453-UOK and unrestricted grants from Hope for Vision and Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD. Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci. 2003;44:3622–8. doi: 10.1167/iovs.03-0030. [DOI] [PubMed] [Google Scholar]
- Betz UA, Bloch W, van den Broek M, Yoshida K, Taga T, Kishimoto T, Addicks K, Rajewsky K, Muller W. Postnatally induced inactivation of gp130 in mice results in neurological, cardiac, hematopoietic, immunological, hepatic, and pulmonary defects. J Exp Med. 1998;188:1955–65. doi: 10.1084/jem.188.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Collinson JM, Hill RE, West JD. Analysis of mouse eye development with chimeras and mosaics. Int J Dev Biol. 2004;48:793–804. doi: 10.1387/ijdb.041885jc. [DOI] [PubMed] [Google Scholar]
- Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem. 2004;279:19064–73. doi: 10.1074/jbc.M309881200. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–42. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–6. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Transgenic animals. Science. 1988;240:1468–74. doi: 10.1126/science.3287623. [DOI] [PubMed] [Google Scholar]
- Jimeno D, Feiner L, Lillo C, Teofilo K, Goldstein LS, Pierce EA, Williams DS. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest Ophthalmol Vis Sci. 2006;47:5039–46. doi: 10.1167/iovs.06-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Y, Ash JD, Al-Ubaidi MR, Chen Y, Ma J, Anderson RE. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004;10:1011–1018. [PubMed] [Google Scholar]
- Le Y, Sauer B. Conditional gene knockout using cre recombinase. Methods Mol Biol. 2000;136:477–85. doi: 10.1385/1-59259-065-9:477. [DOI] [PubMed] [Google Scholar]
- Le Y, Zheng L, Zheng W, Agbaga M, Zhu M, Ash JD, Anderson RE. Mouse opsin promoter controlled expression of Cre recombinase in transgenic mice. Mol Vis. 2006;12:389–398. [PubMed] [Google Scholar]
- Le Y, Zheng W, Rao P, Zheng L, Anderson RE, Esumi N, Zack DJ, Zhu M. Inducible expression of Cre recombinase in the retinal pigmented epithelium. Invest Ophthalmol Vis Sci. 2008;49:1248–1253. doi: 10.1167/iovs.07-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–14. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease) Hum Mol Genet. 1998;7:1517–25. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–7. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- Quiambao AB, Peachey NS, Mangini NJ, Rohlich P, Hollyfield JG, al-Ubaidi MR. A 221-bp fragment of the mouse opsin promoter directs expression specifically to the rod photoreceptors of transgenic mice. Vis Neurosci. 1997;14:617–25. doi: 10.1017/s095252380001258x. [DOI] [PubMed] [Google Scholar]
- Rajala A, Tanito M, Le YZ, Kahn CR, Rajala RV. Loss of neuroprotective survival signal in mice lacking insulin receptor gene in rod photoreceptor cells. J Biol Chem. 2008;283:19781–92. doi: 10.1074/jbc.M802374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala RV, McClellan ME, Chan MD, Tsiokas L, Anderson RE. Interaction of the retinal insulin receptor beta-subunit with the p85 subunit of phosphoinositide 3-kinase. Biochemistry. 2004;43:5637–50. doi: 10.1021/bi035913v. [DOI] [PubMed] [Google Scholar]
- Rucker EB, 3rd, Dierisseau P, Wagner KU, Garrett L, Wynshaw-Boris A, Flaws JA, Hennighausen L. Bcl-x and Bax regulate mouse primordial germ cell survival and apoptosis during embryogenesis. Mol Endocrinol. 2000;14:1038–52. doi: 10.1210/mend.14.7.0465. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–70. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci U S A. 2000;97:13702–7. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Ueki Y, Wang J, Chollangi S, Ash JD. STAT3 activation in photoreceptors by leukemia inhibitory factor is associated with protection from light damage. J Neurochem. 2008;105:784–96. doi: 10.1111/j.1471-4159.2007.05180.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Quiambao AB, Roveri L, Pardue MT, Marx JL, Rohlich P, Peachey NS, Al-Ubaidi MR. Degeneration of cone photoreceptors induced by expression of the Mas1 protooncogene. Exp Neurol. 2000;163:207–19. doi: 10.1006/exnr.2000.7370. [DOI] [PubMed] [Google Scholar]
- Yao L, Yokota T, Xia L, Kincade PW, McEver RP. Bone marrow dysfunction in mice lacking the cytokine receptor gp130 in endothelial cells. Blood. 2005;106:4093–101. doi: 10.1182/blood-2005-02-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Anderson RE, Agbaga MP, Rucker EB, 3rd, Le YZ. Loss of BCL-XL in rod photoreceptors: Increased susceptibility to bright light stress. Invest Ophthalmol Vis Sci. 2006;47:5583–9. doi: 10.1167/iovs.06-0163. [DOI] [PubMed] [Google Scholar]