Abstract
Nicotiana tabacum 46-8 cultivar displays an incompatible interaction with race 0 of Phytophthora parasitica var. nicotianae (Ppn), a fungal pathogen of most tobacco cultivars. At the plant level, incompatibility is characterized by the induction of lipoxygenase (LOX, EC = 1.13.11.12) activity and localized hypersensitive cell death before defense gene activation. To evaluate the involvement of LOX in the onset of plant defense, tobacco 46-8 plants were genetically engineered using full-length or partial-length antisense (AS) tobacco LOX cDNA constructs. AS expression strongly reduced elicitor- and pathogen-induced LOX activity. Eight independent AS-LOX lines were selected and assayed for their response to Ppn. After root or stem inoculation with race 0, all AS-LOX lines but one displayed a compatible phenotype whereas control transformed plants, not containing the AS-LOX cassette, showed the typical incompatible reaction. The presence of the fungus in transgenic lines was demonstrated by PCR amplification of a Ppn-specific genomic sequence. A linear relationship was found between the extent of LOX suppression and the size of the lesion caused by the fungus. The AS-LOX plants also showed enhanced susceptibility toward the compatible fungus Rhizoctonia solani. The results demonstrate the strong involvement of LOX in the establishment of incompatibility in plant–microorganism interactions, consistent with its role in the defense of host plants.
Keywords: antisense RNA, defense, signal transduction
Plant resistance to microorganisms ranges from passive and nonspecific protection involving a large set of genes to active and specific incompatibility responses in which specificity is controlled by a single gene in each partner (1, 2). In most specific systems, resistance is associated with the establishment of a hypersensitive reaction (3) and with early induction of defense gene expression. Unraveling the mechanisms underlying resistance is a major goal of plant pathology.
The development of plant resistance and the induction of defense responses are achieved by means of a complex series of events (4) arising from initial mutual recognition of the two partners and subsequent production of secondary signals. Among them, free radicals and active oxygen species might be involved in triggering hypersensitive cell death whereas ethylene, salicylic acid, and jasmonates induce defense gene expression (5). The involvement of jasmonates in plant responses to pathogens (6, 7) has reinforced our interest in plant lipoxygenases (LOXs, EC = 1.13.11.12). LOXs are non-heme dioxygenases that catalyze the oxygenation of polyunsaturated fatty acids (PUFAs), leading to the formation of hydroperoxides. In mammals, LOXs are involved in the arachidonic acid cascade that converts this PUFA into eicosanoids such as leukotrienes and lipoxins (8, 9). These products, in turn, regulate physiopathological processes, including the immune and inflammatory reactions, and the response to infectious pathogens (10). In plants, LOX gene expression is associated with a number of developmental events and induced by environmental changes, notably pathogen challenge (11). PUFA hydroperoxides resulting from the action of LOX are very reactive and may give rise to free radicals that can contribute to promoting cell death (12). The hydroperoxides may also be converted into more stable, active compounds including aldehydes and hydroxy- and epoxy-fatty acids, some of which show antimicrobial activities (13–15), and jasmonic acid (JA), derived from the LOX product 13-hydroperoxy-octadecatrienoic acid.
In the race–cultivar-specific interaction between tobacco (Nicotiana tabacum) and the phytopathogenic fungus Phytophthora parasitica var. nicotianae (Ppn), we have shown that LOX gene expression and activity are induced after root inoculation with zoospores of Ppn (16). This induction occurs as an early event in the incompatible interaction, whereas it appears 1 day later in the compatible interaction (17). Enhancement of LOX expression in response to fungal, bacterial, and viral pathogen ingress appears to be a general feature occurring both in monocots and dicots (11). In tobacco, a glycopeptide elicitor prepared from the cell wall of Ppn also induces LOX and defense gene expression as well as JA accumulation in cultured cells, whereas JA itself appears to participate in the onset of defense reactions (17, 18). Taken together, these data led to the proposal that LOX could be one of the mediators of resistance to biotic and abiotic stresses. However, its actual in vivo function in these processes is as yet unknown.
The purification of LOX from elicited tobacco cells and infected tobacco plants (16) yielded a preparation showing a single band in SDS/PAGE, suggesting that only one LOX isoform might be induced by the pathogen and its elicitors. In vitro characterization of the enzyme activity showed that it possesses both a 9- and 13-LOX specificity, with a predominance for the 9-LOX function. A cDNA encoding the enzyme specifically expressed upon infection or elicitor treatment was isolated (17, 19). To further evaluate the role of the LOX pathway in plant–pathogen interactions, we chose to modify LOX expression in tobacco via a transgenic approach by using an antisense (AS) strategy. As recipient plant, we used the tobacco cultivar 46-8, which shows resistance to race 0 of Ppn.
The present work shows the effects of AS gene expression on LOX activity and on the phenotype of tobacco plants upon inoculation with Ppn. The susceptibility of transgenic tobacco to Rhizoctonia solani, a plant pathogenic fungus with a broad host range, also was investigated.
METHODS
Plant Material and Pathogen Isolates.
Tobacco plant (Nicotiana tabacum L.) isolines 46-8 and 49-10 (20) were grown on vermiculite in the greenhouse. Line 46-8 is resistant to race 0 of Ppn whereas line 49-10 is susceptible to the same race of the fungus. Ppn race 0 was grown on oatmeal agar at 25°C in the dark. Zoospores were obtained as described (16).
Rhizoctonia solani (isolate R92-1) was propagated as a mycelial culture on sterile Malt agar medium (20 g/liter of malt extract, 2 g/liter of yeast extract) and maintained in the dark at 20°C. Seven-day-old cultures were used for inoculations of tobacco plants.
Elicitor Treatment.
Elicitation was performed on 3-week-old seedlings. Plants were depotted and their roots were immediately dipped in a test tube containing either water (control) or a glycopeptide-containing elicitor preparation (30 μg/ml) obtained by autoclaving a cell wall preparation from the mycelium of Ppn (21). After absorption of the elicitor solution, plants were watered with nutrient solution and harvested 30 h after the beginning of the experiment.
Plant Inoculation.
Root inoculation was carried out on 5-week-old plants with freshly prepared zoospores of Ppn race 0 (16), and symptoms were observed 5 days later. For PCR detection of the fungus in tobacco roots, a slightly modified protocol was followed. Care was taken that only the last portion (0.5 cm) of the root tips would be dipped in the zoospore suspension for 5 h. After this step, the entire root system of the plantlet was washed in sterile water, transferred to another test tube, and watered for 3 days. The inoculated portion of the root tip (0.8 cm) then was cut off and discarded to minimize possible contamination of the root material by adsorbed zoospores; the remaining root material, which had never been in contact with the zoospore suspension, was frozen for further DNA extraction.
Stem inoculations were performed on 10-week-old seedlings as described (22). The stem of each plant was sectioned 7 cm below the apex, and a mycelial pad withdrawn from the periphery of a 7-day-old colony of Ppn race 0 was immediately laid on the cut surface. The extent of fungal colonization was assessed 84 h postinoculation by measuring the length of necrotized infected tissues on a longitudinal section of the stem.
Infections with Rhizoctonia solani were conducted in a growth chamber under constant temperature (27°C), high relative humidity (100%), and with a light–dark regime of 14 h of light (220 μmol/m2⋅s Photons) and 10 h of dark. Mycelial discs (6 mm in diameter) were positioned upside-down on wet leaves of 10-week-old tobacco plants. Symptoms developed as radial necrosis around the mycelial disc. The fungal growth was evaluated by measuring the diameter of each necrotic lesion 4 days postinoculation. All inoculations were performed on different plants in a completely randomized manner, and symptoms were recorded independently by two persons.
Construction of Binary Vectors with AS-LOX Cassettes for Plant Transformation.
pIPM0 was made by inserting the 420-bp fragment of the 35S RNA promoter (p35S) of the Cauliflower Mosaic virus (23), the KpnI SmaI BamHI XbaI multiple cloning site (mcs) of pUC19, and the nopaline synthase terminator (tNOS) in the EcoRI-HindIII sites of the binary vector pBIN19 (24). The T-DNA of pIPM0 also contained the plant selectable marker neomycin phosphotransferase II gene (nptII), conferring resistance to kanamycin, fused to the nopaline synthase promoter (pNOS) and terminator (tNOS) elements (Fig. 1). pIPM120 was constructed by cloning the full-length LOX cDNA XbaI/KpnI 2.9-kb fragment (GenBank accession no. X84040) (19) in AS orientation between the KpnI and XbaI sites of pIPM0. pIPM125 was constructed by ligating an internal BglII/BglII 624-bp fragment (nucleotides 1783–2407 of the cDNA) in the compatible BamHI cohesive site of pIPM0. Insert orientation was checked by restriction enzyme digestions. The borders of each construct, overlapping the cloning sites, were checked by direct sequencing before transfer into agrobacteria.
Figure 1.
Construction of AS-LOX cassettes for plant transformation. (A) Full-length tobacco LOX cDNA (19) and derived sequences subcloned in AS orientation in the cloning vector pIPM0. (B) Structure of the T-DNA constructs transferred to plant cells via Agrobacterium tumefaciens. NTR, nontranslated region; LB and RB, left and right borders.
Transformation, Regeneration, and Selection of Plant Material.
pIPM0, pIPM120, and pIPM125 plasmids were mobilized into Agrobacterium tumefaciens LBA4404 by using a heat-shock treatment (25). The presence and structure of the T-DNA in the bacteria were confirmed by restriction enzyme analysis and by hybridization of total DNA to the LOX cDNA probe.
Leaf disc inoculations with A. tumefaciens strains harboring pIPM0, pIPM120, or pIPM125 plasmids were carried out according to established methods (26), with axenic cultures of the tobacco 46-8 line as starting material. After 7–8 weeks on selective MS medium (150 mg/liter of kanamycin, 500 mg/liter of carbenicillin) (27), shoots were dissected away from independent transformed calli and rooted axenically in selective media without growth regulator. Transgenic plantlets then were transferred to soil in a growth chamber under constant temperature (27°C) and humidity (70%), and with a light–dark regime of 14 h of light (220 μmol/m2⋅s Photons) and 10 h of dark.
Screening of the T0 transformants was performed by PCR using oligonucleotide primers situated on or near the margins of the nptII and, when relevant, AS-LOX sequences. Only positive plants were transferred to the greenhouse and continued to be grown until flowering. They were allowed to self-pollinate to obtain the T1 progeny. T1 seeds were sterilized (28) and sown on solid MS medium supplemented with 200 mg/liter of kanamycin. Only the surviving green plantlets, homozygous or hemizygous for the kanamycin-resistance (KanR) trait were transferred to vermiculite and grown under the above-mentioned conditions. Elicitation and inoculation experiments were performed on these KanR plants.
Nucleic Acid Techniques.
Total DNA from recombinant A. tumefaciens was extracted according to described methods (29). Plant DNA was extracted (30) and characterized by restriction enzyme digestion and Southern hybridization analysis (31).
PCR amplifications were performed in a thermocycler (Omnigene TR3, Hybaid, U.K.) by using the DNA polymerase from Thermus brockianus (ExtraPol II, Eurobio, France). Amplification reactions were conducted in 50 μl total volume containing 500 ng DNA, 10 mM Tris⋅HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, 200 μM of each dNTP, 10 pmol of each specific primer, and one unit of DNA polymerase as described (32). The cycle parameters were as follows: denaturation at 94°C for 45 sec, annealing at 63°C for 45 sec, and extension at 72°C for 1 min. The cycles were repeated 35 times, with the exception of an initial denaturation step of 5 min at 95°C and a final extension step of 5 min at 72°C.
Oligonucleotide primers corresponding to the elicitin gene of Phytophthora parasitica (33) were complementary to a portion of the promoter (5′- GCTCTCATCCACACCCACCCC-3′) and of the 3′ region overlapping the end of the coding sequence (5′-CCCGCTTACAGTGACGCGCACG-3′). The expected amplified product was 410 bp in size.
Total RNA was extracted from tobacco following established procedures (34). Samples (20 μg each) were denatured and separated on 1% agarose gels by using the glyoxal-phosphate buffer technique (31). Northern blot hybridization was carried out in high-SDS buffer (35). The probes were radiolabeled by random priming using [α-32P]dCTP.
Protein Extraction and LOX Assays.
Plant tissues were ground in liquid nitrogen and homogenized in 100 mM Tris⋅HCl buffer (pH 6.8) 10 mM EDTA, 5 mM DTT in the proportion of 1 ml extraction buffer per 0.5 g of tissue. The homogenate was centrifuged at 10,000 × g for 10 min at 4°C. Aliquots of the supernatant were used to measure LOX activity by polarography (16). The protein content of the crude extract was determined by the method of Bradford (36). LOX specific activity was expressed in nKat/mg protein.
Statistical Analysis of the Results.
Results collected from the various experiments were analyzed by using sas 6.11 software (37). Repeated-measures ANOVA and Tukey’s studentized range tests (HSD) were performed by using the sas/stat “PROC.GLM” package (SAS Institute, Cary, NC) according to the software specifications.
RESULTS
Integration of AS-LOX Cassettes into the Tobacco Genome.
The previously characterized cDNA encoding an early-induced, defense-associated tobacco LOX was used to design two AS-LOX cassettes (Fig. 1). Recombinant agrobacteria bearing these constructs or the control LOX-free plasmid pIPM0 were used to transform 46-8 leaf discs. Kanamycin-resistant 46-8 independent T0 primary transformants were selected, and 10 IPM120-T0 lines, 10 IPM125-T0 lines, and 5 IPM0-T0 controls were regenerated.
These transformants then were analyzed for the presence of the KanR cassette and the AS-LOX cassette, when relevant, by PCR and Southern blot experiments. Only plants containing nonrearranged copies of the constructs or the empty vector IPM0 were transferred to soil in the greenhouse for seed setting.
At all stages of development, these plants were indistinguishable from wild-type (WT) untransformed plants, notably in terms of length and size of leaves, stems, and roots; color and shape of photosynthetic tissues; number and shape of flowers; and seed content of capsules.
Further analysis of the transformants was undertaken on their T1 progeny. T1 segregation of the KanR trait was determined by sowing 500 sterilized seeds per transgenic line on selective medium. After 3 weeks, the ratio of “green KanR ” to “white KanS” plants was established and statistically compared by using χ2 tests (P < 0.05) to the theoretical 3:1, 15:1, and 63:1 ratios expected in the case of one, two, or three independent insertions, respectively.
Southern blot experiments and T1 progeny analysis allowed selection of nine unambiguous transgenic lines for subsequent experiments: four full-length IPM120 AS-LOX lines containing either one (120-2, 120-3, and 120-4) or three (120-1) T-DNA insertions; four partial-length IPM125 AS-LOX lines containing either one (125-2), two (125-4), or three (lines 125-1 and 125-3) T-DNA insertions; and one LOX-free T-DNA (IPM0) line, bearing one insertion of pIPM0, as a control.
Transgenic AS-LOX Plants Display a Dramatically Reduced LOX Activity.
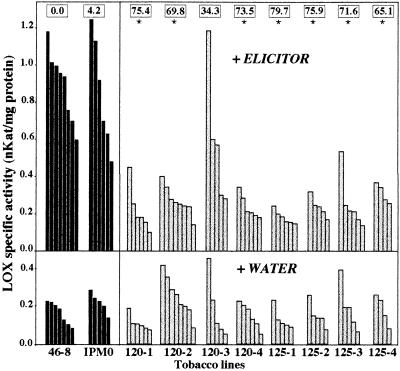
Because the LOX cDNA used in our study had been isolated from an elicitor-induced cDNA library of tobacco, we first determined the LOX activity of individual T1 KanR whole plants of the different transgenic lines in response to the elicitor of Ppn and compared it with the activity of nonelicited, water-treated, plants. Between four and eight seedlings per treatment and per AS-LOX transgenic line were assayed. The experiment included elicited and nonelicited controls, corresponding to WT 46-8 and KanR IPM0 plants. Fig. 2 Lower shows that the LOX activity of untreated plants was low and did not differ statistically between controls and transgenic lines. Elicitor treatment (Fig. 2 Upper), which has no visible effect on plant phenotype, provoked a 4- to 5-fold increase in LOX activity for both the 46-8 WT and IPM0 controls, indicating that there was no significant effect of transformation and regeneration on enzyme activity. In contrast, elicitor-induced LOX activity was very low in most transgenic AS-LOX plants, and, except for line 120-3, the recorded values were close to the basal LOX activity level of noninduced plants.
Figure 2.
Measurements of LOX activity in AS-LOX (gray bars) and control (black bars) tobacco plants whose roots were allowed to absorb either elicitor or water (Upper and Lower, respectively). Each bar represents the LOX activity of an individual plant. Tobacco lines correspond to: 46-8, WT control; IPM0, transgenic 46-8 line bearing the LOX-free T-DNA; 120-1 to -4 and 125-1 to -4, transgenic 46-8 AS-LOX plants. Boxes along the top of the graph indicate the average reduction (%) of LOX activity of each line upon elicitation, as compared with the WT 46-8 control. Significance was determined by repeated-measures ANOVA and Tukey’s HSD test. ∗, Significant difference from the controls, P < 0.001.
We then investigated the response of transgenic tobaccos to inoculation by race 0 of the fungus. This study was conducted on inoculated stems of four lines (120-1, -2, -4, and 125-1). Stem inoculation was used because it allowed easy localization of the infected area, therefore minimizing dilution by healthy tissues during extraction. Inoculated WT 46-8 plants showed a 6.8-fold increase of LOX activity over uninoculated control plants. In comparison, the LOX activity of inoculated AS-LOX lines was only induced by a factor of 2.2–4.8. Thus, the pathogen-induced enzyme activity was reduced by 30–70% compared with that of WT plants (data not shown). These differences were shown to be significant for lines 120-2, 120-4, and 125-1 and therefore confirmed the efficiency of the AS strategy.
LOX transcripts and the balance between sense and AS transcript populations were quantified by Northern hybridization to the LOX cDNA probe. Total RNA from the noninoculated WT 46-8 line did not show any hybridization to this probe, whereas RNA from inoculated WT plants displayed a band at 2.9 kb corresponding to the LOX transcript (Fig. 3). In six of the eight independently transformed plants, endogenous LOX sense transcripts were not detected. Transcripts were detected in low amounts in line 120-2 (lane 4) and in higher amounts in line 120-3 (lane 5). Because of utilization of a cDNA probe and the fact that AS and sense RNAs should have the same size (2.9 kb) in the transgenic IPM120 lines, the band observed for these plants might contain either one or both populations of RNA. This was not the case for the IPM125 plant lines where the expected AS transcript (0.63 kb) was distinguishable from the 2.9-kb sense RNA.
Figure 3.
(A) Northern blot analysis of total RNA isolated from stems of control and transgenic lines 12 h after inoculation with the mycelium of Ppn race 0 (lanes 2–10). RNA samples (20 μg per lane) corresponded to: WT uninoculated (lane 1) and inoculated (lane 2) 46-8 tobacco; inoculated transgenic tobacco IPM120-1 to 120-4 (lane 3–6), and IPM125-1 to 125-4 (lanes 7–10). The radiolabeled full-length LOX cDNA was used as a probe. (B) Ethidium bromide staining of the gel before blotting, showing amounts of RNA loaded.
In most plant lines, AS RNAs were produced in sufficient amounts to counteract the LOX mRNAs induced upon fungal inoculation. Tobacco lines in which both sense and AS transcript populations were absent, probably resulting from complete degradation of both species (120-1, 120-4, 125-1), and tobaccos in which only the 0.63-kb AS transcript was detected (125-2, 125-3, 125-4) exhibited the lowest LOX activity, both upon elicitation through the root system and after stem inoculation (lines 120-4 and 125-1). The higher transcript level observed in transgenic line 120-3 was consistent with the lower average reduction in elicitor-induced LOX activity of the same line (i.e., 34.3% vs. 70–80% in the other transgenic lines). There was no apparent correlation between the number of IPM120 or IPM125 cassettes in the genome of the different lines and the reduction of LOX activity.
Transgene Expression Modifies the Disease Phenotype and Enables the Pathogen to Spread Inside the Roots of Transgenic Tobacco Plants.
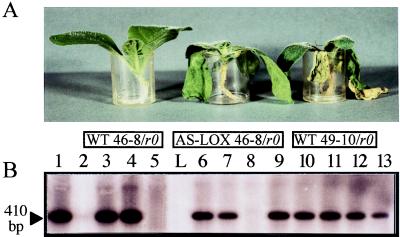
The effect of suppressing LOX induction on the phenotype of inoculated plants first was assessed at the whole-plant level. Transgenic plants and WT-resistant 46-8 and susceptible 49-10 tobacco isolines were inoculated by dipping their roots in a zoospore suspension of Ppn race 0. Uninoculated plants were dipped in nutrient solution only. Inoculations were performed on three to five plants per line and were repeated three times independently. Five days postinoculation, inoculated WT 46-8 and IPM0 plants were devoid of externally visible symptoms, as were their noninoculated counterparts (Fig. 4A). In contrast, inoculated susceptible WT 49-10 plants exhibited a dark-brown color of the whole root system, severe black necrosis of the collar, and complete wilting of the leaves.
Figure 4.
Comparison of the phenotypes of WT and transgenic plants upon inoculation with Ppn race 0 (A). The transgenic IPM120-4 tobacco line was retained as an example. Plants were treated with nutrient solution (controls) or inoculated with a zoospore suspension (2 ml at 106 zoospores/ml) and were photographed 5 days postinoculation in the plastic beaker in which they were watered. From left to right: WT 46-8/r0, inoculated WT plant resistant to race 0; AS-LOX 46-8/r0, inoculated transgenic 120-4 plant; WT 49-10/r0, inoculated WT plant susceptible to race 0. (B) Detection of Ppn in the roots of inoculated plants by PCR amplification of the elicitin gene of the fungus. Three days postinoculation, total DNA was extracted from the upper part of the roots and subjected to PCR amplification. Ten microliters of the amplification products was separated on a 1.8% agarose gel and hybridized with 200 ng of the radiolabeled 410-bp elicitin product as a probe. Control experiments were performed on the following templates: lane 1, race 0 DNA (250 pg); lane 2, 46-8 WT plant DNA (500 ng); lane 3, 46-8 WT plant DNA (500 ng) plus race 0 DNA (250 pg). PCR assays on inoculated material were performed on DNA (500 ng) extracted from plant lines 49-10 WT (lane 4), 46-8 WT (lane 5), 120-1 to -4 (lanes 6–9), and 125-1 to -4 (lanes 10–13). L, molecular weight marker.
Transgenic plants derived from the resistant 46-8 line developed a pronounced browning of the root system, a light-brown collar, and extended wilting of the leaves. These symptoms were observed in 70–90% of the plants in all transgenic lines except line 120-3. The remaining 10–30% showed milder symptoms with less, though still obvious, wilting of the leaves and limited browning of the roots. Transgenic line 120-3 essentially behaved like WT-resistant 46-8 line.
To correlate the symptoms observed after inoculation to the actual colonization of the roots by the fungus, PCR-based detection of Ppn was undertaken. Oligonucleotide primers were designed to target a DNA sequence present in the fungal genome but missing in the plant genome. This sequence corresponded to the elicitin gene of Phytophthora parasitica (33), which encodes a polypeptide belonging to a family of low-molecular-weight toxic molecules secreted by most Phytophthora species. The results of the PCR experiments (Fig. 4B) indicated that the marker sequence was indeed fungus-specific because a product was amplified from Ppn DNA (lane 1) but not from WT plant DNA (lane 2). A reconstruction experiment in which Ppn and plant DNA were mixed showed that even a small amount of fungal DNA could be detected in the presence of excess plant DNA (lane 3). PCR amplifications performed on template DNA extracted from inoculated tobacco isolines showed that the marker gene was not amplified from the DNA of resistant plants (lane 5). On the contrary, the elicitin sequence was amplified strongly from the DNA of susceptible tobacco roots (lane 4). Amplification carried out in the same conditions on DNA extracted from transgenic AS-LOX plants revealed that the fungus was detected in seven out of the eight transgenic lines having a resistant background but expressing an AS-LOX sequence (lanes 6–13). The fungus was not detected in line 120-3 plants, consistent with the phenotype of these plants after inoculation, and with the only slight reduction of pathogen-induced LOX activity. Histological staining of the fungus in inoculated roots with cotton blue also showed that the mycelium of Ppn race 0 colonizes the roots of AS-LOX plants and differentiates sporangia as it does in control susceptible tobacco (data not shown). The fungus was undetectable in IPM0 plants and barely detectable in 120-3 plants. These results fully confirmed that AS-mediated inhibition of LOX allows Ppn race 0 to grow inside the root system of the 46-8 tobacco cultivar in which it does not normally develop.
Transgenic Lines Have Lost the Ability to Block Pathogen Colonization in Stems.
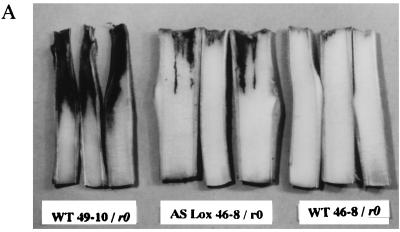
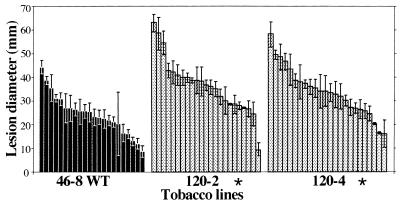
The ability of Ppn race 0 to colonize plant tissues was quantified after stem inoculation by measuring, on longitudinal stem sections, the length of the necrotic lesions caused by the fungus 84 h postinoculation. WT-resistant plants developed a small necrotized lesion typical of a hypersensitive response, averaging 3.4 ± 0.43 mm in length (mean ± SEM, n = 11, Fig. 5B), in which pathogen growth was arrested rapidly (Fig. 5A). In comparison, large lesions developed on susceptible plants (Fig. 5A), with an average length of 38.8 ± 1.76 mm (mean ± SEM, n = 13, data not shown). IPM0 plants did not show statistically significant differences compared with WT-resistant plants (Fig. 5B). When transgenic AS-LOX plants were inoculated, necrotic lesions ranging from 4 to 28 mm in length developed in the stem. These lesions were essentially similar to those observed in susceptible WT plants, displaying maceration, browning, and collapse of the central tissues of the stem (Fig. 5A). Statistical analysis revealed that three lines showed a highly significant difference (P < 0.001) and two lines showed a significant difference (P < 0.05) in lesion length compared with the resistant control 46-8 plants (Fig. 5B). Among the three other lines, two (line 120-1 and 120-2) scored very close to the significance limit whereas line 120-3 did not show any significant difference. Again, absence of lesions in this line correlated with a slight reduction in LOX activity, no symptoms upon inoculation of the plant, and no detectable fungal growth in the roots. A linear correlation between the expression of LOX and of incompatibility scored with a highly significant r (r = −0.87, Bartlett’s χ2 = 149, df = 1, P < 0.001).
Figure 5.
(A) Stem inoculation of WT and transgenic plants by Ppn mycelium. Ten-week-old plants were cut off 7 cm below the apex, and the cut stem section was inoculated with a pad of Ppn race 0 mycelium. After 84 h, the stems were sectioned longitudinally and the lesions caused by the fungus were observed. From left to right: inoculated WT-susceptible 49-10 stems showing large, dark-brown rotted lesions; transgenic AS-LOX 46-8 stems (IPM125-1 retained as an example) displaying a susceptible phenotype; inoculated WT-resistant 46-8 stems, devoid of lesions and only exhibiting faint localized necrosis. (B) Measurement of stem lesions. Ten-week-old control (46-8 WT and IPM0, black bars) and transgenic (IPM120-1 to -4 and 125-1 to -4, gray bars) plants were stem-inoculated and the lesion length was measured on longitudinal stem sections. Each bar represents the size of the lesion measured on one individual. Significance was determined by repeated-measures ANOVA and Tukey’s HSD test. Significant difference from the controls: ∗, P < 0.05; ∗∗, P < 0.001.
Susceptibility of Transgenic AS-LOX Tobacco Plants to Rhizoctonia solani.
To evaluate the effect of reducing LOX induction on the interaction of tobacco with other fungal pathogens, transgenic AS-LOX plants (lines 120-2 and 120-4) were challenged with Rhizoctonia solani, a compatible fungal pathogen causing severe diseases in a wide range of host plants, including tobacco. Lesions were observed in the leaf parenchyma of AS-LOX plants as well as in control WT plants. The diameter of the lesions was measured 4 days postinoculation (Fig. 6). Both AS-LOX lines displayed enhanced susceptibility to the pathogen, the difference with the untransformed 46-8 line being highly significant (P < 0.001).
Figure 6.
Infection of control and transgenic AS-LOX lines by Rhizoctonia solani. Plants were inoculated on the leaf surface. Eight pads per plant were distributed on four different leaves. Four days postinoculation, the average lesion diameter was determined for each plant. Each bar represents the mean lesion size on a plant ± SEM. Significance was determined by repeated-measures ANOVA and Tukey’s HSD test. ∗, Significant difference from the WT 46-8 line at P < 0.001.
DISCUSSION
The occurrence of race–cultivar-specific interactions between plants and microorganisms offers advantages for studying the molecular events underlying resistance and defense induction in the host, allowing comparison of compatible and incompatible situations. The hypersensitive, localized cell death of the host that typifies incompatibility has proved the most efficient way for the plant to block and often destroy the pathogen. Despite recent progress in the characterization of the resistance and avirulence genes, which govern incompatibility (38), no general overview of the transduction pathways by which interaction of their products triggers the hypersensitive response has emerged. It has been proposed that lipid signals derived from the action of LOX on PUFAs might be involved in such a process. The most direct way to check this hypothesis was to produce LOX mutants and look for their phenotype upon inoculation with fungi. To this end, we chose to use an AS strategy.
The tobacco–Ppn interaction is particularly well suited for this purpose because of (i) the low number of LOX genes, probably not exceeding three in the tobacco genome, in contrast to most other plants where multiple LOX isoforms and genes have been reported; (ii) the early induction of one transcript and one isoform during incompatibility and after elicitor treatment, suggesting that only one gene is induced; (iii) the very low basal activity and expression of LOX in the vegetative organs of tobacco; and (iv) the occurrence of race–cultivar-specific interactions in this system. The cultivars studied here display clearly distinguishable phenotypes when inoculated by race 0 of the fungus: the resistant 46-8 line is devoid of externally visible symptoms, whereas the isogenic susceptible 49-10 line shows the black shank disease symptoms. The 46-8 cultivar therefore was chosen as target plant material to apply the AS strategy.
Two AS-LOX constructs were introduced into the plant genome. Examination of the resulting regenerants and T1 progeny showed that healthy transgenic plants displayed a normal phenotype, indicating that the presence of the AS-LOX transgene did not interfere with other general metabolic pathways that might have involved the other LOX enzymes. In particular, the seeds resulting from self-pollination had normal germination rates. Further analysis showed that the induction of LOX that normally occurs in response to the pathogen was decreased strongly in all transgenic lines but one, averaging values close to the basal constitutive level. This decrease was linked to the production of AS RNAs because endogenous sense LOX mRNAs were not detected in inoculated plants of most transgenic lines. Comparison of transgenic AS-LOX lines with the controls showed that transgene expression resulted in a dramatically modified phenotype upon inoculation: whereas the WT 46-8 parent was symptomless, the AS-LOX 46-8 plants displayed the symptoms of a compatible interaction. Thus, this work revealed that suppressing LOX activity was sufficient to turn an incompatible phenotype into a compatible one. Statistical analysis of the data confirmed that suppression of incompatibility was effective in all transgenic lines but one, whatever the mode of inoculation, the length of the AS-LOX sequence construct, and the number of copies in the plant genome. Enhanced susceptibility was confirmed further by PCR detection of Ppn in transgenic plants whereas in WT-resistant plants no fungal signal was amplified. Interestingly, the transgenic line that was not impaired significantly in LOX activity also retained the incompatible phenotype. These data unequivocally demonstrate that a specific inducible LOX is essential for the expression of resistance in tobacco.
Diverse mechanisms might account for the suppression of incompatibility. Understanding whether they act directly or indirectly requires the PUFA hydroperoxides and their metabolites to be precisely determined in WT and transgenic AS-LOX plants. In tobacco, LOX gene expression is induced and JA is produced in the very first hours after elicitor treatment (17, 18). However, the production of JA is only transient, whereas induction of LOX lasts for several hours, suggesting that additional compounds are formed. One might expect incompatibility to result from the complementary effects of these diverse compounds and to be highly compromised if their levels are lowered in AS-LOX plants. Owing to the pleiotropic effects of JA when exogenously supplied to tobacco, attempts to reverse the AS phenotype by JA treatment were not undertaken.
In addition to jasmonates, a number of compounds may serve as endogenous messengers, notably hydrogen peroxide, ethylene, and salicylic acid (39, 40). The question as to whether their respective pathways share common regulatory steps and are somehow connected to the LOX pathway will need to be answered.
In mammals, recent reports emphasized a role of LOX products in regulating apoptotic cell death and resistance to pathogenic agents (10). These findings and the data reported in this work suggest that some proteins and functions involved in programmed cell death related to defense against pathogenic agents might be common to plants and animals. The availability of AS-LOX plants provides an efficient tool to further investigate the function of LOX in plant–microbe interactions.
Acknowledgments
We acknowledge G. Freyssinet, M.-C. Grosjean-Cournoyer, and S. Axiotis for helpful discussions, and Prof. J.-H. Weil for critical review of the manuscript. We are very much indebted to Prof. J. Lauga and Dr. J. Saint-Pierre for help in statistical analyses and to C. DuChatenet and E. Radondy for their technical skills in screening the transformants. We thank R. Delon and B. Cailleteau from SEITA for providing the Rhizoctonia solani isolate. I.R. is a doctorate fellow of the Ministère de la Recherche et de la Technologie. This work was supported in part by the Bio Avenir program (Rhône Poulenc-French Ministries of Research and of Industry).
ABBREVIATIONS
- AS
antisense
- JA
jasmonic acid
- KanR
kanamycin resistance
- LOX
lipoxygenase
- Ppn
Phytophthora parasitica var. nicotianae
- PUFA
polyunsaturated fatty acid
- WT
wild type
References
- 1.Flor H. Annu Rev Phytopathol. 1971;9:276–296. [Google Scholar]
- 2.Keen N. Plant Mol Biol. 1992;19:109–122. doi: 10.1007/BF00015609. [DOI] [PubMed] [Google Scholar]
- 3.Dangl J, Dietrich R, Richberg M. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keen N. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 5.Hammond-Kosak K, Jones J. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creelman R, Mullet J. Proc Natl Acad Sci USA. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasternack C, Parthier B. Trends Plant Sci. 1997;2:302–307. [Google Scholar]
- 8.Samuelson B, Dahlàn S-E, Lindgren J Å, Rouzer C, Serhan C. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 9.Nicolaou K, Ramphal J, Petasis N, Serhan M. Angew Chem Int Ed Engl. 1991;30:1100–1116. [Google Scholar]
- 10.Bailie M, Standiford T, Laichalk L, Coffey M, Strieter R, Peters-Golden M. J Immunol. 1996;157:5221–5224. [PubMed] [Google Scholar]
- 11.Rosahl S. Z Naturforsch. 1996;51c:123–138. doi: 10.1515/znc-1996-3-401. [DOI] [PubMed] [Google Scholar]
- 12.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 13.Kato T, Yamagushi Y, Uyehara T. Naturwissenschaften. 1983;70:200–201. [Google Scholar]
- 14.Verninghi A, Einhorn J, Kunesch G, Malosse C, Ramiandrasoa F, Ravise A. C R Acad Sci. 1985;301:743–748. [Google Scholar]
- 15.Croft K, Juttner F, Slusarenko A. Plant Physiol. 1993;101:13–24. doi: 10.1104/pp.101.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fournier J, Pouénat M, Rickauer M, Rabinovitch-Chable H, Rigaud M, Esquerré-Tugayé M-T. Plant J. 1993;3:63–70. [Google Scholar]
- 17.Véronesi C, Rickauer M, Fournier J, Pouénat M, Esquerré-Tugayé M-T. Plant Physiol. 1996;112:997–1004. doi: 10.1104/pp.112.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickauer M, Brodschelm W, Bottin A, Véronési C, Grimal H, Esquerré-Tugayé M-T. Planta. 1997;202:155–162. [Google Scholar]
- 19.Véronesi C, Fournier J, Rickauer M, Marolda M, Esquerré-Tugayé M-T. Plant Physiol. 1995;108:1342. [Google Scholar]
- 20.Helgeson J, Kemp J, Haberlach G, Maxwell D. Phytopathol. 1972;62:1439–1443. [Google Scholar]
- 21.Pélissier B, Thibaud J, Grignon C, Esquerré-Tugayé M-T. Plant Sci. 1986;46:103–109. [Google Scholar]
- 22.Stavely J. USDA Tech Bull. 1979;1586:87–110. [Google Scholar]
- 23.Gardner R, Howarth A, Hahn P, Brown-Luedi M, Shepherd R, Messing J. Nucleic Acids Res. 1981;9:2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bevan M. Nucleic Acids Res. 1984;12:8711–8722. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holsters M, Dewaele D, Depicker A, Messenf E, Van Montagu M, Schell J. Mol Gen Genet. 1978;136:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 26.Horsch R, Fry J, Hoffmann N, Eicholtz D, Rogers S, Fraley R. Science. 1985;227:1229–1231. [Google Scholar]
- 27.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 28.Rancé I, Tian W, Mathews H, Kochko A, Beachy R, Fauquet C. Plant Cell Rep. 1994;13:647–651. doi: 10.1007/BF00232938. [DOI] [PubMed] [Google Scholar]
- 29.Herrera-Estrella L, Simpson J, editors. Plant Molecular Biology, A Practical Approach. Oxford: IRL; 1988. p. 141. [Google Scholar]
- 30.Dellaporta S, Wood J, Hicks J. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 31.Sambrook J, Fritsch E, Maniatis T, editors. Molecular Cloning, A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.D’aquila R, Bechtel L, Videler J, Eron J, Gorczyca I, Kaplan J. Nucleic Acids Res. 1991;19:3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamoun S, Klucher K, Coffey M, Tyler B. Mol Plant–Microbe Interact. 1993;6:573–581. doi: 10.1094/mpmi-6-573. [DOI] [PubMed] [Google Scholar]
- 34.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 35.Church G, Keiffer-Higgins S. Science. 1988;240:184–188. doi: 10.1126/science.3353714. [DOI] [PubMed] [Google Scholar]
- 36.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.SAS Institute. (1996) Statistical Analysis Systems, Version 6.11.
- 38.Parker J, Coleman M. Trends Biol Sci. 1997;22:291–296. doi: 10.1016/s0968-0004(97)01089-x. [DOI] [PubMed] [Google Scholar]
- 39.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 40.Delaney T, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]