Abstract
Aggregation plays an integral role in multivalent protein-carbohydrate interactions, in Alzheimer’s and other amyloid-related diseases, and in infection response. Efforts have been made to apply controlled-aggregation in toxin sensors. We have developed a label-free intrinsic fluorescence lifetime assay that uniquely can monitor aggregation processes in real time without interference from precipitation. Fluorescence decay curves were measured with high precision at one-second time intervals following addition of a glycodendrimer to a lectin-containing solution. Changes in the fluorescence intensity and lifetime signified formation of complexes. However, these changes are not associated with the initial lectin-sugar binding events. Rather, they appear to be caused by clustering and a subsequent conformational rearrangement of the lectins. Studies were conducted with mannose-functionalized PAMAM dendrimers of the second through the sixth generations with Concanavalin A. The apparent rate constant, when expressed on a per mannose basis, increased with dendrimer generation, particularly in going from the fourth to the sixth generation. However, the identical fluorescence decay waveforms for saturating amounts of dendrimer suggest that all glycodendrimer generations studied reach a comparable state of aggregation. Although self-quenching of tryptophan resonances that was induced by clustering was monitored in this study, the reported method is not limited to such and is viable for numerous binding studies.
Aggregation plays an integral role in many cellular pathways, one of the most important being mediating the infection and proliferation potential of tumors and pathogens.1 Protein aggregation has also been implicated in pathological conditions such as Alzheimer’s and other amyloid-related diseases.2 Owing to the importance of multivalently displayed carbohydrates on cell surfaces, sugar-induced aggregation has drawn considerable attention.3,4 Sensor strategies based on controlled aggregation have been reported for the detection of toxins and other biologically relevant compounds.5 Multivalent interactions often involve multiple weak monovalent binding events. An in-depth understanding of aggregation in complex systems requires studies that go beyond measuring the monovalent association constants. Particularly valuable would be methods capable of characterizing aggregation events in real time.
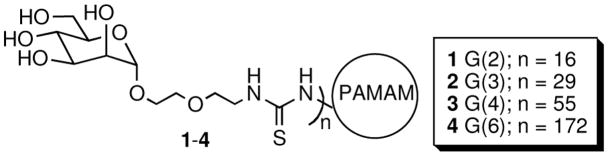
We present here important new information about the aggregation of the mannose-specific lectin Concanavalin A (Con A) by glycodendrimers 1-4 (Figure 1, prepared as described in ref. 6). Glycodendrimers are very well suited for studying the formation and mediation of multivalent interactions. We have reported that binding and inhibition efficacies depend on size and functionalization in several previous studies on the Con A-glycodendrimer system.6 In this case, we apply Con A intrinsic fluorescence in a novel time-domain format.
Figure 1.
Mannose-functionalized PAMAM dendrimers 1–4.
Intrinsic fluorescence is attractive for its label-free aspects, but light scattering and inner filter effects associated with the extensive precipitation that often accompanies aggregation hampers steady-state approaches. Fluorescence lifetime approaches are often stated as immune to the precipitation problems, because the desired information can be extracted from the shape of the decay curve rather than the intensity.7,8 However, conventional lifetime technology is too slow for our needs because we are studying reactions with half-lives as short as just a few seconds. The data reported here were collected with a prototype instrument that increases the rate at which fluorescence lifetime data can be collected by approximately a factor of 100. (See supporting material for details.)
The experimental configuration is straightforward. Known amounts of glycodendrimer solution were added to 2000 μL of 100 μg/mL Con A in a well-stirred cuvette at 25 °C. The baseline of Con A fluorescence (295 nm excitation wavelength, 335 nm emission wavelength, 5 nm bandpass) was established for 30 seconds before the glycodendrimer aliquot was added. Fluorescence decay waveforms were measured once per second for the next 130 to 220 seconds.
The term “fluorescence lifetime” is used for convenience in this Communication. However, we are not following the usual practice of reducing each raw decay curve (“waveform”) to a set of lifetimes and amplitude factors via iterative reconvolution. As explained in the supplementary material, we instead fit the waveforms as a linear combination of free and complexed Con A basis waveforms. (Figures 2 S3) In essence the fluorescence decay waveforms are treated as spectra, in which the independent variable is time after excitation instead of wavelength. It should also be noted that the complex formation and precipitation has minimal effect on the wavelength spectral distribution of the Con A fluorescence.
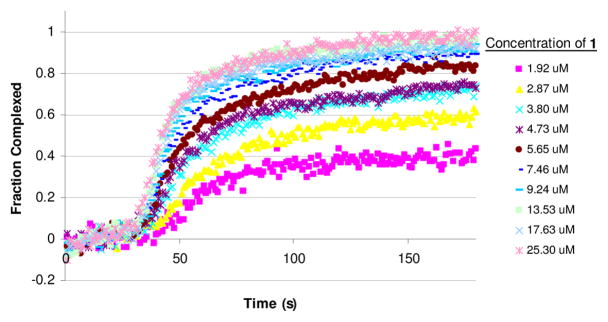
Figure 2.
Complex formation upon addition of 1 to Con A (100 ug/mL).
Control experiments showed that the primary sugar-Con A binding events are not the source of the observed fluorescence changes. Several mM concentration of α O-methyl mannoside (Me-man) (Figures S4 and S10) had minimal effect on the Con A fluorescence and did not result in precipitate formation. To ensure that the dendrimer framework itself is not the source of the Con A quenching, a galactose-functionalized dendrimer that does not bind to Con A was added under the same conditions as the mannose-functionalized dendrimers 1–4. Again, no precipitation was observed visually, and non-specific binding was minimal compared to baseline drift and noise (Figures S5 and S10).
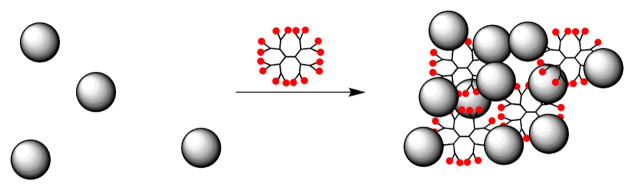
The fluorescence changes are most likely associated with a Con A-Con A protein-protein interaction orchestrated by binding to the glycodendrimer framework. Figure 3 illustrates our hypothesis that a reversible protein-protein interaction between proximal Con A lectins causes the quenching. The primary role of the dendrimers is to hold the Con A lectins in close proximity, thereby increasing their effective concentration. A protein-protein interaction, presumably a conformational change affecting the environment of one of more Con A tryptophans, can then occur.
Figure 3.
Glycodendrimer-mediated lectin aggregation. Left: uncomplexed Con A. Right: cross-linked state.
All generations of glycodendrimer showed saturation behavior, i.e., the fluorescence changes reached a plateau at sufficiently high dendrimer concentration. Furthermore, the fluorescence decay curves at saturation were identical to within experimental uncertainty for all dendrimer generations. Thus, all fluorescence decay curves for the entire range of dendrimer concentrations and all generations, could be fit to a linear combination of just two waveforms, which correspond to the “free” and “complexed” Con A. We conclude that the environment of the complexed Con A is apparently not affected by the generation of dendrimer. (Figure S16).
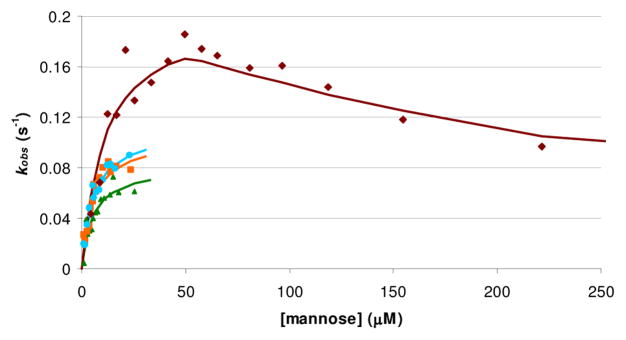
However, the rate of complex formation is sensitive to the dendrimer generation and concentration. The kinetics traces (Figures S14–S15) often appeared bi-phasic with the much faster first step generally accounting for >90% of the total change. Except for very low dendrimer concentrations, a first-order kinetic model fit the data for the first 70 seconds after dendrimer addition very well and yielded an apparent rate constant kobs (Figure 4 and Figures S14–15).
Figure 4.
Representative kobs for Con-A complex formation. 1 ( ), 2 (
), 2 ( ), 3 (
), 3 ( ), 4 (
), 4 ( ).
).
Although the mechanism is undoubtedly much more complicated than a simple first-order process, kobs is a convenient way to depict how differences in dendrimer generation affect glycodendrimer mediated protein aggregation. When comparing results across dendrimer generations, the apparent rate constant increases with increasing dendrimer generation expressed on a per mannose basis. At low concentration of added dendrimer, there is a fixed pool of Con A lectins competing for a small number of glycodendrimers. The rate limiting step in achieving a complex that changes the fluorescence decay waveform may be two Con A lectins becoming cross linked into close proximity by a small number of glycodendrimers. Thus, the overall apparent rate constant increases as the concentration of glycodendrimer increases. As expected, the rate constant approaches a saturation value at sufficiently high dendrimer concentration at which the rate-limiting step becomes the Con A-Con A mutual conformational change.
The multivalent binding in 4 boosts the kinetics over what is observed for the lower generation dendrimers because it also serves to increase the Con A residence time on a sugar binding site. However, at very high dendrimer concentration, we see a slowing of the kinetics for very high dendrimer concentration in G6. The slower kinetics at higher G6 concentration can be understood as a consequence of some fraction of the Con A lectins initially being too far apart to undergo the protein-protein interaction (each Con A has more sugar binding sites to choose from) and the multivalent binding impeding their ability to move to new sites.
Because the same final degree of complex formation is achieved, we suggest there is a slower process by which the Con A’s migrate into adjacent positions on the dendrimer framework, which is the most stable configuration. Brewer and coworkers have proposed a model in which lectin molecules bind and jump from carbohydrate epitope to epitope.9 We are proposing an attractive interaction between Con A molecules on adjacent glycodendrimer binding sites such that their residence time substantially increases. Note that once the entropic penalty of Con A-sugar binding has been paid, only a small enthalpic stabilizing force is necessary for the protein-protein interaction. It is also interesting to speculate on a possible connection between our observations and the selectivity switching reported by the Whitesides group.10
In summary, fast and precise fluorescence lifetime experiments were performed using unlabeled lectin to characterize glycodendrimer-mediated protein aggregation. Lifetime measurements were used in these experiments to explain self-quenching phenomena induced by aggregation states, but this method is not limited to such and is viable for numerous binding studies. For example, intrinsic fluorescence can be used for the study of protein-protein interactions, protein-small molecule interactions, vesicle and micelle formation, oligomerization events, and protein folding. Labeled compounds can be studied as well using the methods described here.
Supplementary Material
Acknowledgments
Support from NIGMS R01 62444 and the Montana Board of Research and Commercialization Technology (Grant 08-48) is gratefully acknowledged.
Footnotes
Supporting Information Available: Methods, description of instrumentation, and fluorescence data for methyl mannose, galactose-functionalized dendrimers, and 1–4 with Con A. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nangia-Makker P, Balan V, Raz A. Cancer Micro. 2008;1:43–51. doi: 10.1007/s12307-008-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood SJ, Maleeff B, Hart T, Wetzel R. J Mol Biol. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 3.(a) Gorelik E, Galili U, Raz A. Cancer Metastasis Reviews. 2001;20:245–277. doi: 10.1023/a:1015535427597. [DOI] [PubMed] [Google Scholar]; (b) Rudd PM, Wormald MR, Dwek RA. TRENDS in Biotech. 2004;22:524–530. doi: 10.1016/j.tibtech.2004.07.012. [DOI] [PubMed] [Google Scholar]; (c) Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed Engl. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; (d) Lee RT, Lee YC. Glycoconjugate J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 4.(a) Segal DM, Weiner GJ, Weiner LM. Curr Opin Immunol. 1999;11:558–562. doi: 10.1016/s0952-7915(99)00015-1. [DOI] [PubMed] [Google Scholar]; (b) Singh RS, Tiwary AK, Kennedy JF. Crit Rev Biotechnol. 1999;19:145–178. [Google Scholar]; (c) Gestwicki JE, Strong LE, Cairo CW, Boehm FJ, Kiessling LL. Chem Biol. 2002;9:163–169. doi: 10.1016/s1074-5521(02)00102-3. [DOI] [PubMed] [Google Scholar]
- 5.(a) Swager T, Satrijo A. J Am Chem Soc. 2008;129:16020–16028. doi: 10.1021/ja075573r. [DOI] [PubMed] [Google Scholar]; (b) Maynor MS, Nelson TL, O’Sullivan c, Lavigne JJ. Org Lett. 2007;9:3217–3220. doi: 10.1021/ol071065a. [DOI] [PubMed] [Google Scholar]; (c) Liu B, Pu K. Macromolecules. 2008;41:6636–6640. [Google Scholar]; (d) Wang M, Zhang D, Zhang G, Tang Y, Wang S, Zhu D. Anal Chem. 2008;80:6443–6448. doi: 10.1021/ac801020v. [DOI] [PubMed] [Google Scholar]; (e) Chen K, Yang S, Hwang C, Fang J. Org Lett. 2008;10:4401–4404. doi: 10.1021/ol8014418. [DOI] [PubMed] [Google Scholar]
- 6.(a) Woller EK, Walter ED, Morgan JR, Singel DJ, Cloninger MJ. J Am Chem Soc. 2003;125:8820–8826. doi: 10.1021/ja0352496. [DOI] [PubMed] [Google Scholar]; (b) Woller EK, Cloninger MJ. Org Lett. 2002;4:7–10. doi: 10.1021/ol016568+. [DOI] [PubMed] [Google Scholar]; (c) Wolfenden ML, Cloninger MJ. J Am Chem Soc. 2005;127:12168–12169. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]
- 7.Rao CV. Immunology: A Textbook. 1. Alpha Science Intl Ltd; Oxford, UK: 2005. p. 65. [Google Scholar]
- 8.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. New York: Kluwer Academic; 1999. p. 698. [Google Scholar]
- 9.Dam TK, Gerken TA, Brewer CF. Biochem. 2009;48:3822–3827. doi: 10.1021/bi9002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horan N, Yan L, Isobe H, Whitesides GM, Kahne D. Proc Nat Acad Sci. 1999;96:11782–11786. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.