Abstract
We have reported previously that adipose tissue-derived stem cells (ADSC) could be induced by isobutylmethylxanthine (IBMX) to differentiate into neuron-like cells and such differentiation was mediated by insulin-like growth factor I (IGF-I) signaling. In the present study we show that both IBMX and IGF-I upregulated neural markers β-III-tubulin and paired-like homeobox transcription factor (Pitx3) in ADSC. Because Pitx3 is important for the maturation and function of dopaminergic neurons and has been shown to be regulated by microRNA miR-133b, we also examined the effects of IBMX and IGF-I on miR-133b expression. The results show that both IBMX and IGF-I upregulated miR-133b expression. When miR-133b was overexpressed in ADSC through transfection of a synthetic microRNA mimic, it caused a downregulation of β-III-tubulin as evidenced by immunofluorescence staining and western blot analysis. Overexpression of miR-133b also downregulated Pitx3 and IGF-1 receptor (IGF-IR), and all such downregulation occurred at the protein but not the RNA level. The apparent posttranscriptional regulation was subsequently found to be exerted through a potential miR-133b target in the 3’-untranslated region of IGF-IR, as evidenced by luciferase assay. Thus, IGF-I signaling and miR-133b co-regulate potential neural differentiation of ADSC through a feedback mechanism, in which IGF-I upregulates miR-133b while miR-133b in turn downregulates IGF-IR.
Keywords: adipose tissue-derived stem cells, neuron-like differentiation, IGF-I, IBMX, MicroRNA, miR-133b
Introduction
Adipose tissue-derived stem cells (ADSC) originate from the vasculature of adipose tissue and are comparable to bone marrow stem cells (BMSC) in differentiation potential (Lin et al., 2008; Lin et al., 2008). Several studies including ours have shown that ADSC could be induced to undergo neuron-like differentiation in vitro (Safford et al., 2002; Ashjian et al., 2003; Ning et al., 2006; Ning et al., 2008). Specifically, we showed that treatment of ADSC with isobutylmethylxanthine (IBMX) led to neuron-like morphological changes and increased expression of neuronal proteins (Ning et al., 2006). Furthermore, we showed that this neuron-like differentiation was mediated by the insulin growth factor (IGF-I) signaling pathway (Ning et al., 2008). Whether this IBMX-inducible, IGF-I-mediated neuron-like differentiation of ADSC is regulated by microRNAs (miRNAs) is the subject of the present study.
During animal development, multiple genes must be expressed coordinately at precise levels both spatially and temporally. Recent studies have shown that such coordinated gene expression is in part regulated by miRNAs, which are non-protein coding, single-stranded RNAs of ~23 nucleotides in length (Sun and Tsao, 2008). These small RNAs destabilize target mRNAs or suppress their translation by binding to complementary sequences in the 3’ untranslated regions (3’-UTRs) (Bartel, 2009). To date, there are more than 500 experimentally verified human miRNAs (Griffiths-Jones et al., 2008). While most of these miRNAs are of unknown function, some are known to be specifically expressed in developing and mature nervous systems (Gao, 2008). For example, miR-124a, which is 100% conserved at the nucleotide level from worms to humans and is expressed throughout the embryonic and adult central nervous systems of different species, has been shown to have a role in the suppression of non-neural, particularly “anti-neural”, genes (Lim et al., 2005; Visvanathan et al., 2007). More recently, miR-124a was shown to promote neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing (Makeyev et al., 2007). Another miRNA, miR-133b, has recently been shown to regulate the maturation and function of midbrain dopaminergic neurons within a negative feedback circuit that includes the paired-like homeodomain transcription factor Pitx3 (Kim et al., 2007).
miRNAs have also been shown to play important roles in regulating stem cell self-renewal and differentiation by repressing the translation of selected mRNAs in stem cells and differentiating daughter cells (Gangaraju and Lin, 2009). Particularly, miR-124a and miR-9 have been found to affect neural differentiation of embryonic stem cells (Krichevsky et al., 2006). In addition, miRNA-26a has been shown to modulate late osteoblasts differentiation of human ADSC by targeting the SMAD1 transcription factor (Luzi et al., 2008). In the present study we show that IBMX-induced neuron-like differentiation of ADSC was regulated by miR-133b within a negative feedback circuit that includes IGF-IR and Pitx3.
MATERIALS AND METHODS
ADSC Isolation and culture
Rat adipose tissue was excised from the inguinal region of male Sprague-Dawley rats, as approved by the Institutional Animal Care and Use Committee. These tissue samples were processed for ADSC isolation as described previously (Ning et al., 2006; Ning et al., 2008). Briefly, the tissue was rinsed with phosphate-buffered saline (PBS) containing 1% penicillin and streptomycin, minced into small pieces, and then incubated in a solution containing 0.075% collagenase type IA (Sigma-Aldrich, St. Louis, MO, USA) for 1 hour at 37°C with vigorous shake. The top lipid layer was removed and the remaining liquid portion was centrifuged at 220×g for 10 minutes at room temperature. The pellet was treated with 160 mM NH4Cl for 10 minutes to lyse red blood cells. The remaining cells were suspended in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), filtered through a 40-µm cell strainer (BD Biosciences, Bedford, MA, USA), and plated at a density of 1 ×106 cells in a 10-cm dish. After reaching 80% confluence, the cells were harvested and stored in liquid nitrogen at a density of 5 × 105 cells per milliliter of freezing media (DMEM, 20% FBS, and 10% dimethylsulfoxide [DMSO]). Cells were thawed and cultured as needed in DMEM supplemented with 10% FBS, 1% nonessential amino acid, 10,000 units/mL penicillin, 10,000 mcg/mL streptomycin SO4, 0.025 mg/mL fungizone, and 110 mg/mL sodium pyruvate. Culture incubator was set at 37°C with 5% CO2. Cells in passages four to eight were used in this study.
Treatment with IBMX, IGF-I, picropodophyllin (PPP), and miRNAs
For treatment with IBMX, ADSC were seeded in 6-well plates or 10-cm dishes at 40–60% confluence in DMEM containing 10% FBS. The next day, the cells were washed 3 times with PBS and the medium changed to DMEM supplemented with 500 µM of IBMX (Sigma-Aldrich, St. Louis, MO). For treatment with IGF-I in the presence or absence of PPP, ADSC were seeded in 6-well plates or 10-cm dishes at 70% confluence in DMEM containing 10% FBS. The next day, the medium was changed to in DMEM containing 1% FBS. Twenty-four h later, the cells were treated with or without 2.5 µM of PPP (cat. # 407247, EMD Biosciences, La Jolla, CA) for 1 h and then 100 ng/ml of IGF-I (cat. # 4121-20, Biovision, Mountain View, CA) for another 3 h. For treatment with miRNAs, ADSC were seeded into 6-well plates or 10-cm dishes at 70% confluence in DMEM containing 10% FBS. The next day, the cells were transfected with 50 nM of miR-133b, miR-124a (cat. AM17100, Ambion Inc, Austin, TX) or miRNA negative control (cat. # AM17120, Ambion) using HiPerfect transfection reagent (cat. # 301705, Qiagen, Valencia, CA). Forty-eight h later, the medium was changed to DMEM containing 1% FBS. Another 24 h later, the cells were treated with or without 2.5 µM of PPP for 1 h and then 100 ng/ml of IGF-I or 500 µM of IBMX for another 3 h. Each experiment was repeated three times.
RT-PCR and Real-time PCR
Primers for these experiments are listed in Table 1. RT-PCR was performed for the detection of IGF-IR and β-III-tubulin mRNA as previously described (Lin et al., 2002). Real-time PCR was performed for the quantification of Pitx3 mRNA as following. The reaction consisted of 4 µl cDNA, 4 µl primers (500 nM), 2 µl H2O and 10 µl SYBR Green PCR master mixture and was run in ABI7300 PCR System (Applied Biosystems, Foster City, CA) with settings of 1 cycle at 95 °C for 10 min, 1 cycle at 95 °C for 15 s and 60 °C for 1 min, 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 95 °C for 15 s. Real-time PCR was also performed for the quantification of miR-133b as following. MicroRNAs were isolated with an isolation kit (cat. # AM1561, Ambion) and reverse transcribed into cDNA using a reverse transcription kit (cat. # RA620A-1, System Biosciences Inc. [SBI], Mountain View, CA). The PCR reaction was composed of 1 µl cDNA, 1 µl of forward primer of miR-133b (cat. CSRA640B-1, SBI) or U6 (internal control), 0.5 µl of 3’-Universal reverse primer, 10 µl of H2O and 12.5 µl of SYBR® Green powerful PCR master mixture. The reaction was run in ABI 7300 PCR System (Applied Biosystems, Foster City, California) with settings of 1 cycle at 95 °C for 10 minutes, 1 cycle at 95 °C for 15 seconds and at 60 °C for 1 minute, and 40 cycles at 95 °C for 15 seconds, 60 °C for 30 seconds and 95 °C for 15 seconds. Each experiment was repeated three times.
Table 1.
Primer sequences
| Gene | Primer sequence | |
|---|---|---|
| RT-PCR | IGF-I R | F: ACATTCGCACCAACGCTTCAGTTC |
| R: ATCACCACCGCAGACTTCTGTCTT | ||
| β-III tubulin | F: AGTACCCTGACCGCATCATGAACA | |
| R: TGGTAGCAGACACAAGGTGGTTGA | ||
| β-actin | F: TCTACAATGAGCTGCGTGTG | |
| R: AATGTCACGCACGATTTCCC | ||
| Real time PCR | Pitx3 | F: ATTGTCAGATGCAGGCACTCCAC |
| R: TTCTTCTTCAGGGAGCCATCCTCA | ||
| GAPDH | F: ATGATTCTACCCACGGCAAG | |
| R: CTGGAAGATGGTGATGGGTT |
Construction of miRNA target reporter vectors
Nucleotide sequence of the 3’-untranslated region (3’-UTR) of rat IGF-IR was retrieved from GenBank (Accession # NM_052807). The sequence was uploaded to the RegRNA website (http://regrna.mbc.nctu.edu.tw/html/prediction.html) for the identification of potential miRNA target sites. One site was identified and its sequence was used to design oligonucleotides for the construction of reporter vectors. The oligonucleotides of the wildtype version were: sense, 5’-ctagtagatcttgtgtgctcactcggtgggcggaggggggagcaggttgtaacaaa; antisense, 5’-agcttttgttacaacctgctcccccctccgcccaccgagtgagcacacaagatcta. The oligonucleotides of the mutant version (as negative control) were: sense, 5’-ctagtagatcttgtgtgctcactcggtgggcggagggggggttgtaacaaa; antisense, 5’-agcttttgttacaaccccccctccgcccaccgagtgagcacacaagatcta. The complementary oligonucleotides were annealed and inserted between the HindIII and SpeI restriction sites of pMIR-Report plasmid (cat. # AM5795, Ambion). The resultant vectors were designated pMIR-IGF1R-3’-UTR and pMIR-IGF1R-3’-UTR-M for the wildtype and mutant, respectively.
Luciferase assay
We have previously used luciferase assay to analyze gene promoter activities (Lin et al., 2002). In the present study we employed the same technique to determine whether miR-133b suppressed IGF-IR expression through binding to the predicted binding site in the 3’-UTR. To accomplish this goal, we conducted two types of experiments, as described in the following. In the first set of experiments, miR-133b was introduced into the system by transfection. Briefly, COS7 cells (Lin et al., 2002) were seeded into 12-well plates in DMEM at 70% confluence. The next day, cells were transfected with 200 ng of pMIR-IGF1R-3’UTR (or pMIR-IGF1R-3’UTR-M), 30 nM of miR-133b (or miR-negative control), and 100 ng of pRL-TK (internal control for transfection efficiency, Promega, Inc., Madison, WI), using Superfectin transfection reagent (Qiagen). Forty-eight h later, the transfected cells were assayed for firefly luciferase activity (pMIR-IGF1R-3’UTR activity) and Renilla luciferase activity (pRL-TK activity) with a Dual-Luciferase Reporter Assay System and TD-20/20 luminometer (Turner BioSystems, Sunnyvale, CA). In the second set of experiments, miR-133b was introduced into the system by treatment of ADSC with IBMX or IGF-I. Briefly, ADSC were seeded into 12-well plates at 70% confluence. The next day, the cells were transfected with 200 ng of pMIR-IGF1R-3’UTR and 100 ng pRL-TK with Superfectin. Forty-eight h later, the cells were treated with 100 ng/ml of IGF-I or 500 µM of IBMX for 3 h. Thereafter, the cells were assayed for luciferase activity as described above. Each experiment was repeated three times.
Immunofluorescence staining
ADSC were seeded into 6-well plates at 70% confluence and transfected the next day with precursor molecules of miR-124a, miR-133b (Cat. # AM17100, Ambion) or miR-negative control (Cat. # AM17110, Ambion) using HiPerfect transfection reagent (Cat. #301705, Qiagen). Forty-eight hours later, the cells were induced with 500 µM IBMX for 3 hours. The cells were then fixed with ice-cold methanol for 8 min, permeabilized with 0.05% Triton X-100 for 5 min, and blocked with 5% normal horse serum in PBS for 1 hr at room temperature. The cells were then incubated with an anti-β-III-tubulin antibody (ab14545, Abcam, Cambridge, MA) for 1 hr at room temperature. After three washes with PBS, the cells were incubated with a secondary antibody (FITC-conjugated goat anti-mouse or anti-rabbit IgG) for 1 hr at room temperature. After three washes with PBS, the cells were further stained with 40,6-diamidino-2-phenylindole (DAPI, for nuclear staining) for 5 min, examined under a fluorescence microscope, and photographed.
Western blot analysis
Cells were seeded in 10-cm dishes and transfected and induced as above. The cells were then lysed in a lysis buffer containing 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, aprotinin (10 µg/ml), leupeptin (10 µg/ml), and PBS. Cell lysates containing 20 µg of protein were electrophoresed in SDS-PAGE and then transferred onto a PVDF membrane (Millipore Corp., Bedford, MA). The membrane was stained with Ponceau S to verify the integrity of the transferred proteins and to monitor the unbiased transfer of all protein samples. Detection of protein on the membrane was performed with the ECL kit (Amersham Life Sciences Inc., Arlington Heights, IL) using anti-β-III-tubulin, anti-IGF-IR (#3027, Cell signaling, Danvers, MA), anti-pitx3 (ab30734, Abcam, Cambridge, MA), or anti-β-actin antibody (A5441, Sigma-Aldrich). Before re-probing with anti-β-actin antibody, the membrane was stripped in 62.5 mM Tris-HCl, pH 6.7, 2% SDS, 10 mM 2-mercaptoethanol at 56 °C for 30 min and then washed four times in 1×TBST.
Statistical analysis
All the statistical data were analyzed with Prism 4 (GraphPad Software Inc., San Diego, CA). T-test was used to compare each pair of treatment. P value <0.05 was considered significant.
RESULTS
IGF-I upregulates β-III-tubulin expression
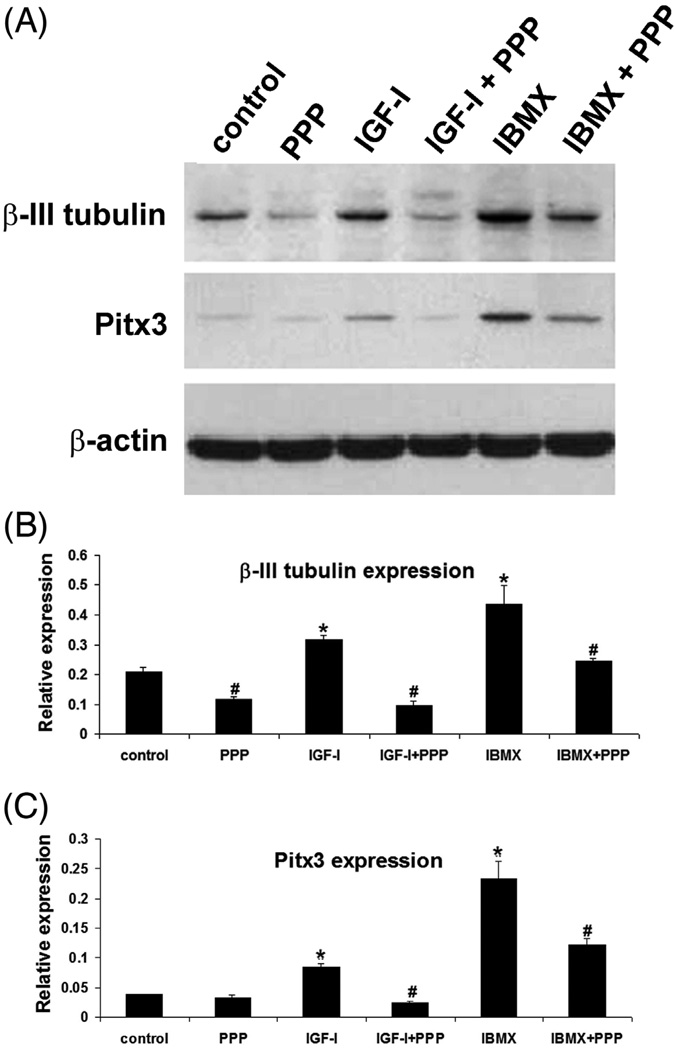
We have shown previously that ADSC treated with 500 µM IBMX assumed a neuronal morphology and expressed higher levels of neural markers such as β-III-tubulin (Ning et al., 2006). We have also shown that this neuron-like differentiation could be suppressed by IGF-IR inhibitor PPP (Ning et al., 2008). In the present study, we confirmed the effects of IBMX and PPP (Fig. 1). More importantly, we for the first time show that IGF-I also induced higher levels of β-III-tubulin expression, and this was suppressed by PPP as well (Fig. 1).
Figure 1. IGF and IBMX upregulate β-III-tubulin and Pitx3 expression.
ADSC were treated with the indicated reagents at top and then analyzed by western blotting for β-III-tubulin and Pitx3 expression, with β-actin serving as control. The experiment was repeated 3 times and each protein band analyzed by densitometry. Statistical analysis of the densitometric data is shown in panels B and C. “Relative expression” denotes ratio of β-III-tubulin or Pitx3 expression versus β-actin expression. * indicates significant difference (P <0.05) when compared to control. # indicates significant difference (P <0.05) when compared to corresponding treatment without PPP.
IGF-I and IBMX upregulate Pitx3 expression
One of the proposed applications for stem cells-derived neurons is treatment of Parkinson’s disease (PD) (Rosser et al., 2007). Toward this goal several studies have shown that stem cells-derived neurons expressed paired-like homeobox transcription factor Pitx3, which is critical for the maturation of dopaminergic neurons (Correia et al., 2007; Kim et al., 2007; Trzaska et al., 2007). In the present study, we observed that both IBMX and IGF-I upregulated Pitx3 expression in ADSC and PPP suppressed both (Fig. 1).
IGF-I and IBMX upregulate miR-133b expression
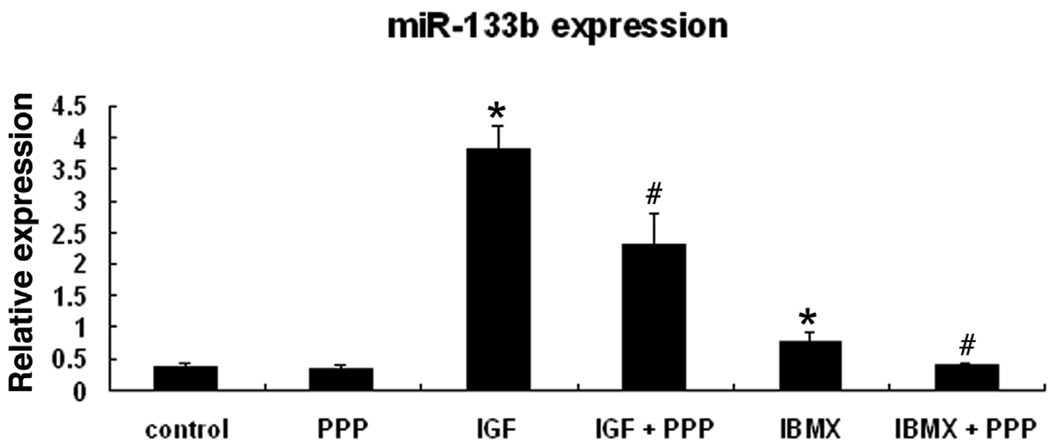
Kim et al (Kim et al., 2007) have shown that the maturation and function of dopaminergic neurons were regulated by miRNA miR-133b within a feedback circuit that includes Pitx3. Specifically, the authors found that miR-133b was expressed at lower levels in the midbrain of PD patient when compared to the midbrain of healthy individuals. In the present study we found that miR-133b was significantly upregulated in IBMX-treated or IGF-I-treated ADSC and the upregulation was suppressed by PPP (Fig. 2).
Figure 2. IGF and IBMX upregulate miR-133b expression.
ADSC were treated with the indicated reagents at bottom and then analyzed by real-time PCR for miR-133b expression, which is presented as a ratio (relative expression) versus U6 RNA expression, based on 3 independent experiments. * indicates significant difference (P <0.05) compared to control. # indicates significant difference compared to corresponding treatment without PPP.
Overexpression of miR-133b downregulates neuron-like differentiation
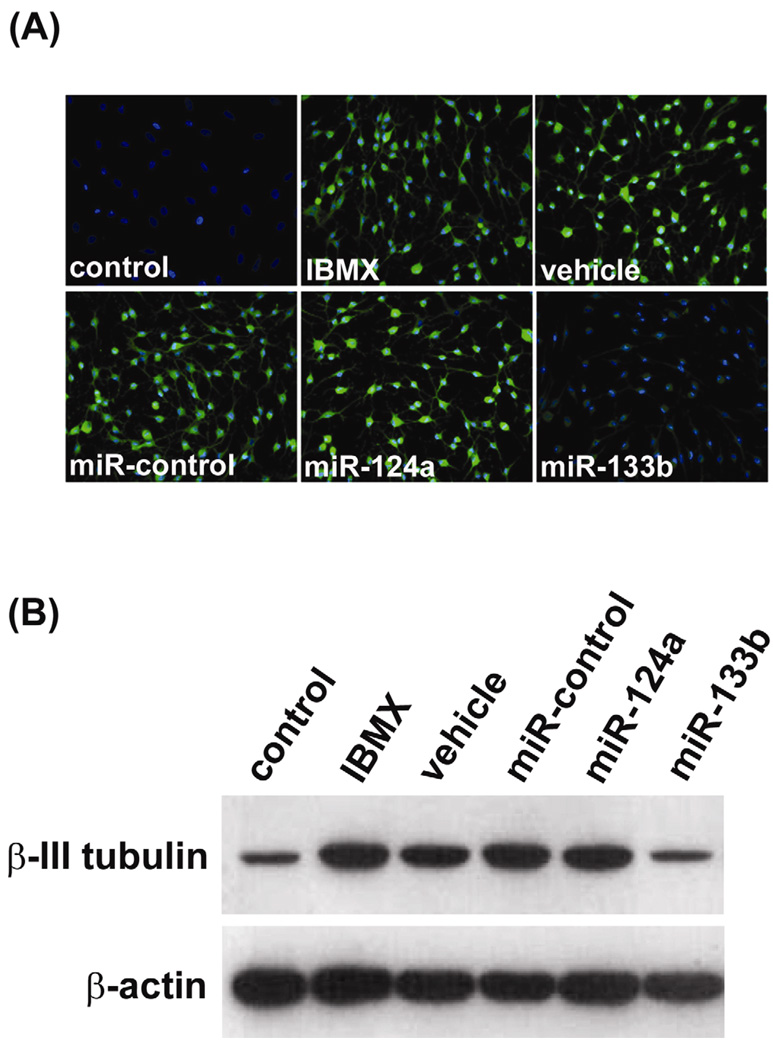
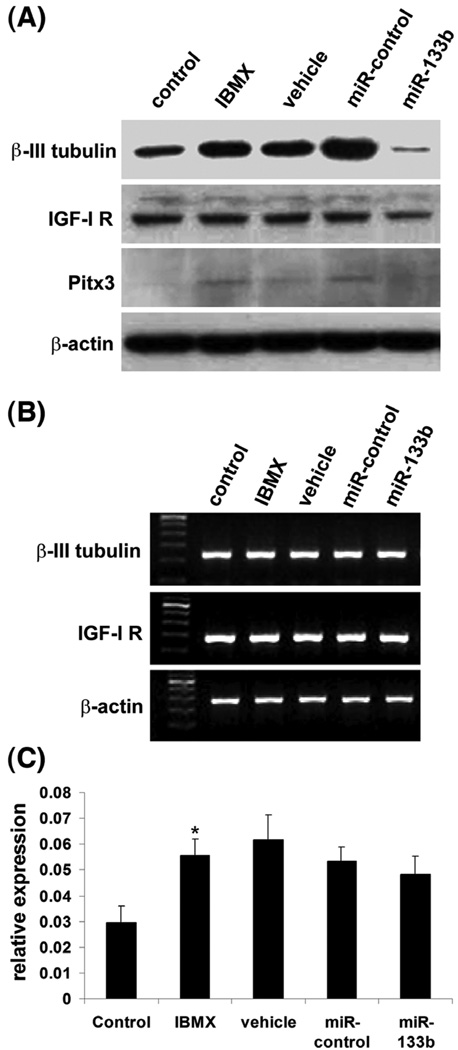
Kim et al (Kim et al., 2007) have also shown that overexpression of miR-133b suppressed Pitx3 activity as well as the maturation/function of dopaminergic neurons. In the present study we examined the effects of miR-133b overexpression on several aspects of ADSC’s neuron-like differentiation. First, we found that transfection of miR-133b suppressed the IBMX-induced upregulation of β-III-tubulin (Fig. 3). This suppressive effect of miR-133 was specific because neither a negative control miRNA nor miR-124a (important for neuronal differentiation (Makeyev et al., 2007; Visvanathan et al., 2007)) exhibited such effect (Fig. 3). Second, we found that transfection of miR-133b downregulated not only β-III-tubulin but also Pitx3 and IGF-IR (Fig. 4A). Third, we found that downregulation of β-III-tubulin, Pitx3 and IGF-IR by miR-133b occurred at the protein but not the mRNA level (Fig. 4B&C).
Figure 3. miR-133b overexpression downregulates β III-tubulin expression.
(A) ADSC were uninduced (control) or induced with IBMX after being transfected with the indicated miRNAs. “Vehicle” denotes transfection reagent. The cells were then analyzed by immunofluorescence staining for β-III-tubulin expression (green). Cell nuclei were identified by DAPI staining (blue). Repeated 3 times. (B) ADSC were uninduced (control) or induced with IBMX after being transfected with the indicated miRNAs. “Vehicle” denotes transfection reagent. The cells were then analyzed by western blotting for β-III-tubulin expression, with β- actin serving as control. Repeated 3 times.
Figure 4. miR-133b overexpression downregulates β-III-tubulin, IGF-IR, and Pitx3 protein expression.
(A) ADSC were uninduced (control) or induced with IBMX after being transfected with the indicated miRNAs. “Vehicle” denotes transfection reagent. The cells were then analyzed by western blotting for β-III-tubulin, IGF-IR and Pitx3 expression, with β-actin serving as control. Repeated 3 times. (B) ADSC were uninduced (control) or induced with IBMX after being transfected with the indicated miRNAs. “Vehicle” denotes transfection reagent. The cells were then analyzed by RT-PCR for β-III-tubulin and IGF-IR expression, with β-actin serving as control. Repeated 3 times. (C) ADSC were uninduced (control) or induced with IBMX after being transfected with the indicated miRNAs. “Vehicle” denotes transfection reagent. The cells were then analyzed by real-time PCR for Pitx3 expression, which is presented as a ratio (relative expression) versus NADPH expression, based on 3 independent experiments. * indicates significant difference (P <0.05) compared to control.
IGF-IR is a miR-133b target
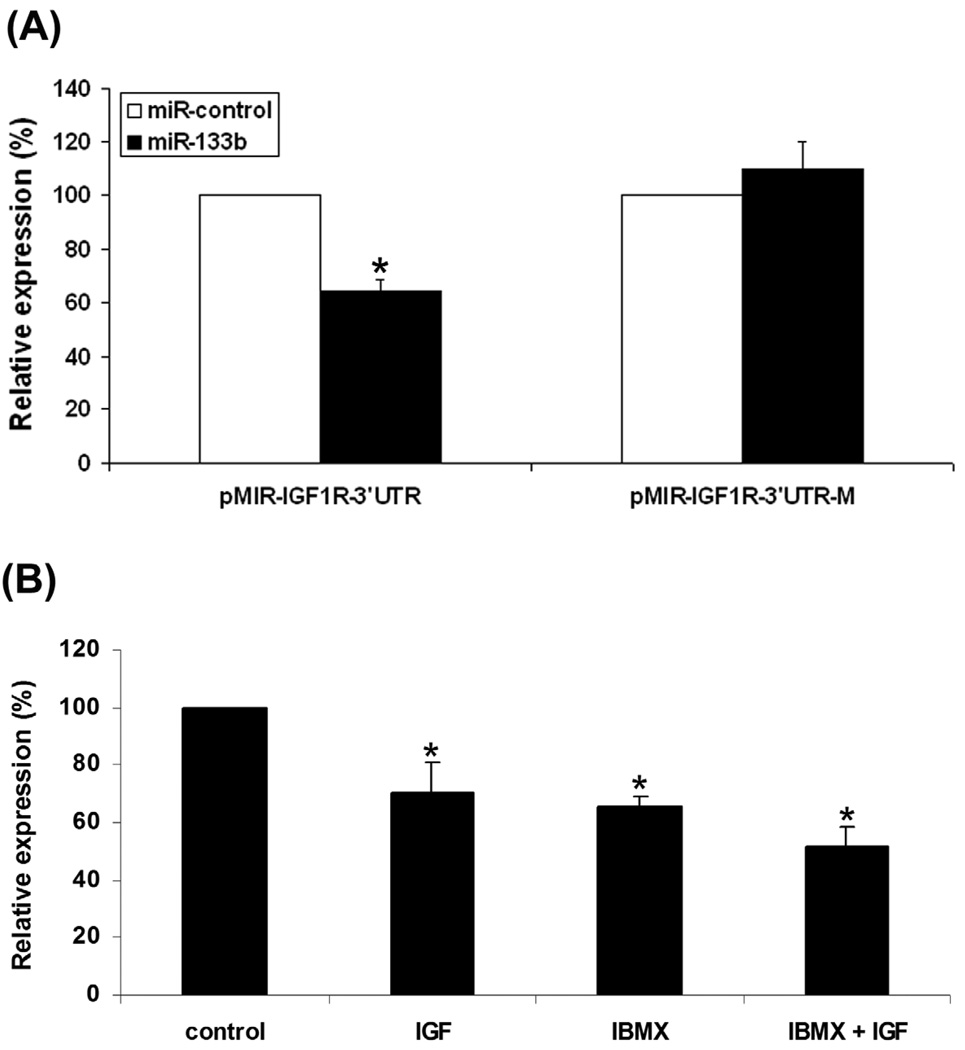
miRNAs are known to regulate the expression of numerous targets posttranscriptionally (Bartel, 2009). By using miRNA target prediction programs on the Internet (Bartel, 2009), we identified one potential miR-133b target in the 3′-untranslated region (3′ UTR) of IGF-IR. When placed downstream of a luciferase reporter gene, this target sequence, but not its mutant version, was indeed repressed by miR-133b (Fig. 5A). When ADSC transfected with the wildtype reporter construct were subjected to treatment with IGF-I or IBMX, its activity was also repressed (Fig. 5B). Thus, it appears that both IGF-I and IBMX upregulated miR-133b, and which in turn suppressed IGF-IR expression.
Figure 5. miR-133b overexpression downregulates IGF-IR posttranscriptionally.
(A) COS7 cells were transfected with luciferase reporter vectors carrying wildtype (pMIR-IGF1R-3’UTR) of mutant (pMIR-IGF1R-3’UTR-M) version of the potential miR-133b target sequence in the 3’UTR of IGF-IR gene. The cells were also transfected with miR-133b (black bar) or miRcontrol (white bar) and pRL-TK (internal control expressing Renilla luciferase activity). The translational regulatory activity of the wildtype or mutant IGF-I 3’UTR was presented as a ratio (relative expression) between Firefly and Renilla luciferase activities, based on 3 independent experiments. * indicates significant difference (P <0.05) compared to miR-control. (B) ADSC were transfected with pMIR-IGF1R-3’UTR and pRL-TK, treated with the indicated reagents at bottom, and then analyzed for Firefly and Renilla luciferase activities. The translational regulatory activity of the 3’UTR of IGF-IR gene was presented as a ratio (relative expression) between Firefly and Renilla luciferase activities, based on 3 independent experiments. * indicates significant difference (P <0.05) compared to untreated control.
DISCUSSION
Several studies including ours (Ning et al., 2006; Ning et al., 2008) have shown that ADSC could be induced to undergo neuron-like differentiation in vitro. Evidence for such differentiation is generally based on morphological changes and expression of neural markers, not functional properties such as electric conductivity. While deficient in this regard, this line of study offers an experimental system in which the underlying molecular mechanisms may overlap with those of true neuronal differentiation. Indeed, in our hands, the IBMX-induced neuron-like differentiation and its associated phenotypical changes are reproducible and mimic those identified with proven neuronal differentiation. For example, while we have reported that IBMX-induced neuron-like differentiation and its associated increase of β-III-tubulin expression were mediated by IGF-I signaling (Ning et al., 2008), a recent study confirmed that IGF-I promotes neuronal differentiation of neural stem cells with increased expression of β-III-tubulin (Choi et al., 2008). Additionally, a recent review article summarized the importance of IGF-I in brain development (D'Ercole and Ye, 2008).
In our previous study we used PPP as probe to uncover the IGF-I signaling pathway as an underlying mechanism for ADSC’s neuron-like differentiation (Ning et al., 2008). In the present study we further showed that IGF-I itself was able to induce upregulated expression of β-III-tubulin, and this was accompanied by upregulated expression of another neural markers Pitx3. The importance of Pitx3 for the maturation and function of dopaminergic neurons has been well documented (Jacobs et al., 2009). Recent studies have also demonstrated the protective effects of IGF-I on dopaminergic neurons (Ebert et al., 2008; Quesada et al., 2008). Thus, our data point out the possibility that IGF-I might assert its protective effects on dopaminergic neurons through activation of Pitx3.
The activity of Pitx3 in dopaminergic neurons has been shown to be regulated by miR-133b (Kim et al., 2007). While miR-133b expression is downregulated in PD patients, overexpression of miR-133b led to downregulation of Pitx3. Thus, the authors proposed: “miR-133b functions within a feedback loop, as Pitx3 specifically induces transcription of miR-133b, and Pitx3 activity is downregulated by miR-133b posttranscriptionally.” Consistent with this proposal, several more recent studies have uncovered similar feedback regulatory mechanisms for various miRNAs. For example, Xia et al (Xiao et al., 2008) reported that cardiac potassium current expression was regulated by miR-133b through a feedback mechanism. Likewise, Luzi et al (Luzi et al., 2008) reported that, while miR-26a was upregulated in osteogenic differentiation of ADSC, inhibition of miR-26a expression actually promoted osteogenic differentiation of ADSC. In the present study, we found that IGF-I signaling led to increased expression of miR-133b, and which in turn downregulated the expression of IGF-IR. Such a feedback regulatory mechanism may provide a safeguard against overexpression of key molecules in cellular differentiation and other biological processes, as summarized in a recent review article (Bartel, 2009).
ACKNOWLEDGEMENTS
This work was supported by grants from the Arthur Rock Foundation and the National Institutes of Health (DK64538, DK045370, and DK069655).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashjian PH, Elbarbary AS, Edmonds B, DeUgarte D, Zhu M, Zuk PA, Lorenz HP, Benhaim P, Hedrick MH. In vitro differentiation of human processed lipoaspirate cells into early neural progenitors. Plast Reconstr Surg. 2003;111:1922–1931. doi: 10.1097/01.PRS.0000055043.62589.05. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KC, Yoo DS, Cho KS, Huh PW, Kim DS, Park CK. Effect of single growth factor and growth factor combinations on differentiation of neural stem cells. J Korean Neurosurg Soc. 2008;44:375–381. doi: 10.3340/jkns.2008.44.6.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia AS, Anisimov SV, Roybon L, Li JY, Brundin P. Fibroblast growth factor-20 increases the yield of midbrain dopaminergic neurons derived from human embryonic stem cells. Front Neuroanat. 2007;1:4. doi: 10.3389/neuro.05.004.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ercole AJ, Ye P. Expanding the mind: insulin-like growth factor I and brain development. Endocrinology. 2008;149:5958–5962. doi: 10.1210/en.2008-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Beres AJ, Barber AE, Svendsen CN. Human neural progenitor cells over-expressing IGF-1 protect dopamine neurons and restore function in a rat model of Parkinson's disease. Exp Neurol. 2008;209:213–223. doi: 10.1016/j.expneurol.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin C-S, Chow S, Lau A, Tu R, Lue TF. Human PDE5A gene encodes three PDE5 isoforms from two alternate promoters. Int J Impot Res. 2002;14:15–24. doi: 10.1038/sj.ijir.3900802. [DOI] [PubMed] [Google Scholar]
- Lin CS, Xin ZC, Deng CH, Ning H, Lin G, Lue TF. Recent advances in andrology-related stem cell research. Asian J Androl. 2008;10:171–175. doi: 10.1111/j.1745-7262.2008.00389.x. [DOI] [PubMed] [Google Scholar]
- Lin G, Garcia M, Ning H, Banie L, Guo YL, Lue TF, Lin CS. Defining stem and progenitor cells within adipose tissue. Stem Cells Dev. 2008;17:1053–1063. doi: 10.1089/scd.2008.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning H, Lin G, Fandel T, Banie L, Lue TF, Lin CS. Insulin growth factor signaling mediates neuron-like differentiation of adipose tissue-derived stem cells. Differentiation. 2008;76:488–494. doi: 10.1111/j.1432-0436.2007.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning H, Lin G, Lue TF, Lin CS. Neuron-like differentiation of adipose tissue-derived stromal cells and vascular smooth muscle cells. Differentiation. 2006;74:510–518. doi: 10.1111/j.1432-0436.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- Quesada A, Lee BY, Micevych PE. PI3 kinase/Akt activation mediates estrogen and IGF-1 nigral DA neuronal neuroprotection against a unilateral rat model of Parkinson's disease. Dev Neurobiol. 2008;68:632–644. doi: 10.1002/dneu.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser AE, Zietlow R, Dunnett SB. Stem cell transplantation for neurodegenerative diseases. Curr Opin Neurol. 2007;20:688–692. doi: 10.1097/WCO.0b013e3282f132fc. [DOI] [PubMed] [Google Scholar]
- Safford KM, Hicok KC, Safford SD, Halvorsen YD, Wilkison WO, Gimble JM, Rice HE. Neurogenic differentiation of murine and human adipose-derived stromal cells. Biochem Biophys Res Commun. 2002;294:371–379. doi: 10.1016/S0006-291X(02)00469-2. [DOI] [PubMed] [Google Scholar]
- Sun BK, Tsao H. Small RNAs in development and disease. J Am Acad Dermatol. 2008;59:725–737. doi: 10.1016/j.jaad.2008.08.017. quiz 738–740. [DOI] [PubMed] [Google Scholar]
- Trzaska KA, Kuzhikandathil EV, Rameshwar P. Specification of a dopaminergic phenotype from adult human mesenchymal stem cells. Stem Cells. 2007;25:2797–2808. doi: 10.1634/stemcells.2007-0212. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Xiao J, Luo X, Lin H, Wang Z, Nattel S. Feedback remodeling of cardiac potassium current expression: a novel potential mechanism for control of repolarization reserve. Circulation. 2008;118:983–992. doi: 10.1161/CIRCULATIONAHA.107.758672. [DOI] [PubMed] [Google Scholar]