Summary
Recent models of lipid-free apolipoprotein A-I, including a cross-link/homology model and an x-ray crystal structure have identified two potential functionally relevant “patches” on the protein surface. The first is a hydrophobic surface patch composed of leucine residues 42, 44, 46, and 47 and the second a negatively charged patch composed of glutamic acid residues 179, 191, and 198. To determine if these domains play a functional role, these surface patches were disrupted by site-directed mutagenesis and the bacterially expressed mutants were compared with respect to their ability to bind lipid and stimulate ABCA1-mediated cholesterol efflux. It was found that neither patch plays a significant functional role in the ability of apoA-I to accept cholesterol in an ABCA1-dependent manner, but that the hydrophobic patch did affect the ability of apoA-I to clear DMPC liposomes. Interestingly, contrary to previous predictions, disruption of the hydrophobic surface patch enhanced the lipid binding ability of apoA-I. The hydrophobic surface patch may be important to the structural stability of the lipid binding regions of apoA-I, or may be a necessary permissive structural element for lipid binding.
Keywords: apolipoprotein A-I, ATP binding cassette transporter A1, functionality, surface patch, lipid-free model, lipid-binding
Introduction
Cardiovascular disease (CVD) is the leading cause of death in the industrialized world [1]. Numerous prospective studies have demonstrated an inverse correlation between high density lipoprotein (HDL) plasma levels and every major manifestation of CVD [2,3]. The cardioprotective effects of HDL may be due in part to its role in the reverse cholesterol transport system, the process by which excess cholesterol is packaged into HDL and transported from the periphery to the liver for excretion as bile salts [4,5]. This represents the body’s primary method of cholesterol excretion. Many studies have been performed to better understand the factors controlling the production of HDL. It has become clear that HDL biogenesis is dependent on the proper function of the membrane transport protein, ATP-binding cassette transporter A1 (ABCA1) [6-9]. The lack of functional ABCA1 leads to Tangier disease, characterized by a near absence of plasma HDL, the accumulation of cholesterol esters in the liver, spleen, lymph nodes, and macrophages, and an increased risk of coronary artery disease [8-10]. It is thought that HDL is formed when ABCA1 facilitates the efflux of phospholipids and cholesterol to lipid-free apolipoprotein A-I (apoA-I) [11], but, the details of the interaction between apoA-I and ABCA1 are unclear. Recently, Ajees, et al. published an x-ray crystallography model of full length lipid-free apoA-I [12]. This model generally resembles an earlier model of lipid-free apoA-I developed using a combination of chemical cross-linking following by mass spectroscopy and in silico sequence threading [13]. Upon analysis of the crystal structure, Ajees, et al. identified clusters of both charged and hydrophobic residues on the protein surface. The first is a hydrophobic surface patch composed of leucine residues 42, 44, 46, and 47. These residues are located at the turn between the first and second helix in the N-terminal four helix bundle in both the Ajees crystal structure and in the Silva cross-linking model. This arrangement is unusual in that such a high concentration of hydrophobic residues might be expected to be sequestered in the interior of the helical bundle. Ajees et al. suggested that the solvent exposed hydrophobic patch might be an initial lipid interaction site which triggered the structural transition of apoA-I from its compact lipid-free state to an open, lipid-bound state. The second domain is a negatively charged patch composed of glutamic acid residues 179, 191, and 198 (and possibly 183). This patch is located between the helical bundle and the C-terminal lipid-binding domain in both models, although the patch appears more localized in the crystal structure, owing to the higher state of organization of the C-terminus in the crystal. It has been speculated that the charged surface patches identified might be important for a direct interaction between apoA-I and ABCA1, which has been speculated to occur prior to the lipid transfer event [14,15]. The functional significance of these apoA-I surface patches for HDL formation is unknown.
To determine if these surface patches have functional importance with respect to lipid binding and ABCA1-mediated cholesterol efflux, we generated three point mutants of apoA-I which disrupt these sites, shown in Figure 1. The mutants apoA-I L(42,44)D and apoA-I L(46,47)D each have two of the four Leu residues comprising the putative hydrophobic patch replaced by a negatively charged Asp of relatively similar molecular volume. For functional studies of the negatively charged surface patch, the central glutamic acid (198) was changed to a lysine residue, thus separating the remaining two negatively charged residues and in effect eliminating this surface patch. We found that none of the mutations introduced in this study altered cholesterol efflux via ABCA1 from murine macrophages. Interestingly, and in contrast to previous predictions, introduction of negative charges into the hydrophobic patch actually increased the ability of apoA-I to solubilize synthetic dimyristoyl phosphotidylcholine liposomes.[12].
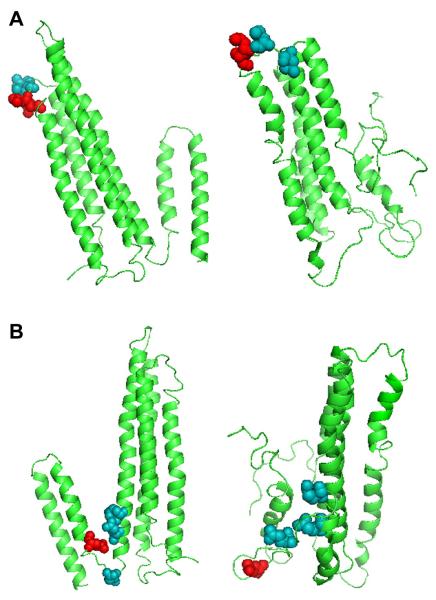
Figure 1.
A) Hydrophobic surface patch of apoA-I displayed on the X-ray crystal structure model (left) and lipid-free apoA-I homology model (right) [21][12]. The models depict residues 42 and 44, which composed mutant L42,44D as space-filled light (teal) and residues 46 and 47, which composed mutant L46,47D as space-filled dark (red). B) Negatively-charged surface patch of apoA-I displayed on X-ray crystal structure model (left) and lipid-free apoA-I homology model (right), with residue 183 included for completion [21][12]. The models depict residues 179, 183, and 191 as space-filled light (teal), and residue 198 as space-filled dark (red).
Materials and Methods
2.1 Cells and Reagents
Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Fisher (Pittsburgh, PA). IgA protease was obtained from Mobitech (Gottingen, Germany). His bind resin was obtained from Novagen (Madison, WI). Dimyristoyl-phosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids (Alabaster, AL). Dulbecco’s Modified Eagle Medium (DMEM) and fetal bovine serum (FBS) were acquired from Invitrogen (Carlsbad, CA). [1,2-3H(N)]-cholesterol was supplied by Amersham Biosciences (Piscataway, NJ). 8-bromoadenosine 3′,5′-cyclic monophosphate sodium salt (8-bromo-cAMP) and gentamicin were purchased from Sigma (St. Louis, MO). The Bis(sulfosuccinimidyl) suberate (BS3) cross-linker was obtained from Pierce (Rockford, IL). The RAW264.7 macrophages used in the cholesterol efflux assay were obtained from the American Type Culture Collection (Manassas, VA), and were maintained in DMEM supplemented with 10% FBS and 10 μg/mL gentamicin. Radiolabeling and efflux measurements were performed in DMEM supplemented with 0.2% fatty-acid free bovine serum albumin (Calbiochem,Gibbstown, NJ). All other reagents were the highest quality available.
2.2 Point mutagenesis of human apoA-I
DNA manipulation of apoA-I cDNA was performed in a PET30 construct which contains an N-terminal histidine-tag (his-tag) separated from the human apoA-I cDNA by an IgA protease cleavage site [16]. Point mutations were generated using PCR-based site-directed mutagenesis (Quick-Change, Stratagene, La Jolla, CA). Complementary primers were synthesized which matched the target sequence except at the codon they were intended to change. Additionally, all the primers were designed to add a silent restriction enzyme site to the construct for screening purposes. When completed, all mutant sequences were verified by the Cincinnati Children’s Hospital Sequencing Core on an Applied Biosystems 3730 DNA Analyzer.
2.3 ApoA-I Expression and Purification
ApoA-I was expressed and purified in Escherichia coli (E. coli) as previously described [16]. Briefly, BL-21 E. coli cells transformed with our construct of choice were grown to an absorbance of 0.7-0.8 at 600 nm. IPTG was added to a final concentration of 0.5 mM. After 2 hr, cells were pelleted, and stored dry at −80°C. Cells were lysed using probe sonication, and the apoA-I was purified using His bind resin followed by a phenyl sepharose hydrophobic interaction column (GE Healthcare). The his-tag was cleaved from the apoA-I with IgA protease, and the apoA-I was purified away from the his-tag using a Superdex 200 size exclusion chromatography column (GE Healthcare). Later sets of proteins were expressed with a tobacco etch virus cleavable his-tag. This purification protocol omitted the phenyl sepharose column, as previously described [17]. All protein samples remained in solution during the entire purification procedure and when purified were stored in standard Tris buffer (STB: 150 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.02% sodium azide). When used for a control, human plasma apoA-I was isolated as previously described [18], and stored in STB.
2.4 Far UV spectral analysis by circular dichroism
All samples were freshly dialyzed against 20 mM phosphate buffer (pH 7.4), and diluted to 100 μg/ml. All measurements were made on a Jasco J-715 spectropolarimeter. A background scan of phosphate buffer was subtracted from each sample scan. The mean residual ellipticity of each sample was calculated as described by Woody [19], and the formula of Chen et al. [20] was used to calculate the fractional helical content using the mean residual ellipticity at 222 nm. This measurement was performed on two independently prepared sets of proteins on different days. Percent helicity for each mutant was normalized to WT apoA-I for each set of proteins. Normalized trial values were averaged, and compared to WT apoA-I using an unpaired, two-tailed student’s t-test, with a p-value of 0.05 or less indicating a significant change in percent helicity compared to WT apoA-I.
2.5 Tertiary Structural Analysis Using Tryptophan Fluorescence and BS3 Cross-linking Analysis
All fluorescence measurements were performed on a Photon Technology International Quantamaster spectrometer. The four tryptophan residues in apoA-I were excited at 295 nm to minimize tyrosine fluorescence interference. The emission spectra were collected from 305-380 nm at room temperature. The protein samples were 100 μg/ml in 20 mM phosphate buffer (pH 7.4), and a phosphate buffer blank was measured and subtracted from each sample scan. Each sample was measured three times, and the wavelength of maximum fluorescence determined was the average of the maximum wavelength of each scan. We performed this measurement on two independently prepared protein sets and the average wavelength of maximum fluorescence of each mutant was compare to that of WT apoA-I using an unpaired, two-tailed student’s t-test, with a p-value of 0.05 or less indicating a significant change in tertiary structure compared to WT apoA-I.
In order to assess the self-association characteristics of each mutant compared to WT apoA-I, a BS3 cross-linking analysis was used. Each lipid-free mutant was incubated for 24 hr at 4°C with the cross-linker BS3 at a mole:mole ratio of 10:1 (BS3: protein) in phosphate buffered saline (pH=7.4). The final protein concentration was 1 mg/mL. Self-associated complexes were assessed with SDS-PAGE.
2.6 Assessing lipid binding using the DMPC clearance assay
The dimyristoyl-phosphatidylcholine (DMPC) lipid clearance assay was performed as previously described [21]. Proteins in STB were added to lipid vesicles at a mass:mass ratio of 2.5:1 (DMPC:protein) and the absorbance at 325 nm was recorded every 30 sec for 20 min on an Amersham Biosciences Ultraspec 4000 UV/visible spectrophotometer. Samples were measured three times and absorbances averaged. The solution temperature was held at 24.5°C during the duration of the assay using a temperature controlled cuvette. Although lipid binding and solubilization is a complex, multistep process, this assay is widely accepted in the field as a means of approximating the lipid-binding ability of an apolipoprotein. Two independently prepared protein sets were measured using different preparations of multilamellar vesicles. The average rate constant of each mutant was compared to that of WT apoA-I using an unpaired, two-tailed student’s t-test, with a p-value of 0.05 or less indicating a significant change in the rate of lipid binding.
2.7 ABCA1-dependent cholesterol efflux assay
RAW 264.7 macrophages were grown to 80% confluency, then incubated with media containing 1.0 μCi/mL [1,2-3H(N)]-cholesterol +/− 0.3 mM 8-bromo-cAMP. After a thorough wash, media containing a given concentration of apoA-I +/− 0.3 mM 8-bromo-cAMP was applied for 8 hr. Media alone and with 10 μg/mL of human plasma isolated apoA-I were included as experimental controls. After 8 hr the media was filtered and sampled to determine the amount of labeled cholesterol transferred to the media by scintillation counting. Three trial wells were treated with each protein sample, with the percent efflux calculated for each sample as the average of its three wells. Percent efflux was calculated by dividing the counts in the media by the total internalized counts per well. The total internalized counts were determined by dissolving the media-only treated cells with isopropanol at the end of the experiment, drying down this solution, resolubilizing the sample in toluene, and counting. Two sets of independently prepared protein samples were measured using cells prepared on separate days. The percent efflux of each mutant was normalized to WT apoA-I. In dose response experiments, the percent efflux of each mutant was normalized to the highest dose of WT apoA-I. Each mutant’s average normalized percent efflux from the two trials was compared to that of the corresponding WT apoA-I dose using an unpaired, two-tailed student’s t-test, with a p-value of 0.05 or less indicating a significant change in the ability of the mutant to stimulate cholesterol efflux compared to WT apoA-I.
Results
3.1 Structural Characterization of ApoA-I Variants
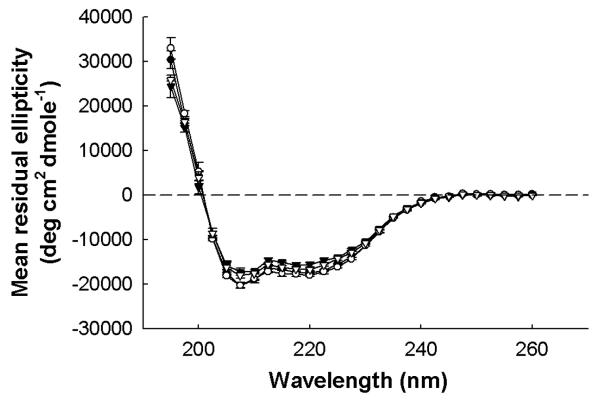
To understand the effect on secondary structure of sequence changes associated with the mutant apoA-I protein, each protein was analyzed using circular dichroism spectroscopy. As seen in Figure 2, each mutant generated a spectrum with minima in the mean residual ellipticity at 208 and 222 nm, the hallmark of a predominantly α-helical structure. Table 1 shows the calculated percent helical contents of the various mutants. Most mutants exhibited helical contents that were statistically indistinguishable from WT apoA-I, with the exception of the L46,47D mutant, which showed a slightly lower value. This suggests that disruption of these residues in the hydrophobic surface patch leads to small changes in the secondary structure of the protein. Since the difference in percent helicity and thus secondary structure of the L46,47 mutant was small compared to WT apoA-I however, this mutant was included in the functional studies discussed below. To further evaluate any structural changes associated with the mutations, the wavelength of maximum tryptophan fluorescence (WMF) was measured, as shown in Table 2. The WMF varies depending on the chemical environment of the four tryptophan residues in apoA-I. If these residues are buried within the hydrophobic core of the protein, the WMF is characteristically blue shifted to about 333 nm compared to tryptophan in an aqueous environment, which fluoresces around 355 nm [22]. All the mutants displayed a tryptophan fluorescence pattern similar to that of WT apoA-I except L46,47D, which showed a slightly higher wavelength of maximum tryptophan fluorescence, again suggesting that the average tryptophan environment in the L46,47D mutant is more exposed to the aqueous environment than WT apoA-I. Together the CD and tryptophan fluorescence data indicate that disruption of the hydrophobic patch at residues 46 and 47 leads to small changes in the lipid-free structure of apoA-I, suggesting these residues may play a minor role in the overall stability of this protein. Finally, to assess the extent of self-association of each mutant compared to WT apoA-I, each mutant was cross-linked in solution with BS3, and the extent of self-association analysed by SDS-PAGE. At 1 mg/mL, all mutants showed comparable proportions of monomer and dimer in solution as WT apoA-I (data not shown).
Figure 2.
Representative circular dichroism spectra of WT apoA-I (•), apoA-I E198K (○), apoA-I L42,44D (▼), and apoA-I L46,47D (▽). Far UV (195-260 nm) spectra of each protein at 0.1 mg/mL in 20mM phosphate buffer, pH 7.0. Every fifth data point is plotted. Points are the mean of three replicate scans from 260 to 190 nm at 100 nm/min with a 0.5 nm step size and 0.5 sec response, using a bandwidth of 1 mm and a slit width of 500 μm. Bars represent one SD.
Table 1.
Helical Content of WT and mutant apoA-I
| ApoA-I Mutant | Normalized % α-helical content1 | Standard Error |
|---|---|---|
| WT | 1.000 | 0.000 |
| E198K | 1.079 | 0.179 |
| L42,44D | 0.8443 | 0.057 |
| L46,47D | 0.9484* | 0.020 |
Average of two sets of triplicate measurements made on independent preparations of proteins. Values normalized to WT.
p<0.05 compared to WT by unpaired, 2-tailed student’s t-test
Table 2.
Tryptophan Fluorescence of WT and mutant apoA-I.
| ApoA-I Mutant | Maximum λ of fluorescence1 | Standard Error |
|---|---|---|
| WT | 332 | 0.5 |
| E198K | 333 | 0.5 |
| L42,44D | 335 | 0.8 |
| L46,47D | 336* | 0.3 |
Average of two sets of triplicate measurements made on independent preparations of proteins.
p<0.05 compared to WT by unpaired, 2-tailed student’s t-test
3.2 Lipid Clearance by ApoA-I Variants
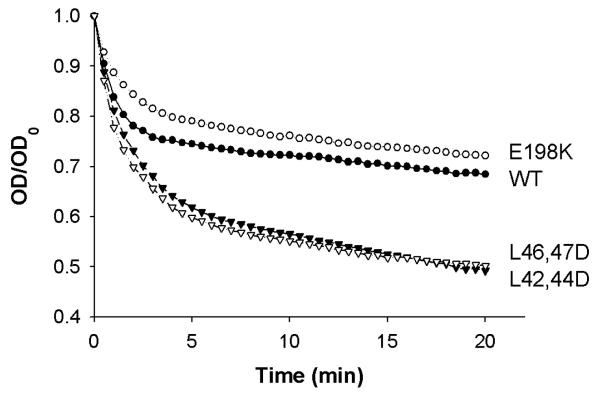
The rate at which each apoA-I variant can bind to and solubilize multilamellar DMPC vesicles (MLV) was measured. Figure 3 shows the decrease in turbidity over time as the protein binds and emulsifies the MLVs. Table 3 presents the rate constants of lipid clearance for the first 5 minutes of the reaction assuming the binding event occurs under pseudo-first order kinetics with excess MLV present. All k-values are normalized to WT apoA-I. It is clear that apoA-I E198K cleared liposomes similarly to WT apoA-I. However, introduction of negatively charged residues into the putative hydrophobic surface patch in apoA-I L(42,44)D and apoA-I L(46,47)D resulted in a significant increase in the rate of liposome clearance compared to WT apoA-I.
Figure 3.
Representative Curves from Liposome Clearance Assay. WT apoA-I (•), apoA-I E198K (○), apoA-I L42,44D (▼), and apoA-I L46,47D (▽). The experiment was performed as described in Materials and Methods. All points are the mean of three replicates. The fractional absorbance was calculated as optical density (OD) divided by the initial optical density (OD0). A rate constant, k (min-1), was calculated for each mutant by fitting the fractional absorbance at 325 nm vs time to a monoexponential decay equation for the first 5 min of the reaction using Microsoft Excel. (See Table 2)
Table 3.
Rate constants for pseudo-first order reaction of DMPC clearance assay.
| ApoA-I Mutant | k (min−1) | Standard Error |
|---|---|---|
| WT | 1.00 | 0.00 |
| E198K | 0.767 | 0.23 |
| L42,44D | 1.59* | 0.07 |
| L46,47D | 1.77* | 0.09 |
Rate constants calculated by fitting first 5 min of clearancetime courses to pseudo -first order equation. All k values normalized to WT. The k-values shown are an average of two trials of independently prepared protein sets.
p<0.02 compared to WT by unpaired -2-tailed student’s t-test
3.3 ABCA1-dependent cholesterol efflux by ApoA-I Variants
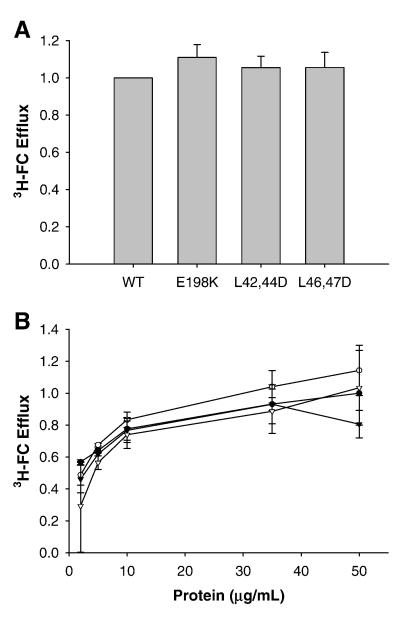
The ability of each apoA-I variant to promote ABCA1-dependent cholesterol efflux from RAW 264.7 macrophages labeled with 3H-cholesterol was measured. In this cell system, treatment with cAMP leads to increased expression of ABCA1 on the cell surface [23-25]. In these cells, the increased cellular cholesterol effluxed to apoA-I with cAMP treatment is due solely to this increase in ABCA1 on the cell surface. As seen in Figure 4A, in the presence of ABCA1, all the lipid-free apoA-I mutants promoted comparable magnitudes of cholesterol efflux as the WT apoA-I when compared at 10 μg/ml. To ensure that the cholesterol efflux was not saturated at this concentration, a range of apoA-I concentrations were investigated (Fig. 4B). It is clear that all mutants showed a concentration-dependent promotion of cholesterol efflux leading to saturation that was similar to WT apoA-I. Together, these data suggest that, despite the disruption of the putative hydrophobic or negatively charged surface patches, apoA-I can still fully participate in cholesterol efflux, and thus in HDL biogenesis.
Figure 4.
Efflux of cholesterol to lipid-free apoA-I mutants. As described in Materials and Methods, RAW264.7 cells were labeled for 18 h with tritiated cholesterol in the presence of cAMP to upregulated ABCA1. After removal of labeling media and washing of the cells, medium containing lipid-free acceptor, again in the presence of cAMP unless otherwise noted, was added to the cells for 8 h. The free cholesterol (FC) efflux was calculated as percentage of total cell cholesterol, calculated as the sum of efflux media counts and intracellular counts after the cells were solubilized in isopropanol. A) Cholesterol efflux to lipid free acceptors. B) Dose curve of cholesterol efflux to lipid-free acceptors. Percent efflux is normalized to corresponding dose of WT. Bars represent mean + SE from two independent trials of three replicates. WT apoA-I (•), apoA-I E198K (○), apoA-I L42,44D (▼), and apoA-I L46,47D (▽).
Discussion
Due to its highly dynamic nature, the determination of a detailed three-dimensional structure of lipid-free apoA-I has proven difficult. Only in the last few years have attempts been successful in producing reasonably detailed models. The first of these was generated by using a fragment sequence threading approach guided by 21 experimental distance constraints provided by chemical cross-linking [13,16]. This structure depicted a loosely associated four helical bundle reminiscent of higher resolution structures determined for a fragment of apoE [26] and intact lipophorin III [27-29]. The C-terminus was predicted to be almost completely lacking in stable secondary structure, consistent with assertions that stabilization of an amphipathic helix in this domain triggers lipid binding. Ajees et al were successful at producing an X-ray crystal structure of full length lipid-free apoA-I under conditions of high concentration and with the additive chromium tris-acetylacetonate [12]. This structure also depicted an N-terminal four helical bundle and a separate C-terminal domain. However, the C-terminus was more highly organized than predicted in the homology model. Although an important advance, this structure probably represents a highly stabilized conformation that does not reflect its in-solution conformation, as it is not consistent with a significant number of previous experimental observations. For a detailed discussion of both the homology and X-ray crystal structures, please see [30].
Although both models of lipid-free apoA-I have their limitations, a striking feature of both is the relatively exposed patch of hydrophobic Leu residues at the turn of the first and second helix in the N-terminal bundle. Palgunachari et al. have speculated that apoA-I binds to lipid surfaces by first anchoring its 1st and 10th amphipathic helix into lipid surface defects followed by subsequent cooperative binding of the central helices [31]. It is clear from both the homology and crystal structures that helix 10 (the most C-terminal helix) could be positioned to participate in such a reaction. However, the availability of the N-terminal helix, associated with the helical bundle in both models, is less clear. One could speculate, as Ajees et al. did, that the unusual exposure of the Leu surface patch might act as an initial interaction site which triggers an opening of the bundle to allow the N-terminal sequence to bind lipid. Following this logic, we hypothesized that disrupting this putative patch with negative charges should decrease the tendency of apoA-I to interact with lipids. Our results shown in Figure 3 with apoA-I L(42,44)D and apoA-I L(46,47)D clearly do not support this hypothesis. In fact, we saw the opposite result, an apparently enhanced ability to bind lipids for both mutants. Given that the introduction of such polar residues should have disrupted the hydrophobic nature of the patch, we conclude that this site is not likely to participate in initial interactions with lipid. One possible explanation for the increase in lipid affinity is that our introduced mutations destabilized the already tenuous thermodynamic stability of the apoA-I helical bundle. It has been suggested that apoA-I lipid-binding may be driven by a decrease in free energy provided by lipid contact [32]. Thus, factors that increase the thermodynamic stability of the lipid-free apoA-I are expected to decrease the free energy savings gained by lipid binding, i.e. the protein is less likely to bind lipid. It follows that the decrease in the thermodynamic stability of the lipid-free protein would have the opposite effect and tend to drive the equilibrium toward the lipid-bound state. Thus, the hydrophobic patch may play a role in overall stability of the lipid-free protein.
It is interesting to note that, despite clear changes in lipid affinity, the surface patches did not exhibit differences in ABCA1-mediated cholesterol efflux. Phillips et al. have proposed that apoA-I solubilizes membrane domains that are generated by ABCA1 activity in addition to direct binding and stabilization of ABCA1 at the cell surface [14]. They demonstrated a distinct correlation between solubilization of membrane-like lipid preparations and the degree of cholesterol efflux. In earlier work, we suggested a similar relationship, though this only held for mutations made in the C-terminal lipid binding helix of apoA-I. Indeed, a “helix-swap” mutant was produced in other regions of apoA-I that exhibited normal ABCA1 cholesterol efflux, but impaired DMPC clearance. Again in this work it is shown that mutations made in the N-terminal bundle can have significant effects on DMPC lipid clearance, but no effect on ABCA1-mediated cholesterol efflux. Clearly, further work is needed to address the complex relationship of lipid affinity and cholesterol efflux under control of ABCA1.
Our results unambiguously showed that introduction of a positive charge into the putative negative charge patch had no effect on lipid-binding or cholesterol efflux. In contrast to the hydrophobic patch described above, the existence of such a charged patch is less clear. It is readily apparent in the crystal structure (Fig. 1), as the residues are oriented by the relatively organized C-terminal helical domains apparent in the model. In the homology model, these residues are predicted to exist in a region of random coil and it seems unlikely that such a charge cluster could occur in such a dynamic part of the molecule. Nevertheless, we introduced a positive charge in the middle of the “patch” as indicated in the crystal structure. Our structural and functional experiments were unable to detect any consequence of this substitution. We suggest that, even if such a patch truly exists in lipid-free apoA-I, that it does not play a major role in lipid binding or ABCA1-mediated cholesterol efflux.
Acknowledgements
This work was supported by grant HL67093. The authors thank Dr. Amy Ghering for help with methodology, Dr. Apryll Stalcup for generous donation of time on her spectropolarimeter, and Stacy Martin and Jamie Morris for help with protein expression.
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- apoA-I
apolipoprotein A-I
- BS3
Bis(sulfosuccinimidyl) suberate
- CD
circular dichroism
- CVD
cardiovascular disease
- DMPC
dimyristoyl-phospatidylcholine
- E. coli
Escherichia coli
- FBS
fetal bovine serum
- FC
free cholesterol
- his-tag
histidine-tag
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
Luria broth
- Leu
leucine
- OD
optical density
- OD0
initial optical density
- STB
standard Tris buffer
- Trp
tryptophan
- WMF
wavelength of maximum fluorescence
- λ
wavelength
References
- [1].Thom T, Haase N, Rosamond W, Howard VJ, Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O’Donnell C, Kittner S, Lloyd-Jones D, Goff DC, Hong YL, Adams R, Friday G, Furie K, Gorelick P, Kissela B, Marler J, Meigs J, Roger V, Sidney S, Sorlie P, Steinberger J, Wasserthiel-Smoller S, Wilson M, Wolf P. Heart disease and stroke statistics - 2006 update - A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;113:E85–E151. doi: 10.1161/CIRCULATIONAHA.105.171600. [DOI] [PubMed] [Google Scholar]
- [2].Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- [3].Assmann G, Schulte H, vonEckardstein A, Huang YD. High-density lipoprotein cholesterol as a predictor of coronary heart disease risk. The PROCAM experience and pathophysiological implications for reverse cholesterol transport. Atherosclerosis. 1996;124:S11–S20. doi: 10.1016/0021-9150(96)05852-2. [DOI] [PubMed] [Google Scholar]
- [4].Groen AK, Elferink R.P. Oude, Verkade HJ, Kuipers F. The ins and outs of reverse cholesterol transport. Ann. Med. 2004;36:135–145. doi: 10.1080/07853890310020635. [DOI] [PubMed] [Google Scholar]
- [5].Tall AR, Wang N. Tangier disease as a test of the reverse cholesterol transport hypothesis. J. Clin. Invest. 2000;106:1205–1207. doi: 10.1172/JCI11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- [7].Marcil M, Brooks-Wilson A, Clee SM, Roomp K, Zhang LH, Yu L, Collins JA, van Dam M, Molhuizen HO, Loubster O, Ouellette BF, Sensen CW, Fichter K, Mott S, Denis M, Boucher B, Pimstone S, Genest JJ, Kastelein JJ, Hayden MR. Mutations in the ABC1 gene in familial HDL deficiency with defective cholesterol efflux. Lancet. 1999;354:1341–1346. doi: 10.1016/s0140-6736(99)07026-9. [see comments] [DOI] [PubMed] [Google Scholar]
- [8].Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, Assmann G. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- [9].Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- [10].Assman G, Eckardstein A, Brewer BJ. The Online Metabolic & Molecular Bases of Inherited Disease: Chapter 122: Familial Analphalipoproteinemia: Tangier Disease. 2001.
- [11].Zannis VI, Chroni A, Krieger M. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. Journal of Molecular Medicine. 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]
- [12].Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: Insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [13].Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A Three-Dimensional Molecular Model of Lipid-Free Apolipoprotein A-I Determined by Cross-Linking/Mass Spectrometry and Sequence Threading. Biochemistry. 2005;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- [14].Vedhachalam C, Duong PT, Nickel M, Nguyen D, Dhanasekaran P, Saito H, Rothblat GH, Lund-Katz S, Phillips MC. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 2007;282:25123–25130. doi: 10.1074/jbc.M704590200. [DOI] [PubMed] [Google Scholar]
- [15].Yancey PG, Bortnick AE, Kellner-Weibel G, Llera-Moya M, Phillips MC, Rothblat GH. Importance of Different Pathways of Cellular Cholesterol Efflux. Arterioscler. Thromb. Vasc. Biol. 2003;23:712–719. doi: 10.1161/01.ATV.0000057572.97137.DD. [DOI] [PubMed] [Google Scholar]
- [16].Panagotopulos SE, Witting SR, Horace EM, Nicholas MJ, Sean DW. Bacterial expression and characterization of mature apolipoprotein A-I. Protein Expr. Purif. 2002;25:353–361. doi: 10.1016/s1046-5928(02)00020-7. [DOI] [PubMed] [Google Scholar]
- [17].Tubb MR, Smith LE, Davidson WS. Purification of recombinant apolipoproteins A-I and A-IV and efficient affinity tag cleavage by tobacco etch virus protease. J. Lipid Res. 2009;50:1497–1504. doi: 10.1194/jlr.D900003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: A mass spectrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- [19].Woody RW. Circular dichroism. [Review] [195 refs] Methods in Enzymology. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- [20].Chen YH, Yang JT, Martinez HM. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 1972;11:4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- [21].Panagotopulos SE, Witting SR, Horace EM, Hui DY, Maiorano JN, Davidson WS. The Role of Apolipoprotein A-I Helix 10 in Apolipoprotein-mediated Cholesterol Efflux via the ATP-binding Cassette Transporter ABCA1. J. Biol. Chem. 2002;277:39477–39484. doi: 10.1074/jbc.M207005200. [DOI] [PubMed] [Google Scholar]
- [22].Weinberg RB. Exposure and electronic interaction of tyrosine and tryptophan residues in human apolipoprotein A-IV. Biochemistry. 1988;27:1515–1521. doi: 10.1021/bi00405a018. [DOI] [PubMed] [Google Scholar]
- [23].Abe-Dohmae S, Suzuki S, Wada Y, Aburatani H, Vance DE, Yokoyama S. Characterization of apolipoprotein-mediated HDL generation induced by cAMP in a murine macrophage cell line. Biochemistry. 2000;39:11092–11099. doi: 10.1021/bi0008175. [DOI] [PubMed] [Google Scholar]
- [24].Oram JF, Lawn RM, Garvin MR, Wade DP. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 2000;275:34508–34511. doi: 10.1074/jbc.M006738200. [DOI] [PubMed] [Google Scholar]
- [25].Smith JD, Miyata M, Ginsberg M, Grigaux C, Shmookler E, Plump AS. Cyclic AMP induces apolipoprotein E binding activity and promotes cholesterol efflux from a macrophage cell line to apolipoprotein acceptors. J. Biol. Chem. 1996;271:30647–30655. doi: 10.1074/jbc.271.48.30647. [DOI] [PubMed] [Google Scholar]
- [26].Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- [27].Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-A resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- [28].Fan D, Zheng Y, Yang D, Wang J. NMR solution structure and dynamics of an exchangeable apolipoprotein, Locusta migratoria apolipophorin III. J. Biol. Chem. 2003;278:21212–21220. doi: 10.1074/jbc.M208486200. [DOI] [PubMed] [Google Scholar]
- [29].Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Davidson WS, Thompson TB. The structure of apolipoprotein a-I in high density lipoproteins. J. Biol. Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- [31].Palgunachari MN, Mishra VK, Lund-Katz S, Phillips MC, Adeyeye SO, Alluri S, Anantharamaiah GM, Segrest JP. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Implications for HDL assembly. Arteriosclerosis, Thrombosis & Vascular Biology. 1996;16:328–338. doi: 10.1161/01.atv.16.2.328. [DOI] [PubMed] [Google Scholar]
- [32].Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]