Abstract
Somatic mitochondrial DNA alterations have been found in all types of cancer. To better understand the role of mitochondria and their involvement in the pathogenic mechanisms of cancer development, the effects of cancer mitochondria were investigated in a defined nuclear background using a transmitochondrial cybrid system. Our results demonstrated that cancer mitochondria confer a significant reduction in cell growth when cells are metabolically stressed in a galactose medium. Activities of the respiratory chain complexes, cellular oxygen consumption, and ATP synthesis rates were found to be much lower in breast cancer cells, than those in normal breast epithelial cells of MCF-10A (10A). These results suggest that there is reduced mitochondrial function in the studied breast cancer cell lines. Similarly reduced mitochondrial function was observed in cybrids containing cancer mitochondria. Novel tRNA mutations were also identified in two breast cancer cell lines, possibly responsible for the observed mitochondrial dysfunction. We conclude that altered mitochondria in cancer cells may play a crucial role in tumor development.
Keywords: Breast cancer, Transmitochondrial cybrids, Mitochondrial tRNA mutation, Defective oxidative phosphorylation, P53
1. Introduction
Cancer cells adapt to hypoxic and acidic conditions generated during progressive tumor cell growth by shifting the burden of energy metabolism from mitochondrial oxidative phosphorylation to glycolysis [1]. Although the molecular pathogenic mechanism is currently unknown, it is expected that mitochondria play a crucial role in this transition.
The major function of mitochondria is the generation of ATP, the energy currency of the cell, by oxidative phosphorylation. During this process reactive oxygen species (ROS) are produced. ROS can cause oxidative DNA damage and increase tumorigenicity [2, 3] as well as metastatic ability [4–6]. Thus, mitochondria could play an important role in multiple steps of tumor development. Mitochondrial dysfunction, one of the most notable features of cancer cells, can be caused by molecular defects in mitochondrial or nuclear genes that encode proteins involved in mitochondria biogenesis; respiratory chain assembly and function; maintenance of membrane potential; and energy metabolic or signal transducing pathways.
Somatic alterations in mitochondrial DNA (mtDNA) are commonly found in 30–100% of all tumors studied, including breast cancer [7–11]. However, most of these somatic mtDNA alterations are neutral variants without any functional relevance [7–11]. These mtDNA alterations may simply reflect the genomic instability of tumor cells. On the other hand, cancerous mitochondria may contain a collection of somatic mtDNA mutations, each present at a level of heteroplasmy below current detection limits. Nonetheless, a small number of deleterious mtDNA mutations that result in altered mitochondrial function have been found [12–15].
Despite the large volume of reports documenting somatic mtDNA alterations in cancer cells, studies of the roles of the altered mitochondrial genome in tumor development have been scarce. In order to investigate the contribution of defective mitochondria to cancer, the effect of nuclear genes must be excluded [16, 17]. Using transmitochondrial cybrids, mitochondria carrying a homoplasmic pathogenic point mutation (m.8993T>G in ATP6) were studied in prostate cancer cell lines [18, 19]. This mutant mtDNA also conferred an advantage in the early stage of tumor growth when transplanted into nude mice [18, 19]. A recent report demonstrated that defective oxidative phosphorylation in thyroid oncocytic carcinoma was associated with combined complex I/III deficiency by the identification of mtDNA mutations in genes encoding a complex I subunit and cytochrome b [15]. Meanwhile, Matoba et al. have shown that P53, one of the most frequently mutated genes in cancers, modulates the balance between the two major energy production pathways: aerobic mitochondrial respiration and anaerobic glycolysis [20]. The modulation by P53 was mediated by a protein responsible for the assembly of complex IV [20]. In P53−/− cells, cytochrome c oxidase activity was found to be reduced [20, 21]. These observations suggest that the down regulation of mitochondrial respiratory function in cancer cells may be achieved by mutations in the nuclear genome, in the mitochondrial genome or in both genomes.
To further understand the role of mitochondria in cancer, we characterized the mitochondrial function in breast caner cell lines and in their transmitochondrial cybrids using biochemical and molecular methods.
2. Material and methods
2.1. Material
Osteosarcoma-derived cell line 143B.TK− (143B), immortalized mammary epithelial cell line MCF-10A (10A), the breast cancer cell lines MDA-MB-231 (231), MDA-MB-436 (436), and MDA-MB-453 (453) were procured from the American Type Culture Collection (Manassas, VA). Carbonyl cyanide, p-(trifluoro-methoxy) phenylhydrazone, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), rotenone, actinomycin D, ferric cyanide, NADH, glutamate, malate, succinate, ADP, ATP monitoring kit and mouse β-actin antibody were purchased from Sigma Chemical Co. (St. Louis, MO). Dulbecco’s modified Eagle’s medium (DMEM), and fetal bovine serum were from Invitrogen (Carlsbad, CA, USA). Antibody for P53 was purchased from Cell Signaling Technology (Danvers MA). Antibodies for mitochondrial proteins were from MitoSciences (Eugene, Oregon). SCO2 antibody was from Abeam, Inc. (Cambridge, MA). Secondary antibodies for rabbit and mouse were from Bio-Rad (Hercules, CA)..
2.2. Cell culture and the generation of ρ0 and cybrid cell lines
All breast cancer cell lines and transmitochondrial cybrid cell lines were maintained in DMEM medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mg/ml bromodeoxyuridine (BrdU) according to published protocols [22]. The 206 rho0 cell line derived from osteosarcoma (143B) used as the mitochondria acceptor cell line was kept in the same DMEM medium with the addition of 50 μg of uridine/ml and 1 mM pyruvate [23]. Normal breast epithelial 10A cells were cultured in mammary epithelial growth medium (Clonetics, San Diego, CA) supplemented with 100 ng/mL cholera toxin (Calbiochem, La Jolla, CA) [24]. All cell cultures were grown in a humidified incubator at 37°C with 5% CO2. Medium was replenished every 2 days.
Rho0 cell lines were generated by treating 143B cells with ethidium bromide according to published procedures [23]. The depletion of mtDNA was confirmed by real time quantitative PCR. Once the rho0 condition is established, rho0 clones were maintained in medium without ethidium bromide as described above. Cybrids were generated by the fusion of rho0 cells with enucleated mitochondria in 45% polyethylene glycol (MW 1450; Sigma cat P-5402) for 60 seconds according to published procedures [25]. Following fusion, cells were plated in selective medium lacking uridine. The cybrids generated containing mitochondria derived from 10A, 231, 436, and 453 cell lines are named as 10A/143B, 231/143B, 436/143B, and 453/143B, respectively.
2.3. Sequence analysis of the entire mitochondrial genome and the quantification of tRNAThr and tRNASer using northern blot analysis
Total genomic DNA was extracted from cultured cells using the phenol/chloroform method according to standard protocols. Sequence analysis was performed using BigDye terminator (version 3.1) cycle sequencing reagent kit on an ABI 3730 XLl DNA Analyzer (Foster City, CA) as previously described [26]. Results from DNA sequencing were compared with the published Cambridge reference sequences from http://www.mitomap.org [11, 27].
Isolation of total RNA and Northern blot analysis was carried out according to published procedures [28]. Approximately 30 μg of total RNA was subjected to Northern blotting and subsequently hybridized with a 32P-end-labeled oligonucleotides probe specific to tRNAThr (5′-TCATCTCCGGTTTACAAGACTG -3′) and tRNASer(UCN) (5′-GTTGGCTTGAAACCAGC -3′). The amount of tRNAThr and tRNASer(UCN) was normalized to the amount of nuclear-encoded 5S ribosomal RNA (the probe specific for 5S rRNA was 5′-GGGTGGTATGGCCGTAGAC-3′ complementary to the 3′ region).
2.4. Cell viability measurement
The tetrazolium salt MTT was used to determine the cell viability as previously described [29, 30]. Briefly, cells (5×103/well) were seeded into 96-well plates and incubated in DMEM glucose-free medium supplemented with 5 mmol/L galactose, 5 mmol/L Na-pyruvate, and 10% FBS (DMEM-galactose medium) for 48 h. Absorbance of solubilized formazan salts was measured with a Tecan Infinite M200 microplate reader (Mannedorf, Switzerland) at 570 nm.
2.5. Oxygen consumption
Cells were cultured for 3 days until approximately 80–90% confluent. Cells were then fed fresh medium 2 hours before harvesting, then trypsinized, washed in phosphate buffered saline (PBS), suspended in growth medium and counted. Oxygen consumption was determined by the Instech 600BH chamber system (Plymouth Meeting, PA, USA) and Oxygen monitor 5300A (Yellow Springs, Ohio, USA). Typically, 5×106 cells were incubated in 1 ml growth medium at 37°C. Oxygen consumption was measured for each cell line. Respiration rates (nmol atomic oxygen/min/million cells) were reported after normalization of the polarographic oxygen consumption rates to cell number [31].
2.6. ATP synthesis
ATP synthesis was measured by a luciferin/luciferase assay [15, 32]. Briefly, after the cells were trypsinlized and resuspended (7 × 106/mL) in buffer A [10 mmol/L KCl, 25 mmol/L Tris-HCl, 2 mmol/L EDTA, 0.1% bovine serum albumin, 10 mmol/L potassium phosphate, 0.1 mmol/L MgCl2 (pH 7.4)]. Cells were kept for 15 minutes at room temperature, followed by incubation in buffer A with 50 μg/mL digitonin for 1 minute. After quick centrifugation at low speed, the cell pellet was resuspended in 100μl of buffer A and 10 μl aliquots were incubated with 5 mmol/L malate plus 5 mmol/L glutamate (complex I–driven substrates), or with 10 mmol/L succinate (complex II–driven substrate) plus 2 μg/mL rotenone, and 0.2 mmol/L ADP for 3 minutes in the presence or absence of 10 μg/ml oligomycin. The rate of ATP synthesis is expressed as nmol ATP/min/mg protein.
2.7. Electron transport chain (ETC) analysis
Cells were sonicated and then used for electron transport chain (ETC) enzyme assays. ETC enzymes were assayed at 30° C using a t emperature-controlled spectrophotometer; Ultraspec 6300 pro, Biochrom Ltd., (Cambridge, England). Each assay was performed in triplicate. The activities of complex I (NADH : ferricyanide reductase), complex II (succinate dehydrogenase), total and rotenone sensitive complex I+III (NADH : cytochrome c reductase), complex II+III (succinate : cytochrome c reductase), complex III (cytochrome c reductase), complex IV (cytochrome c oxidase), and CS (citrate synthase) were measured using appropriate electron acceptors/donors according to published procedures [33, 34]. The increase or decrease in the absorbance of cytochrome c at 550 nm was measured for complex I+III, II+III, III, and complex IV. The activity of complex I was measured by oxidation of NADH at 340 nm. For complex II, the reduction of 2, 6-dichloroindophenol (DCIP) at 600 nm was measured. Citrate synthase (CS) was used as a marker for mitochondrial content and was measured by reduction of Ellman’s reagent at 412 nm. Enzyme activities are expressed both relative to total protein as nmol/min/mg protein and as a ratio of CS activity.
2.8. Western blotting
Cells were harvested and then collected by centrifugation at 600 × g for 10 min at 4°C. Cell pellets were washed once in PBS and then re-suspended in radioimmuno precipitation assay (RIPA) buffer. Total protein were lysed as described elsewhere [35]. Samples were then separated on 10% SDS-polyacrylamide electrophoresis gels and electroblotted onto nitrocellulose membranes. Respiratory chain proteins were detected by a monoclonal antibody at a dilution of 1:200. P53 and SCO2 antibody were diluted by 1:1000. Mouse monoclonal antibody for β-actin was used at a dilution of 1:3000. Horseradish peroxidase-labeled secondary antibody (1:3000) was detected by enhanced chemiluminescence.
2.9. Statistical analysis
All analyses and experiments were repeated at least three times. Results are presented as the mean ± SD. Statistical analysis was performed using Student’s t test.
3. Results
3.1. Cancer cell viability reduced under metabolic stressful condition, suggesting mitochondrial defects
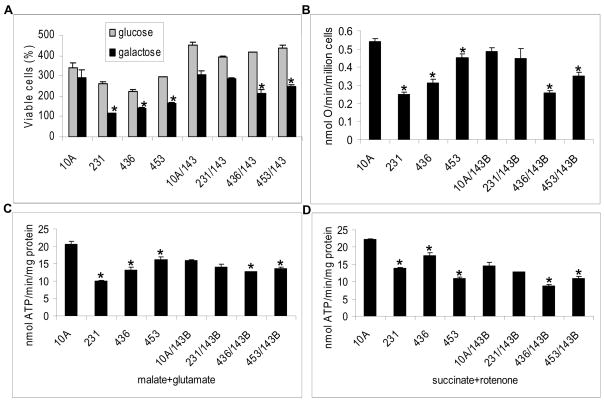
To assess the efficiency of the utilization of oxidative phosphorylation, normal (10A) and breast cancer (231, 436, and 453) cells and their respective cybrids were grown in galactose medium, a metabolically stressful condition. As depicted in Fig. 1A, the number of viable cells is significantly reduced when grown in galactose medium compared to glucose medium except in normal epithelial 10A cells (P<0.01). While in galactose medium, the number of viable cells in cell lines containing cancer mitochondria, including parental and cybrid cells, are significantly reduced (P<0.01) when compared to 10A parental cells or cybrids, with the exception of the 231/143B cybrids, in which the number of viable cells is lower but not statistically significant. Cell viability assays was also performed by measuring protease activities in the living cells and the results are similar to MTT assays (data not shown). Since cybrid cells contain the same nuclear genome and the only difference is in the origin of the mitochondria, it can be inferred that the inability to utilize galactose as the sole energy source by the mitochondrial oxidative phosphorylation pathway is attributed to the altered mitochondria derived from breast cancer cell lines.
Fig. 1.
A, Cell viability. Cells were incubated in glucose (grey) or galactose medium (black). Cell viability was then determined after 48 hours of growth in the indicated medium using the MTT assay. MTT absorbance value at time 0 was considered as the 100% value of viable cells. The number of viable cells was the average of three measurements. B, Oxygen consumption rates (nmol O/min/million cells) normalized to protein concentration. C&D, The rate of mitochondrial ATP synthesis was determined in the cells. ATP synthesis driven by complex I (glutamate plus malate) or complex II (succinate with rotenone as inhibitor for complex I) substrates with or without oligomycin was determined. The rates of mitochondrial ATP synthesis (nmol ATP/min/mg protein) represent the oligomycin sensitive reaction. The asterisk, * (P<0.05), indicates that the significant difference in cells containing cancer mitochondria, including cancer parental cells or their cybirds, when compared to 10A parental cells or its cybrids. Each measurement was performed in triplicate.
3.2. Oxygen consumption rates are reduced in cancer cells and their cybrids
To further investigate the defective respiratory chain function of cancer mitochondria, oxygen consumption rate was measured (Fig. 1B). It was found that oxygen consumption was reduced in the parental breast cancer cell lines (231, 436 and 453) when compared to 10A cells (P values are 0.00002, 0.013, and 0.0008, respectively), and in cybrids (436/143B or 453/143B) compared to 10A/143B (P values are 0.003 and 0.002). However, in 231/143B cybrids, there was no significant difference in oxygen consumption rates suggesting that the 143B nucleus can partially compensate for the dysfunction of mitochondria derived from the 231 cancer cell line.
3.3. Tumor mitochondria have reduced ATP synthesis rate
The efficiency of oxidative phosphorylation in cancer cells and cybrids was analyzed by measuring ATP synthase activity driven by complex I or complex II substrates. The rate of ATP synthesis driven by complex I or complex II substrates was significantly reduced in the 231, 436, and 453 parental cancer cell lines compared to normal 10A cells (P values are: complex I substrates: 0.00003, 0.003, 0.01 and complex II substrates: 0.000005, 0.002, 0.0003, respectively). For the 436/143B and 453/143B cybrids, the ATP synthesis rates were also significantly lower than that of 10A/143B cybrids (P<0.05) (Fig. 1C–D). The ATP synthesis rate in 231/143B cybrids was not significantly reduced when compared to 10A/143B.
3.4. Tumor mitochondria exhibit reduced respiratory chain complex activities
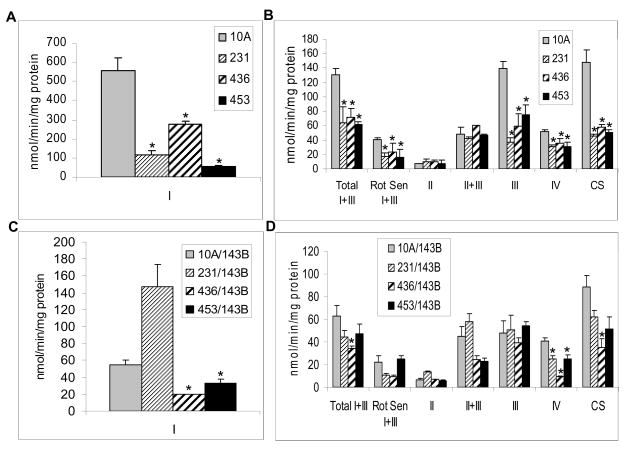
Mitochondrial respiratory chain complex activities were also investigated. As shown in Fig. 2, activities of complexes I, I+III, III, and IV, as well as CS, were dramatically reduced in breast cancer cells compared to those of normal (10A) cells (P<0.05) (Fig. 2A–B). These results are consistent with the defects that would affect overall mitochondrial protein synthesis, for example, mutations in the tRNA genes. Similarly, most complex activities were reduced in the cybrids, but only the reduction in complex IV was statistically significant in all three cybrids containing cancer mitochondria compared to normal 10A mitochondria (Fig. 2C–D). Significant reduction in complex I activity was also observed in 436/143B and 453/143B cybrids (Fig. 2C). Since the activities of most of the complexes containing mitochondrial encoded subunits were reduced in 436/143B cybrids, the results suggested that the mitochondrial dysfunction in the 436 cell line could possibly be caused by molecular defects in the mitochondrial genome. In 231/143B cybrids, some of the mitochondrial complex activities (I, II+III, and III) were partially restored by the donor nucleus of 143B cells. These results suggested that the mitochondria derived from the 231 breast cancer cell line might contain defects that were encoded by nuclear genes. The activity of complex II, which does not contain any subunits encoded by mitochondrial genome, exhibits no significant difference among the different cell lines, parental or cybrids. When the complex activities were normalized to complex II, the results were similar to those normalized to protein concentrations (data not shown). Notably, citrate synthase activity was reduced in the cybrids containing cancer mitochondria when compared to 10A mitochondria, but the reduction was not significant. This was due to fact that the cybrids were all of the same nuclear background, unlike the breast cancer cells where the CS activity was markedly reduced in tumor cells compared to 10A cells.
Fig. 2.
Electron transport chain assay. Complex activities were determined as described in the materials and methods and normalized with protein concentration. A, Activity of complex I in breast cancer cells. B, Activities of complex I+III total, I+III rotenone sensitive, II, II+III, III, IV, and CS in breast cancer cells. C, Activity of complex I in cybrids. D, Activities of complex I+III total, I+III rotenone sensitive, II, II+III, III, IV, and CS in cybrids. Complex I: NADH: Ferricyanide dehydrogenase; Complex I+III: NADH: cytochrome c reductase, including total and Rotenone sensitive; complex II: Succinate dehydrogenase; Complex II+III: Succinate: cytochrome c reductase; Complex III: cytochrome c reductase; Complex IV: Cytochrome c oxidase; CS: Citrate synthase. Columns, mean; Bars, SD; *, P<0.05.
3.5. Identification of molecular defects in mitochondrial DNA
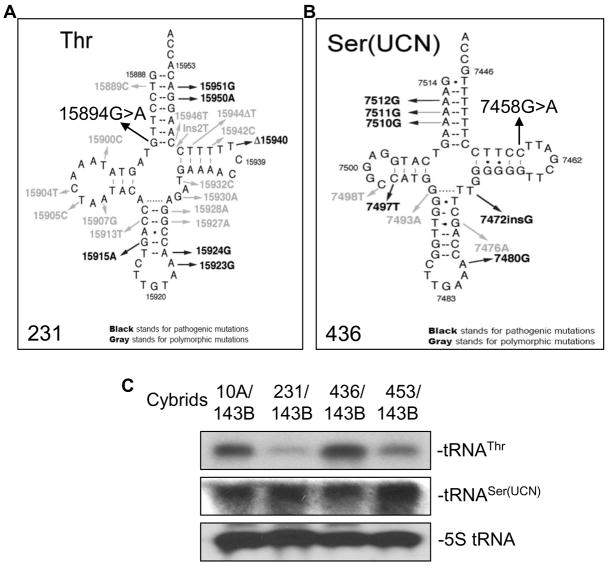
Sequencing of the entire mitochondrial genome of the breast cancer cell lines and their cybrids revealed numerous polymorphisms and neutral variants when compared to the Cambridge Reference Sequence (Table 1). Identical mtDNA variants were also present in the corresponding cybrid cell lines, confirming that the mitochondria in the cybrid cells were indeed derived from the original breast cancer cell lines. Most of these variants have been reported to be benign. However, two novel variants of potential pathogenic effect were identified: a homoplasmic 15894 G>A tRNAThr in the 231 breast cancer cell line and a heteroplasmic 7458 G>A in tRNASer(UCN) in the 436 breast cancer cell line (Fig. 3A–B). Potentially pathogenic mutations were not found in the mitochondrial DNA from the normal breast epithelial 10A cells or the 453 breast cancer cell line (Table 1).
Table 1.
Mitochondrial DNA Sequences of Breast Cancer Cell Lines and Cybrids
| Mutations | Gene | Base Change | Codon | AA Change | 10A | 231 | 436 | 453 | Reported in Mitomap |
|---|---|---|---|---|---|---|---|---|---|
| Haplogroup | HV | X | H | H | |||||
| 72T>C | HV2 | T>C | − | − | + | − | Yes | ||
| 73A>G | HV2 | A>G | + | + | − | + | Yes | ||
| 153A>G | HV2 | A>G | − | + | − | − | Yes | ||
| 185G>A | HV2 | G>A | + | − | − | − | Yes | ||
| 185G>A | HV2 | G>T | − | − | − | + | Yes | ||
| 195T>C | HV2, OH | T>C | − | + | − | − | Yes | ||
| 225G>A | CSB1 | G>A | − | + | − | − | Yes | ||
| 226T>CT | CSB1 | T>C | − | + | − | − | Yes | ||
| 228G>A | CSB1 | G>A | + | − | − | + | Yes | ||
| 241A>G | mtTF1 binding | A>G | − | − | − | − | Yes | ||
| 263A>G | HV2 | A>G | + | + | + | + | Yes | ||
| 295C>T | mtTF1 bidning | C >T | + | − | − | + | Yes | ||
| 300_301het_insCC | + | − | − | − | Yes | ||||
| 310T>C | OH, CSB2 | T>C | + | − | − | − | Yes | ||
| 310_311insC | − | − | + | − | Yes | ||||
| 315_316insC | OH, CSB2 | insC | − | + | − | − | Yes | ||
| 462C>T | d-loop | C >T | + | − | − | − | Yes | ||
| 489T>C | d-loop | T>C | + | − | − | + (ht) | Yes | ||
| 514_515insAC | − | − | + | − | Yes | ||||
| 750A>G | MTRNR1 (12S) | A>G | + | + | + | + | Yes | ||
| 751A>G | MTRNR1 (12S) | A>G | − | − | + | − | novel | ||
| 1320G>AG | MTRNR1 (12S) | G>A | − | − | + | − | novel | ||
| 1438A>G | MTRNR1 (12S) | A>G | + | + | + | + | Yes | ||
| 1719G>A | 16S rRNA | G>A | − | + | − | − | Yes | ||
| 2285T>C | MTRNR2 (16S) | T>C | − | − | + | − | novel | ||
| 2706A>G | MTRNR2 (16S) | A>G | + | + | − | − | Yes | ||
| 3010G>A | MTRNR2 (16S) | G>A | + | − | − | − | Yes | ||
| 3915G>A | ND1 | G>A | GGG>GGA | G203G | − | − | − | + | Yes |
| 3992C>T | ND1 | C>T | ACA>ATA | T229M | − | − | + | − | Yes |
| 4216T>C | ND1 | T>C | TAT>CAT | Y304H | + | − | − | + (ht) | Yes |
| 4769A>G | ND2 | A>G | ATA>ATG | M100M | + | + | + | + | Yes |
| 6221T>C | COI | T>C | CCT>CCC | P106P | − | + | − | − | Yes |
| 6249G>A | COI | G>A | GCT>ACT | A116T | + | − | − | − | novel |
| 6267G>A | COI | G>A | GCA>ACA | A122T | − | + | − | − | novel |
| 6371C>T | COI | C >T | TCC>TCT | S156S | − | + | − | − | Yes |
| 7028C>T | COI | C >T | GCC>GCT | A375A | + | + | − | − | Yes |
| 7458G>AG | MT-TS1 | G>A | − | − | + (ht) | − | novel | ||
| 8269G>A | Non-coding | G>A | − | − | + | − | Yes | ||
| 8860A>G | ATP6 | A>G | ACA>GCA | T112A | + | + | + | + | Yes |
| 9123G>A | MTATP6 | G>A | CTG>CTA | L199L | − | − | + | − | Yes |
| 10044A>G | tRNA Gly | A>G | − | − | + | − | Yes | ||
| 10084T>C | ND3 | T>C | ATC>ACC | I9T | + | − | − | − | Yes |
| 11251A>G | ND4 | A>G | CTA>CTG | L164L | + | − | − | − | Yes |
| 11719G>A | ND4 | G>A | GGG>GGA | G320G | + | + | − | − | Yes |
| 11932C>T | ND4 | C>T | ATC>ATT | I391I | − | + | − | − | novel |
| 12612A>G | ND5 | A>G | GTA>GTG | V92V | + | − | − | − | Yes |
| 12705C>T | ND5 | C>T | ATC>ATT | I123I | − | + | − | − | Yes |
| 13020T>C | ND5 | T>C | GGT>GGC | G228G | − | − | − | − | Yes |
| 13135G>A | ND5 | G>A | GCA>ACA | A267T | − | + | − | − | Yes |
| 13708G>A | ND5 | G>A | GCA>ACA | A458T | + | − | − | − | Yes |
| 13966A>G | ND5 | A>G | ACG>GCG | T544A | − | + | − | − | Yes |
| 14365C>T | ND6 | C>T | GTG-GTA(Rev) | V103V | − | − | + | − | Yes |
| 14470T>C | ND6 | T>A | GGA>GGT | G68G | − | + | − | − | novel |
| 14766C>T | CYTB | C>T | ACT>ATT | T7I | − | + | − | − | Yes |
| 15034A>G | CytB | A>G | CTA>CTG | L96L | − | + | − | − | Novel |
| 15326A>G | CYTB | A>G | ACA>GCA | T194A | − | − | − | + | Yes |
| 15894G>A | MT-TT | G>A | − | + | − | − | novel | ||
| 16069C>T | HV1 | C >T | + | − | − | − | Yes | ||
| 16126T>C | HV1 | T>C | + | + | − | − | Yes | ||
| 16129G>C | HV1 | G>A | − | − | − | + | Yes | ||
| 16189T>A | HV1 | T>A | − | + | − | − | novel | ||
| 16223C>T | HV1 | C>T | − | + | − | − | Yes | ||
| 16278C>T | HV1 | C>T | − | + | − | − | Yes | ||
| 16319G>A | HV1 | G>A | + | − | − | − | Yes | ||
| 16519T>C | D-loop | T>C | − | + | − | + | Yes |
Note: ht: heteroplasmy
Fig. 3.
The tRNA mutation and northern blot analysis. A, Secondary structure of human mtstRNAThr in 231 breast cancer cells. B, Secondary structure of human mt-tRNASer(UCN) in 436 breast cancer cells. C, Northern blot assay of mt-tRNAThr and mt-tRNASer(UCN) for the cybrids, including 10A/143B, 231/143B, 436/143B, and 453/143B.
The effect of 15894G>A and 7458 G>A alterations on the synthesis of tRNAThr and tRNASer(UCN) was investigated by northern blot. The steady-state amounts of the mutant tRNAThr or tRNASer(UCN) was quantified by normalization to the amount of nuclear-encoded 5S rRNA. The results showed reduced tRNAThr in 231/143B cybrids (22%) and tRNASer(UCN) in 436/143B cybrids (79%) compared to 10A/143B cybrids without the tRNAThr or tRNASer(UCN) mutations. It was also found that the steady-state amount of tRNAThr in the 453/143B cybrid cells was about 57% compared to that of the wild-type tRNAThr in the control cybrids 10A/143B (Fig. 3C). The cause for this reduction is not clear. This suggests that other factors might regulate the transcription of tRNAThr in the 453/143B cybrids.
3.6. Western analysis of mitochondrial respiratory chain proteins the parental breast cancer cells and cybrids
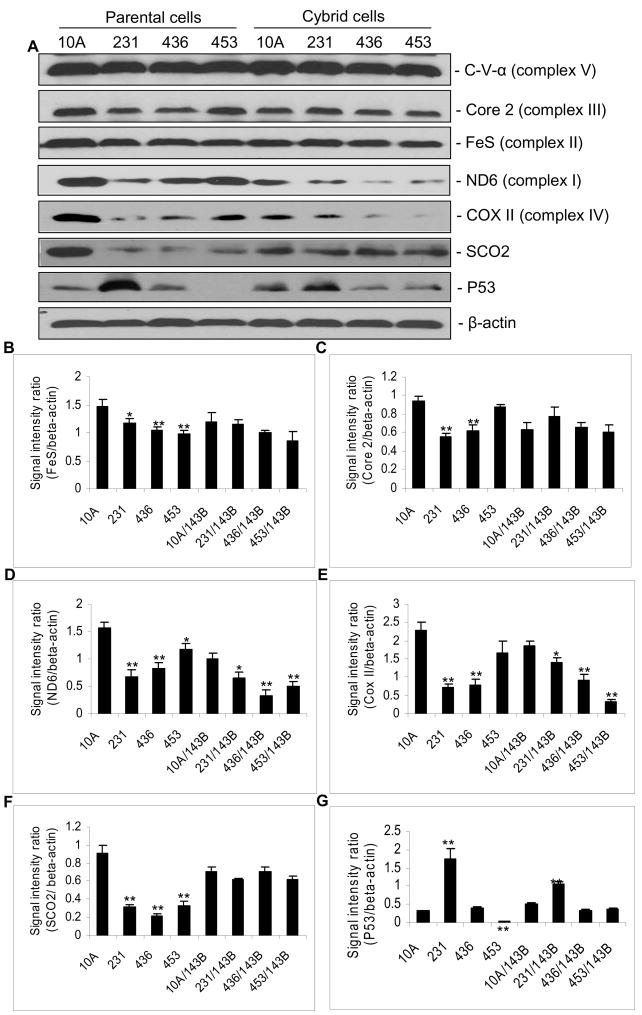
The amount of nuclear encoded mitochondrial proteins:, Core 2 and FeS, was reduced in parental cancer cells when compared to normal 10A cells, suggesting a generalized reduction in mitochondrial respiratory function in cancer cells. However, there is no significant difference in the amount of proteins in the cybrid cells because they share the same nuclear genes (Fig. 4A–C). The mitochondrial encoded proteins, ND6 and COX II (complex IV, cytochrome c oxidase subunit II), are reduced in both the parental cancer cell lines and the cybrids containing mitochondria derived from cancer cell lines, suggesting that mitochondrial protein synthesis is reduced in cancer cells (Fig. 4A, D, E). Interestingly, the expression of SCO2, a nuclear encoded complex IV assembly factor and a downstream target for P53, is markedly reduced in parental cancer cells, while their expression in the cybrids remains unaffected (Fig. 4A, F). It should be noted that P53 is mutated in all three breast cancer cell lines; a dominant negative mutation in 231 cell and frame-shift null mutations in 436 and 453 cell lines. The mutant P53 protein is overly expresis similarly over expressed in 231/143B compared to 10A/143B cybrids (P<0.01). However, P53 expression was reduced in 436/143B and 453/143B cybrids (P<0.05) compared to 10A/143B cybrids (Fig. 4A, G). These results suggest that signals of oncogenic properties from cancer mitochondria can be transmitted to the nucleus to regulate the nuclear gene expression.
Fig. 4.
A, Western blot analysis. The proteins analyzed include C-V-α (complex V), Core 2 (complex III), FeS (complex II), ND6 (complex I), COX II (complex IV), SCO2, P53, and β-actin. B–G, Signal intensities normalized to that of β-actin. Data shown are the Mean ± SD of triplicate assays. The asterisk, * (P<0.05), ** (P<0.01), indicates significant difference of protein expression in cells containing cancer mitochondria, including cancer parental cells or their cybirds, when compared to 10A parental cells or its cybrids.
4. Discussion
Somatic mtDNA alterations have been commonly observed in all types of cancer tissues and cell lines [7–11, 36–40]. However, a majority of these somatic mtDNA changes are in the non-coding D-loop regions or result in silent amino acid changes that do not have any apparent effect on mitochondrial function. Only a few may harbor identifiable pathogenic mutations. In addition, the methods used to identify these somatic mtDNA alterations are limited to the detection of homoplasmic or high heteroplasmic changes. Cancer mitochondria may carry a collection of low heteroplasmic levels of random somatic mtDNA mutations that are not detectable using current mutation detection technologies. Thus, using cybrids containing mitochondria derived from either cancer or normal breast epithelial cells in the same nuclear background is crucial in detecting functional alterations of cancer mitochondria. In this report, we establish that cybrids containing mitochondria derived from breast cancer cells are unable to survive under metabolically stressful conditions and that they exhibit reduced oxygen consumption ability, ATP synthesis, and electron transport chain activities. The reduction in mitochondrial function in cancer cells is also demonstrated by the reduction of viable cells when they are forced to depend on mitochondrial oxidative phosphorylation as the sole source of energy supply under metabolically stressful conditions (Fig. 1A).
Since the cybrids were generated by fusing the mitochondrial depleted rho0 cells with enucleated breast cancer cells, followed by propagation of selected colonies, the observed mitochondrial dysfunction in the cybrids suggest that there may be genomic changes in the cancer mitochondria. Subsequent sequencing analysis indeed identified mutations in the tRNAThr and tRNASer(UCN) genes in 231 and 436 breast cancer cell lines respectively. These mutations were shown to affect the amount of corresponding tRNA and the levels mitochondrial protein synthesis (Fig. 3–4). The accountability of the mitochondrial DNA mutations for the observed mitochondrial dysfunction is further substantiated by the inability of the donor nucleus from 143B cells to fully rescue mitochondrial function. Mutations in the mitochondrial genome were not found in 453 cells. Nevertheless, the amount of tRNAThr and the level of mitochondrial encoded protein synthesis were reduced. Since intact cancer mitochondria were used, this reduction may be due to other unidentified mitochondrial associated factors or epigenetic changes. It is also possible that the observed mitochondrial dysfunction reflects a collection of low heteroplasmic somatic molecular alterations that are beyond the current detection limit.
Interestingly, the replacement of the cancer nucleus with the 143B nucleus partially restored mitochondrial function in the 231/143B cybrids, suggesting that a portion of the observed mitochondrial deficiency may be contributed by nuclear gene alterations. In fact, all three breast cancer cell lines harbor mutations in the P53 gene. Both 436 and 453 cells carry frame-shift truncation mutations; c.205ins7 and c.367del30, respectively [41, 42]. These result in null mutations with no P53 protein produced (Fig. 4A, G) [41, 42]. Whereas a missense dominant negative mutation, p.R280K [41] was found in the P53 protein of 231 cells. This mutant protein is over-expressed (Fig. 4A, G) in 231 cells.
The expression of human SCO2, an assembly factor for complex IV (cytochrome c oxidase, COX), is regulated by P53 [20]. In breast cancers, SCO2 levels are reduced compared to normal 10A cells (Fig. 4A, F). However, with the 143B nucleus in the cybrid cells, the expression of SCO2 is not reduced in cancer cybrids, confirming that the 143B cells express wild type P53 and wild type SCO2 (Fig. 4A, F, G).
Our studies clearly demonstrate the cross-talk between the nuclear and mitochondrial genomes. In the breast cancer cells, regardless of gain-of-function or loss-of-function of P53 mutations, the expression of SCO2 is reduced in cancer cells (Fig. 4A, F, G). However, in cybrid cells with the same wild type nuclear background, levels of SCO2 gene remain constant regardless of their mitochondrial origin (cancer or normal). This stems from the fact that both the wild type SCO2 and P53 are derived from the 143B nuclear background. Nevertheless, P53 protein levels are consistently increased in the 231 cybrids. These results suggest that the changes in the mitochondria of the 231 cell line maintain the oncogenic property of over-expression of the P53 gene, supporting the observation that the mitochondria is communicating with the nucleus. On the other hand, in the cybrids, the over-expressed P53 is the wild type protein that does not directly affect the expression of the SCO2, a mitochondrial COX assembly protein. This regulatory signal is sent from the nucleus to mitochondria. Thus, the dual genome cross-talk is bidirectional. The mechanism requires further investigation.
In conclusion, we have demonstrated that mitochondria in cancer cells have been altered, genetically and functionally. These alterations can send signals to the nucleus, which in turn can regulate the expression of genes involved in energy metabolism and tumorigenic properties.
Acknowledgments
This work was supported by an NIH/NCI (R01 CA-100023) to L. Wong.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki R, Suzuki Y, Yonezawa Y, Ota Y, Okamoto Y, Demizu Y, Huang P, Yoshida H, Sugimura K, Mizushina Y. DNA polymerase gamma inhibition by vitamin K3 induces mitochondria-mediated cytotoxicity in human cancer cells. Cancer Sci. 2008;99:1040–1048. doi: 10.1111/j.1349-7006.2008.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasgupta S, Hoque MO, Upadhyay S, Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008;68:700–706. doi: 10.1158/0008-5472.CAN-07-5532. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, HJ ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 6.Augenlicht LH, Heerdt BG. Mitochondria: integrators in tumorigenesis? Nat Genet. 2001;28:104–105. doi: 10.1038/88800. [DOI] [PubMed] [Google Scholar]
- 7.Bai RK, Wong LJ. Simultaneous detection and quantification of mitochondrial DNA deletion(s), depletion, and over-replication in patients with mitochondrial disease. J Mol Diagn. 2005;7:613–622. doi: 10.1016/S1525-1578(10)60595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 9.Kumimoto H, Yamane Y, Nishimoto Y, Fukami H, Shinoda M, Hatooka S, Ishizaki K. Frequent somatic mutations of mitochondrial DNA in esophageal squamous cell carcinoma. Int J Cancer. 2004;108:228–231. doi: 10.1002/ijc.11564. [DOI] [PubMed] [Google Scholar]
- 10.Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61:5998–6001. [PubMed] [Google Scholar]
- 11.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62:972–976. [PubMed] [Google Scholar]
- 12.Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004;64:1966–1971. doi: 10.1158/0008-5472.can-03-2956. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 14.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 15.Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, Rugolo M, Romeo G. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66:6087–6096. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 16.Krishnamachary B, Berg-Dixon S, Kelly B, Agani F, Feldser D, Ferreira G, Iyer N, LaRusch J, Pak B, Taghavi P, Semenza GL. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–1143. [PubMed] [Google Scholar]
- 17.Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs. Oncogene. 2006;25:4768–4776. doi: 10.1038/sj.onc.1209602. [DOI] [PubMed] [Google Scholar]
- 18.Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- 19.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 21.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 22.Ramljak D, Romanczyk LJ, Metheny-Barlow LJ, Thompson N, Knezevic V, Galperin M, Ramesh A, Dickson RB. Pentameric procyanidin from Theobroma cacao selectively inhibits growth of human breast cancer cells. Mol Cancer Ther. 2005;4:537–546. doi: 10.1158/1535-7163.MCT-04-0286. [DOI] [PubMed] [Google Scholar]
- 23.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh TC, Wijeratne EK, Liang JY, Gunatilaka AL, Wu JM. Differential control of growth, cell cycle progression, and expression of NF-kappaB in human breast cancer cells MCF-7, MCF-10A, and MDA-MB-231 by ponicidin and oridonin, diterpenoids from the chinese herb Rabdosia rubescens. Biochem Biophys Res Commun. 2005;337:224–231. doi: 10.1016/j.bbrc.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Bayona-Bafaluy MP, Manfredi G, Moraes CT. A chemical enucleation method for the transfer of mitochondrial DNA to rho(o) cells. Nucleic Acids Res. 2003;31:e98. doi: 10.1093/nar/gng100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brautbar A, Wang J, Abdenur JE, Chang RC, Thomas JA, Grebe TA, Lim C, Weng SW, Graham BH, WLJ The mitochondrial 13513G>A mutation is associated with Leigh disease phenotypes independent of complex I deficiency in muscle. Mol Genet Metab. 2008;94:485–490. doi: 10.1016/j.ymgme.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Wong LJ, Liang MH, Kwon H, Park J, Bai RK, Tan DJ. Comprehensive scanning of the entire mitochondrial genome for mutations. Clin Chem. 2002;48:1901–1912. [PubMed] [Google Scholar]
- 28.Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275:4251–4257. doi: 10.1074/jbc.275.6.4251. [DOI] [PubMed] [Google Scholar]
- 29.Berridge MV, Tan AS, McCoy KD, Kansara M, Rudert F. CD95 (Fas/Apo-1)-induced apoptosis results in loss of glucose transporter function. J Immunol. 1996;156:4092–4099. [PubMed] [Google Scholar]
- 30.Ghelli A, Zanna C, Porcelli AM, Schapira AH, Martinuzzi A, Carelli V, Rugolo M. Leber’s hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium. J Biol Chem. 2003;278:4145–4150. doi: 10.1074/jbc.M210285200. [DOI] [PubMed] [Google Scholar]
- 31.Herst PM, Berridge MV. Cell surface oxygen consumption: a major contributor to cellular oxygen consumption in glycolytic cancer cell lines. Biochim Biophys Acta. 2007;1767:170–177. doi: 10.1016/j.bbabio.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Zanna C, Ghelli A, Porcelli AM, Martinuzzi A, Carelli V, Rugolo M. Caspase-independent death of Leber’s hereditary optic neuropathy cybrids is driven by energetic failure and mediated by AIF and Endonuclease G. Apoptosis. 2005;10:997–1007. doi: 10.1007/s10495-005-0742-5. [DOI] [PubMed] [Google Scholar]
- 33.Vu TH, Sciacco M, Tanji K, Nichter C, Bonilla E, Chatkupt S, Maertens P, Shanske S, Mendell J, Koenigsberger MR, Sharer L, Schon EA, DiMauro S, DeVivo DC. Clinical manifestations of mitochondrial DNA depletion. Neurology. 1998;50:1783–1790. doi: 10.1212/wnl.50.6.1783. [DOI] [PubMed] [Google Scholar]
- 34.Enns GM, Hoppel CL, DeArmond SJ, Schelley S, Bass N, Weisiger K, Horoupian D, Packman S. Relationship of primary mitochondrial respiratory chain dysfunction to fiber type abnormalities in skeletal muscle. Clin Genet. 2005;68:337–348. doi: 10.1111/j.1399-0004.2005.00499.x. [DOI] [PubMed] [Google Scholar]
- 35.Porcelli AM, Ghelli A, Zanna C, Valente P, Ferroni S, Rugolo M. Apoptosis induced by staurosporine in ECV304 cells requires cell shrinkage and upregulation of Cl- conductance. Cell Death Differ. 2004;11:655–662. doi: 10.1038/sj.cdd.4401396. [DOI] [PubMed] [Google Scholar]
- 36.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 37.Kirches E, Krause G, Warich-Kirches M, Weis S, Schneider T, Meyer-Puttlitz B, Mawrin C, Dietzmann K. High frequency of mitochondrial DNA mutations in glioblastoma multiforme identified by direct sequence comparison to blood samples. Int J Cancer. 2001;93:534–538. doi: 10.1002/ijc.1375. [DOI] [PubMed] [Google Scholar]
- 38.Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. Mitochondrial DNA alteration in esophageal cancer. Int J Cancer. 2001;92:319–321. doi: 10.1002/ijc.1204. [DOI] [PubMed] [Google Scholar]
- 39.Wong LJ, Lueth M, Li XN, Lau CC, Vogel H. Detection of mitochondrial DNA mutations in the tumor and cerebrospinal fluid of medulloblastoma patients. Cancer Res. 2003;63:3866–3871. [PubMed] [Google Scholar]
- 40.Tan DJ, Chang J, Chen WL, Agress LJ, Yeh KT, Wang B, Wong LJ. Novel heteroplasmic frameshift and missense somatic mitochondrial DNA mutations in oral cancer of betel quid chewers. Genes Chromosomes Cancer. 2003;37:186–194. doi: 10.1002/gcc.10217. [DOI] [PubMed] [Google Scholar]
- 41.Runnebaum IB, Nagarajan M, Bowman M, Soto D, Sukumar S. Mutations in p53 as potential molecular markers for human breast cancer. Proc Natl Acad Sci U S A. 1991;88:10657–10661. doi: 10.1073/pnas.88.23.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Concin N, Zeillinger C, Tong D, Stimpfl M, Konig M, Printz D, Stonek F, Schneeberger C, Hefler L, Kainz C, Leodolter S, Haas OA, Zeillinger R. Comparison of p53 mutational status with mRNA and protein expression in a panel of 24 human breast carcinoma cell lines. Breast Cancer Res Treat. 2003;79:37–46. doi: 10.1023/a:1023351717408. [DOI] [PubMed] [Google Scholar]