Abstract
Background
Methicillin-resistant Staphylococus aureus (MRSA) is an important nosocomial and community-associated (CA) pathogen. Recently, a variant of the MRSA USA300 clone emerged and disseminated in South-America causing important clinical problems.
Methods
S. aureus isolates were prospectively collected (2006 to 2008) from 32 tertiary hospitals in Colombia, Ecuador, Peru, and Venezuela. MRSA isolates were subjected to antimicrobial susceptibility testing, pulsed field gel electrophoresis (PFGE), and categorized as healthcare-associated (HA)-like or CA-like clones based on genotypic characteristics and detection of genes encoding the Panton-Valentine leukocidin (PVL) and staphylococcal cassette mec (SCCmec) IV. Additionally, MLST of representative isolates of each major CA-MRSA pulsotype, and detection of USA300-associated toxins and the arcA gene were performed in all isolates categorized as CA-MRSA.
Results
A total of 1570 S. aureus were included; 651 were MRSA (41%), with the highest rates of MRSA isolation in Peru (62%), and lowest in Venezuela (26%) and 71%, 27%, and 2% were classified as HA-like, CA-like, and non-CA/HA-like clones, respectively. Only 9 MRSA isolates were confirmed to have reduced susceptibility to glycopeptides (GISA phenotype). The most common pulsotype (designated ComA) amongst the CA-like MRSA strains was found in 96% of isolates with the majority (81%) having ≤6 bands difference with the USA300-0114 strain. Representative isolates of this clone were ST8 but, unlike the USA300-0114 strain, they harbored a different SCCmec IV subtype and lacked arcA (an indicator of the arginine catabolic mobile element (ACME)).
Conclusion
A variant CA-MRSA USA300 clone has now become established in South America and, in some countries, is endemic in hospital settings.
Keywords: MRSA USA300-ST8, Latin-America, ST923
Methicillin-resistant Staphylococcus aureus (MRSA) has been recognized as a major cause of infections worldwide. Although initially recognized as an important nosocomial pathogen, MRSA is now endemic in the community outside hospitals [1]. Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections are caused by highly virulent strains of S. aureus which can affect healthy individuals [2] and, most worrisome, CA-MRSA strains have now been increasingly reported as an important cause of nosocomial infections indicating that they may become endemic in hospital settings [3]. Another important issue related to the treatment of MRSA infections is the emergence of resistance to vancomycin [4] and the observation that the effectiveness of vancomycin for the treatment of severe infections may be compromised [5].
Diverse molecular typing tools have established that the worldwide dissemination of MRSA is mainly due to a few successful clones [6] with a rather specific geographical pattern [7]. HA-MRSA clones detected in South America have typically belonged to two major genotypes: i) the Brazilian ST239, SCCmec III (MRSA-ST239-III) [8–9], and ii) the Chilean clone (MRSA-ST5-I) [8–12]; the later appears to have replaced the Brazilian clone on the continent. Regarding CA-MRSA, two major clones also have been identified in South America: i) MRSA-ST30-IV, mainly present in the Southern part of the continent [13,14] and ii) MRSA-ST8-IV (and its single locus variant (SLV), MRSA-ST923-IV) which is genetically related to MRSA USA300 and was reported for the first time as the predominant and exclusive genetic lineage in Colombia [15]. In this work, we report the first prospective, multicentre study of the molecular epidemiology of MRSA recovered from four Latin American countries in 2006 to 2008.
METHODS
Study Design
The participating centers included 32 high-level care hospitals which were distributed as follows: i) 22 hospitals in Colombia (located in six cities); ii) 5 hospitals in Ecuador (one city), iii) 3 hospitals in Peru (one city), and 2 hospitals in Venezuela (one city). The S. aureus isolates included in this study were collected prospectively from individual patients (repeated isolates from the same patient were excluded). Each hospital collected 50 to 150 consecutive isolates which were recovered from January 2006 to January 2008 according to a specific protocol and monitored by the local coordinator in each country. Clinical specimens included blood, cerebrospinal fluid (CSF), urine, secretions from complicated skin structures and soft tissue infections (SSTIs), and post-surgical wound infections (after clinical evaluation), pleural fluid, bronchoalveolar lavage, pericardial collection, intra-abdominal, or intra-cerebral abscess, bone tissue from suspected osteomyelitis, arthritis aspirates (in the setting of septic arthritis), and peritoneal fluid (in the setting of peritonitis including those associated with peritoneal dialysis). Isolates recovered from catheters, sputum, and those labeled as “skin” without a clinical justification were excluded. The organisms were identified in the local hospital and, once included in the protocol, were sent to the reference laboratory (Bogota, Colombia) in a transport medium (AMIES with charcoal, BBL, Franklin Lakes, NJ USA). Upon arrival, the isolates were re-identified (see below), and preserved in soy trypticase broth (TSB) with 10% glycerol at −70°C.
Bacterial isolates and antimicrobial susceptibility testing
Confirmation of the identification at the species level of all S. aureus isolates was performed by a multiplex PCR assay [16]. The antimicrobial susceptibility profiles of all isolates were determined by the agar dilution method following the Clinical Laboratory Standards Institute (CLSI) guidelines [17]. Screening for reduced susceptibility to vancomycin was performed on all MRSA isolates according to the CLSI guidelines [17]. In addition, all MRSA isolates with a vancomycin MIC of 1 μg/mL or 2 μg/mL were further screened with two additional agar methods (Mueller-Hinton supplemented with 5 μg/mL of teicoplanin and brain heart infusion (BHI) supplemented with 4 μg/mL of vancomycin) as described previously [18, 19]. MRSA isolates which were positive for at least one of the screening tests were additionally tested by the Etest macromethod (EM) [20]; subsequently, isolates identified as positive by the Etest were further evaluated by the new Etest GRD VA (32-0.5 μg/mL); TP (32-0.5 μg/mL) strip (AB BioMeriux S.A., Marcy l’Etoile, France) [21]. The following reference organisms were used as controls for susceptibility testing, and evaluation of low level glycopeptide resistance (GISA) and heterogeneous glycopeptide resistance (hGISA): S. aureus ATCC 29213, ATCC 700698 (Mu3; hGISA) and ATCC 700699 (Mu50; GISA).
Molecular typing and detection of genes associated with virulence
The SCCmec types (I to IV) and subtypes were evaluated in all MRSA isolates by multiplex PCR according to previously reported assays [22–24]. The MRSA strains USA300-0114 (SCCmec type IVa), USA300 (SCCmec type IVb), MR108 (SCCmec type IVc) [25], JCSC4469 (SCCmec type IVd) [23] and HAR22 (SCCmec IVh) [24] were used as controls for PCR assays. Pulsed field gel electrophoresis (PFGE) was performed on all MRSA isolates, with some modifications of a previously described method [26]. Banding patterns were interpreted according to standard criteria [27]; S. aureus NCTC8325 was used as a molecular size control and representatives of MRSA strains belonging to the Chilean, Brazilian and Pediatric clones [12], MRSA NRS 382 (New York/Japan clone), NRS 123 (MRSA USA400), USA300-0114, and USA300 (carrying the SCCmec IVb) were used as controls for comparisons of PFGE banding patterns. Multilocus sequence typing (MLST) [28] was performed using representatives isolates of each PFGE type and the presence of six CA-MRSA virulence-associated genes (lukS-PV, lukF-PV, arcA, sek, seq, and bsaA) previously reported in the USA300 (ST8) strain was investigated in all CA-MRSA isolates using PCR assays, and primers described previously [29, 30]. The USA300-0114 strain was used as a positive control for the PCR amplifications.
RESULTS
Phenotypic characteristics of MRSA in Latin-American Hospitals
A total of 1890 consecutive S. aureus isolates was collected from 32 hospitals in four countries. Overall, 320 isolates were not included in the study due to protocol violations which most frequently were due to contamination, isolates from the same patient, source not included in the protocol or misidentification. From the included isolates (total of 1570), Colombian hospitals contributed 707 (45%), Ecuador, 309 (20%), Peru, 287 (18%), and Venezuela, 267 (17%) of the isolates. Blood, secretions from surgical wound, and complicated SSTIs were the most common sources of S. aureus isolates accounting for 27%, 26%, and 8%, respectively.
Methicillin resistance in S. aureus was found in 41% of isolates with geographic variations (Peru, 62%; Colombia, 45%; Ecuador, 28%; Venezuela, 26%). Table 1 shows the percentage of resistance to each class of antibiotic in the region and per country in S. aureus and in the subset of MRSA isolates. Overall, MRSA isolates demonstrated high rates of resistance to erythromycin (75%), clindamycin (72%), ciprofloxacin (72%), and gentamicin (69%). The lowest rates of resistance were found to SXT (5%), rifampin (5%), and minocycline (1%). All isolates were susceptible to linezolid and vancomycin (MIC90 1 μg/mL MIC, range 0.5–2 μg/mL) by agar dilution method. Among 651 MRSA isolates evaluated, only 9 isolates (6 from Peru, 2 from Colombia, and 1 from Ecuador) were found to be GISAs after the screening and confirmatory methods were used.
Table 1.
Phenotypic characteristics of S. aureus in the Andean region
| Organism (n) | Country (No. of isolates) | No of resistant isolates (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | ERY | CLI | CIP | GEN | CHL | RIF | TET | MIN | SXT | ||
| S. aureus (1570) | Colombia (707) | 318 (45) | 288 (41) | 227 (32) | 240 (34) | 225 (32) | 5 (1) | 11 (1) | 147 (21) | 1 (0.1) | 5 (1) |
| Peru (287) | 177 (62) | 200 (70) | 178 (62) | 180 (63) | 189 (66) | 69 (24) | 11(4) | 34 (12) | 7(2) | 17 (6) | |
| Ecuador (309) | 87 (28) | 61 (20) | 20 (6) | 32 (10) | 37 (12) | 7 (2) | 15 (5) | 75 (24) | 1 (0.3) | 16 (5) | |
| Venezuela (267) | 69 (26) | 98 (37) | 54 (20) | 66 (25) | 52 (19) | 0 (0) | 11 (4) | 50 (19) | 0 (0) | 3 (1) | |
| Total | 651 (41) | 647 (41) | 479 (30) | 518 (33) | 503 (32) | 81 (5) | 48 (3) | 306(19) | 8 (0.5) | 41(3) | |
| MRSA (651) | Colombia (318) | - | 227 (71) | 221 (69) | 217 (68) | 204 (64) | 3 (1) | 9 (3) | 58 (18) | 0 (0) | 2 (1) |
| Peru (177) | - | 175 (99) | 174 (98) | 176 (99) | 171 (97) | 62 (35) | 10 (6) | 23 (13) | 7 (4) | 16 (9) | |
| Ecuador (87) | - | 27 (31) | 18 (21) | 25 (29) | 23 (26) | 6 (7) | 12 (14) | 39 (45) | 1 (1) | 16 (18) | |
| Venezuela (69) | - | 60 (87) | 53 (77) | 53 (77) | 49 (71) | 0 (0) | 5 (7) | 6 (9) | 0 (0) | 1 (1) | |
| Total | - | 489 (75) | 466 (72) | 471 (72) | 447 (69) | 71 (11) | 36 (5) | 126(19) | 8 (1) | 35 (5) | |
OXA: oxacillin, ERY: erythromycin, CLI: clindamycin, CIP: ciprofloxacin, GEN: gentamicin, CHL: chloramphenicol, RIF: rifampin, TET: tetracycline, MIN: minocycline, SXT: trimethoprim-sulfamethoxazole.
Hospital vs. Community - Associated MRSA in the Andean Region
In order to differentiate the South American MRSA isolates with typical HA pulsotype (HA-like) from those with a CA type (CA-like), the PFGE banding pattern of all isolates was compared to that of representatives isolates of the most common HA clones previously described in South-America (e.g., Chilean, Brazilian, Pediatric, and New York/Japan). Isolates which were not genetically related to any of those clones (> 6 bands difference) were tested for the presence of lukF-lukS genes (encoding PVL) and SCCmec IV, and organisms which yielded a positive result for both were designated as CA-like strains and their PFGE pattern compared to that of representatives of the USA300-0114 strain (SCCmec IVa), a USA300 strain carrying the SCCmec cassette IVb and a representative of USA400 (NRS 123). Using these genotypic criteria, three groups of isolates were clearly identified: i) 461 isolates (71%) were categorized as having a HA-like MRSA type; ii) 174 (27%) isolates were likely to be CA-MRSA (CA-like MRSA) and, iii) 16 isolates (2%) showed patterns not related to the circulating HA or CA clones previously characterized in the region and lacked the genes encoding PVL. All MRSA isolates recovered in Peru had genotypic characteristics (see below) of HA-like MRSA and we were unable to identify any isolate with a CA-like profile. Conversely, 74% of MRSA isolates submitted from Ecuadorian centers had a CA-like genotype. In Venezuela and Colombia, 14% and 31% of MRSA isolates recovered in hospitals had characteristics compatible with CA-like MRSA, respectively (Table 2).
Table 2.
Distribution of HA-like MRSA and CA-like MRSA pulsotypes in the Andean region.
| Country | HA-MRSA pulsotypes % (n) | CA-MRSA pulsotypes % (n) | Non HA/CA-MRSA % (n) |
|---|---|---|---|
| Colombia | 68 (216) | 31 (100) | 1 (2) |
| Peru | 97 (171) | 0 (0) | 3 (6) |
| Ecuador | 24 (21) | 74 (64) | 2 (2) |
| Venezuela | 77 (53) | 14 (10) | 9 (6) |
| Total | 71 (461) | 27 (174) | 2 (16) |
The HA-like MRSA group were mostly recovered from blood (30%) and PFGE analysis revealed that the majority (92%) were clonally related to a major PFGE pulsotype, designated pulsotype A in this work, which is related to the pattern previously observed for the Chilean clone, which harbors the SCCmec type I and belongs to the sequence type 5 (ST5) by MLST. SCCmec typing of 23 representative isolates from this clone recovered in different countries confirmed the presence of SCCmec I and MLST of a representative isolate yielded the ST5, confirming the genetic relatedness with the Chilean clone. Additionally, minor pulsotypes were identified: i) pulsotype B, which corresponds to the Brazilian clone, was detected in 20 isolates (4%), was the predominant HA-like clone found in Ecuadorian hospitals (62%) and was identified in 4% of the HA-like MRSA isolates from Peru. Among isolates belonging to the Brazilian clone (n=20), 90% harbored the SCCmec type III and were resistant to SXT, while 10% harbored the SCCmec I and were susceptible to SXT; ii) pulsotype C, corresponding to the New York/Japan clone, was detected in 18 isolates (4%). Most (72%) of the pulsotype C isolates were associated with SCCmec type II and the remaining isolates carried the SCCmec type IV.
Amongst the CA-like MRSA group (174 isolates), secretions from SSTIs, surgical wound infections and blood were the most common sources accounting for 26%, 25% and 18%, respectively, and we were able to identify four pulsotypes amongst these 174 CA-MRSA isolates. The major pulsotype (designated ComA, figure 1) was detected in 167 (96%) isolates (Table 3) and was the most common type in the four countries of the region, being found in 100%, 98%, and 50% of the CA-like MRSA from Ecuador, Colombia, and Venezuela, respectively. The majority (81%) of CA-like MRSA isolates corresponding to the ComA pulsotype were clonally related (≥6 bands difference) to the USA300 MRSA-ST8-IV epidemic strain and representatives isolates were shown to belong to ST8. However, unlike the USA300-0114 strain, the vast majority of these isolates (158 out of 167) carried a SCCmec subtype other than IVa; when a different set of primers for SCCmec typing [24]was used (which includes specific primers for the J1 region), a band corresponding to subtypes IVc/IVE was found in the majority of these isolates (93%), suggesting that a different SCCmec IV variant was present in CA-MRSA from South-America. The subtype IVa, typical of USA300-0114, was only detected in 9 isolates (5%), and we were unable to determine a subtype (1%) in two isolates. MLST of representative isolates from the pulsotype ComA confirmed that they belonged to ST8 (Table 3) and we detected bsaA, sek, and seq in 99%, 95%, and 91% of these isolates, respectively. Conversely, the majority of CA-like MRSA from the ComA clone lacked the arcA gene (only present in 4% of the isolates), which is an indicator of the presence of the arginine catabolic mobile element (ACME) typical of the USA300-0114 strain. An important characteristic of these CA-like MRSA isolates was the high rates of resistance to tetracycline (41%) found, but not minocycline (typical of the tet(K) determinant). Rates of resistance to erythromycin (10%), clindamycin (4%), ciprofloxacin (5%), gentamicin (2%), SXT (4%), chloramphenicol (1%), and rifampin (1%) were low. Of note, the highest rates of resistance to antibiotics were found in CA-like MRSA isolates from Ecuador (Table 3). Four minor pulsotypes which differed in ≥7 bands from the patterns of isolates USA300 MRSA-ST8-IV were found amongst the rest of the isolates exhibiting CA-like characteristics; these were designated ComB (ST6), ComC (SLV of ST5), ComD (ST22), and ComE (ST923, which had only been described previously in Colombia) [15].
Figure 1.
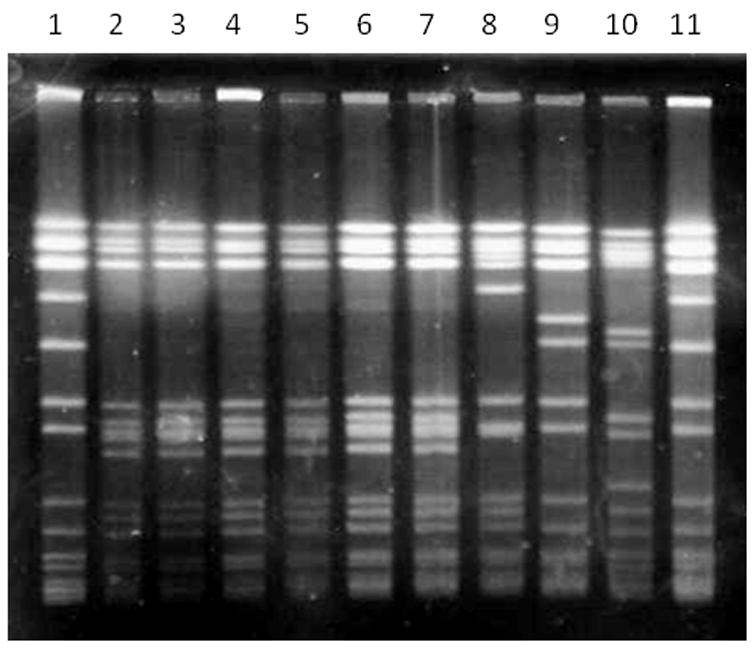
Pulsed field gel electrophoresis (PFGE) of CA-like MRSA isolate representatives of the ComA pulsotype from different countries. Lane 1, S. aureus NCTC 8325; lane 2, Col-177 (Colombia); lane 3, HUV-01 (Colombia); lane 4, CA-12 (Colombia); lane 5, C609 (Colombia), lane 6, V2125 (Venezuela); lane 7, E403 (Ecuador); lane 8, USA300-0114 strain; lane 9, USA300 varrying SCCmec IVb (Nebraska); lane 10, USA400 (N. Dakota); lane 11, S. aureus NCTC 8325.
Table 3.
Molecular and phenotypic characteristics of CA-MRSA clones from hospital laboratories in the Andean Region.
| Country (No. isolates) | PFGE Pulsotype/Clone (%) | ST | SCCmecIV subtypea (%) | Virulence genes (% of isolates) | Resistance profileb (% of isolates) |
|---|---|---|---|---|---|
| Colombia (100) | ComA (98) | 8 | IVc-E (98) | bsaA (99) | TET (43), ERY (9), CLI (6), CIP (4), SXT (2), GEN (1) |
| IVa (1) | sek (93) | ||||
| IVNT (1) | seq (89) | ||||
| arcA (1) | |||||
| ComB (1) | 6 | IVb (100) | bsaA (100) | None | |
| ComC (1) | SLV of ST5 | IVa (100) | Negative | GEN (100), CHL (100) | |
| Ecuador (64) | ComA (100) | 8 | IVc-E (87) | bsaA (100) | TET (39), ERY (9), CIP (5), GEN (5), SXT (6), CHL (2) RIF (2) |
| IVa (11) | sek (98) | ||||
| IVNT (2) | seq (95) | ||||
| arcA (8) | |||||
| Venezuela (10) | ComA (50) | 8 | IVc-E (80) | bsaA (100) | TET (20), ERY (20), CLI (20), CIP (20) |
| IVa (20) | sek (80) | ||||
| seq(80) | |||||
| arcA (20) | |||||
| ComD (10) | 22 | IVa (100) | Negative | GEN (100) | |
| ComE (40) | 923 | IVa (100) | bsaA (100) | ERY (100), TET (100) | |
| sek (100) | |||||
| seq (100) |
SCCmecIV subtyping by multiplex PCR described by Milheirico et al.
Percentage of isolates resistant to ERY (erythromycin), CLI (clindamycin), CIP (ciprofloxacin), GEN (gentamicin), CHL (chloramphenicol), RIF (rifampin), TET (tetracycline), SXT (trimethoprim-sulfamethoxazole) and MIN (minocycline).
IVNT, Non-typeable SCCmec IV
SLV, single locus variant
Among the remaining 16 isolates, whose PFGE pattern was unrelated to that of the HA or CA patterns and which lacked the gene encoding PVL, we identified 12 pulsotypes (designated DifA to DifL). Amongst these isolates, high rates of resistance to quinolones (69%), erythromycin (62%), clindamycin (50%), and gentamicin (62%) were observed and SCCmec types III and IV were found in most.
DISCUSSION
This multicentric study evaluated prospectively and systematically the phenotypic characteristics and population genetics of MRSA in four Latin American countries of the Andean region. The protocol was designed to include patient isolates consecutively submitted to the clinical laboratory in each hospital under specific guidelines and clinical criteria for collection to avoid the recovery of isolates that were likely to represent colonization. The overall rate of isolation of MRSA was 41%, with important differences between countries. In Colombia, the rate of MRSA in the participating 22 hospitals (from 6 cities across the country) was 45%; this value is similar to a previous multicentric surveillance study (rates of 51%) [31], indicating that the prevalence of MRSA in Colombia remains high. In contrast, the participating centers and cities involved in the other countries were fewer, making generalizations regarding rates of MRSA in those countries more difficult and raising the possibility that the molecular epidemiology of the organisms recovered may be skewed by the predominance of a particular strain in a given hospital. In Ecuador, for example (unlike the other countries of the region), the local hospital clinical laboratories also receive and process clinical samples from ambulatory services and outpatient clinics which likely influenced the type of isolates that were collected. In fact, the majorities of clinical samples from Ecuador originated from SSTIs and were likely from patients who were not in the hospital. Therefore, the rates of MRSA and the proportions of HA-like vs. CA-like MRSA circulating in the Ecuadorian hospitals are difficult to estimate. Nonetheless, our aim was to determine the population genetics of consecutive isolates submitted to a hospital clinical laboratory and the findings indicate a high circulation of CA-like MRSA in Ecuador. Moreover, the limitations specified above are common in this type of study which included centers from different countries with heterogeneous populations and varied antibiotic prescribing policies.
The most striking finding of our study was that the highly virulent USA300 MRSA-ST8-IV lineage (which includes the MRSA-ST923-IV, a SLV of ST8) was the predominant and almost exclusive CA-like clone in this region of Latin America accounting for ca. 21% of MRSA isolates. We had previously reported the emergence and dissemination of this USA300 clone variant in Colombia, causing severe skin and soft tissue infections in outpatients with important morbidity and mortality [15]. In this study, we confirmed that the same strain (exhibiting the PFGE banding pattern ComA) has now been established in other Colombian hospitals (accounting for 31% of their MRSA isolates) and also has now been identified in Ecuador and Venezuela (100% and 50% of CA-like MRSA isolates, respectively). The South American USA300 MRSA-ST8-IV has unique characteristics when compared to the USA300-0114 strain: i) it has a different SCCmec subtype cassette; ii) it appears to lack the ACME island and iii) 41% of isolates exhibited resistance to tetracycline (although minocycline remained active) while the rates of resistance to erythromycin were low. Our findings confirm that a USA300-ST8 derivative genetic lineage has now been established in Latin America and support the hypothesis that a highly virulent ancestral USA300-ST8 methicillin-susceptible S. aureus strain, related to the USA300-0114, was likely present in this region of the continent and subsequently acquired the SCCmec independently. Recently, it has been shown that the USA300 lineage is a derivative of a progenitor strain USA500 [32] and it is tempting to speculate that the South American USA300 variant may also be sublineage derivative of USA500. Our results also indicate that the CA-MRSA lineage prevalent in this area of the continent differs substantially from that of isolates found in the southern cone of South America where MRSA-ST30-IV, and MRSA-ST5-IV derivatives appear to predominate [13, 14, 33]. A single MRSA-ST22-IV isolate found in Venezuela (with the ComD pattern, Table 3) belongs to one of the pandemic MRSA clones, referred as EMRSA-15 [7], predominant in United Kingdom hospitals and characterized by a low frequency of multidrug resistance and the presence of SCCmec IV; this clone has also been recently identified in isolates from non-hospitalized patients in Europe [34]. Our results also indicate an important variation in the molecular epidemiology of HA-MRSA in the Andean region. The HA Chilean clone (MRSA-ST5-I) has now been successfully established in Colombia, Peru, and Venezuela. This clone was first identified in Chile in the late 1990s [9], replaced the previously predominant “Pediatric” clone in Colombian hospitals in a span of two years [12] and now is spreading to the rest of the continent [8–11]. Of note, the New York/Japan (MRSA-ST5-II) clone was detected in a few MRSA isolates (3%) from the region, and it is the first time that the presence of this clone is reported in Latin America.
The vancomycin MIC90 of the MRSA isolates from this study was 1 μg/mL, which is identical to that previously reported in Colombia [31], indicating that an obvious “MIC creep” [35] has not occurred in MRSA from this region of Latin-America. We also report, for the first time, the emergence of GISA isolates in the northern area of Latin-America (isolates with reduced susceptibility to vancomycin had been previously reported in Brazil) after following a very strict methodology that included three different methods of screening. Of interest, one of the nine VISA isolates exhibited a CA genotype, supporting the finding that this phenotype may also be present in MRSA USA300, as described previously [36].
In conclusion, we present evidence that MRSA USA300 genetic lineage has now been established as the almost exclusive CA-like clone in the northern region of South America, and has now entered nosocomial settings in some countries.
Acknowledgments
This manuscript is dedicated to the memory of Carlos Carrillo, MD. Some reference isolates were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus Program (NARSA). We are grateful to Dr. Kristina Hulten, Texas Children Hospital, for the gift of strain USA300-0114; Dr. RV Goering for providing MRSA USA300 carrying the SCCmec IVb strain, Dr. Herminia de Lencastre for the reference strain HAR22 (SCCmec IVh), and Dr. Hiramatsu and Dr. Ito for the strain 85/2082 (SCCmec III), WIS [WBG8318]-JCSC3624 (SCCmec V), MR108 (SCCmec IVc) and JCSC4469 (SCCmec IVd). We also want to thank the Universidad El Bosque for their financial support and are indebted to Nicolas Caycedo, Liliana Franco, Ana María Pardo, Doménico Ruocco, Karen Jiménez, Oscar López y Juan Carlos Fernández for technical assistance. We also thank Maria Virginia Villegas for facilitating the coordination of participating centers in Colombia.
The following personnel and hospitals participated in the collection of isolates: Colombia; Bogotá: Fundación Salud Bosque: Claudia Londoño and Martha Herrera; Hospital Simón Bolívar: Constanza Correa; Clínica de Occidente: Norma Montoya; Clínica del Niño: Wilson Daza and Martha Uzeta; Hospital El Tunal: Narda Olarte and Martha Garzón; Hospital Santa Clara: Gloria Gallo; Hospital Occidente de Kennedy: Fernando Peñaloza and Nubia Escobar; Clinica San Pedro Claver: Martha Ruiz; Hospital San Ignacio: Carlos Álvarez, Nidia Torres and Ziomara González; Fundación Santa Fe de Bogotá: Clara Luz Rico; Clínica Infantil Colsubsidio: Giovanni Rodríguez and Deise Rojas; Clínica Saludcoop Jorge Piñeros Corpas: Juan Benavides, Maritza Pérez and Esperanza Guevara; Instituto Nacional de Cancerología: Patricia Arroyo. Cali: Centro Médico Imbanaco: María Virginia Villegas and Beatriz Vanegas; Clínica Saludcoop Occidente Cali: María del Socorro Rojas; Hospital Universitario del Valle: Ernesto Martínez and Nancy Villamarín. Medellín: Hospital Pablo Tobón Uribe: Sergio Jaramillo and Jaime López; Clínica Saludcoop Juan Luis Londoño: Magda Cárdenas. Bucaramanga: Clínica La Foscal: Adriana pinto; Fundación Cardiovascular: Adriana Pinto. Neiva: Hospital Universitario Hernando Moncaleano: Marino Cabrera and Luz Eneyda Quintero. Pereira: Hospital Universitario San Jorge: Carmen Elisa Llanos. Ecuador: Hospital Vozandes, Hospital Eugenio Espejo, Hospital Baca Ortiz, Hospital Carlos Andrade Marín, Hospital General de las Fuerzas Armadas. Perú: Laboratorio Clínico Carlos Carrillo: Gene Martínez Medina and Susana Kuwae de Okuhama; Hospital Nacional Sergio Bernales: Federico Yañez Rojas and Liliana Alvarado; Instituto Nacional de Enfermedades Neoplásicas: Greenlandia Ferreyros Brandon and María Silva; Hospital Nacional Hipólito Unanue: Rosa Avurio Usca and Gladys Patiño Soto. Venezuela: Altagracia Merentes: Centro Medico de Caracas and Hospital Vargas de Caracas.
Financial support
This work was funded in part by an independent research grant from Pfizer, SA. CAA and BEM are supported by a K99/R00 Pathway to Independence Award (1K99-AI72961) and a R37 AI47923 from the National Institute of Allergy and Infectious Diseases, respectively. DP was partially funded by a graduated scholarship from The Instituto Colombiano para el Desarrollo de la Ciencia y Tecnología, “Francisco José de Caldas”, COLCIENCIAS.
Footnotes
Presented in part: 48th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy/Infectious Diseases Society of America 46th Annual Meeting, Washington, DC, 25–28 October 2008 (abstract C2-219).
Potential conflict of interest
Dr. Arias has received lecture fees from Pfizer and Merck and grant support from Pfizer. Dr. Murray has grant support from Johnson & Johnson, Astellas, Palumed and Intercell and has served as consultant for Astellas Pharma US Inc & Theravance Inc., Cubist, Targanta Therapeutics Corporation, Johnson & Johnson, Pfizer, AstraZeneca and Wyeth-Ayerst. Dr. Zurita reports research grants from Pfizer and Wyeth. Dr. Guzman has served as consultant for Pfizer, Merck and Co, Wyeth and Becton and Dickinson. All other authors no conflicts of interest.
References
- 1.Grundmann H, Aires de Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;68:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005;5:275–86. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 3.Maree CL, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG. Community-associated meticillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg Infect Dis. 2007;13:236–42. doi: 10.3201/eid1302.060781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieradzki K, Roberts RB, Haber SW, Tomasz A. The Development of Vancomycin Resistance in a Patient with Meticillin-Resistant Staphylococcus aureus Infection. N Engl J Med. 1999;340:517–23. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Ma XX, Sato K, et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol. 2003;41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nübel U, Roumagnac P, Feldkamp M, et al. Frequent emergence and limited geographic dispersal of meticillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–5. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright MC, Robinson DA, Randle G, et al. The evolutionary history of meticillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci USA. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sola C, Gribaudo G, Vindel A, et al. Identification of a novel meticillin-resistant Staphylococcus aureus epidemic clone in Córdoba, Argentina, involved in nosocomial infections. J Clin Microbiol. 2002;40:1427–35. doi: 10.1128/JCM.40.4.1427-1435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aires De Sousa M, Miragaia M, Sanches IS, et al. Three-year assessment of meticillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J Clin Microbiol. 2001;39:2197–205. doi: 10.1128/JCM.39.6.2197-2205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayor L, Ortellado J, Menacho C, et al. Molecular characterization of meticillin-resistant Staphylococcus aureus isolates collected in Asunción, Paraguay. J Clin Microbiol. 2007;45:2298–300. doi: 10.1128/JCM.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola C, Cortes P, Saka HA, Vindel A, Bocco JL. Evolution and molecular characterization of meticillin-resistant Staphylococcus aureus epidemic and sporadic clones in Cordoba, Argentina. J Clin Microbiol. 2006;44:192–200. doi: 10.1128/JCM.44.1.192-200.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz C, Moreno J, Renzoni A, et al. Tracking meticillin-resistant Staphylococcus aureus clones in Colombian hospitals over 7 years (1996–2003): emergence of a new dominant clone. Int J Antimicrob Agents. 2005;26:457–62. doi: 10.1016/j.ijantimicag.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Benoit SR, Estivariz C, Mogdasy C, et al. Community strains of meticillin-resistant Staphylococcus aureus as potential cause of healthcare-associated infections, Uruguay, 2002–2004. Emerg Infect Dis. 2008;14:1216–23. doi: 10.3201/eid1408.071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro A, Coronado AZ, Silva-Carvalho MC, et al. Detection and characterization of international community-acquired infections by meticillin-resistant Staphylococcus aureus clones in Rio de Janeiro and Porto Alegre cities causing both community- and hospital-associated diseases. Diagn Microbiol Infect Dis. 2007;59:339–45. doi: 10.1016/j.diagmicrobio.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Arias CA, Rincon S, Chowdhury S, et al. MRSA USA300 clone and VREF a U.S.-Colombian connection? N Engl J Med. 2008;359:2177–9. doi: 10.1056/NEJMc0804021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 18th informational supplement M100-S18; Wayne, Pa. 2008. [Google Scholar]
- 18.Wootton M, MacGowan AP, Walsh TR, Howe RA. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J Clin Microbiol. 2007;45:329–32. doi: 10.1128/JCM.01508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiramatsu K, Aritaka N, Hanaki H, et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–3. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 20.Walsh TR, Bolmström A, Qwärnström A, et al. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J Clin Microbiol. 2001;39:2439–44. doi: 10.1128/JCM.39.7.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusof A, Engelhardt A, Karlsson A, et al. Evaluation of a new Etest vancomycin-teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J Clin Microbiol. 2008;46:3042–7. doi: 10.1128/JCM.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in meticillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–61. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in meticillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in meticillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60:42–8. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist update. 2003;6:41–52. doi: 10.1016/s1368-7646(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 26.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–63. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gelelectrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of meticillin-resistant and meticillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 30.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of meticillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 31.Arias CA, Reyes J, Zúñiga M, et al. Multicentre surveillance of antimicrobial resistance in enterococci and staphylococci from Colombian hospitals, 2001–2002. J Antimicrob Chemother. 2003;51:59–68. doi: 10.1093/jac/dkg002. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Diep BA, Villaruz AE, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:5883–8. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sola C, Saka HA, Vindel A, Bocco JL Cordoba MRSA Collaborative Study Group. Emergence and dissemination of a community-associated meticillin-resistant Panton-Valentine leucocidin-positive Staphylococcus aureus clone sharing the sequence type 5 lineage with the most prevalent nosocomial clone in the same region of Argentina. J Clin Microbiol. 2008;46:1826–31. doi: 10.1128/JCM.01949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amorim ML, Faria NA, Oliveira DC, et al. Changes in the clonal nature and antibiotic resistance profiles of meticillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45:2881–8. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakoulas G, Moellering RC. Increasing antibiotic resistance among methicillin resistant Staphylococcus aureus strains. Clin Infect Dis. 2008;46:S360–7. doi: 10.1086/533592. [DOI] [PubMed] [Google Scholar]
- 36.Graber CJ, Wong MK, Carleton HA, et al. Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerg Infect Dis. 2007;13:491–3. doi: 10.3201/eid1303.060960. [DOI] [PMC free article] [PubMed] [Google Scholar]


