Abstract
Thyroid hormone receptors (TRs) are hormone-regulated transcription factors that control multiple aspects of normal physiology and development. Mutations in TRs have been identified at high frequency in certain cancers, including human hepatocellular carcinomas (HCCs). The majority of HCC TR mutants bear lesions within their DNA recognition domains, and we have hypothesized that these lesions change the mutant receptors' target gene repertoire in a way crucial to their function as oncoproteins. Using stable cell transformants and expression array analysis, we determined that mutant TRs isolated from two different HCCs do, as hypothesized, display a target gene repertoire distinct from that of their normal TR progenitors. Only a subset of genes regulated by wild-type TRs were regulated by the corresponding HCC-TR mutants. More surprisingly, the HCC-TR mutants also gained the ability to regulate additional target genes not recognized by the wild-type receptors, and were not simply restricted to repression, but could also activate a subset of their target genes. We conclude that the TR mutants isolated from HCC have sustained multiple alterations from their normal progenitors that include not only changes in their transcriptional outputs, but also changes in the genes they target; both are likely to contribute to neoplasia.
INTRODUCTION
Thyroid hormone receptors (TRs) play key roles in normal physiology and development (Brent, 2000; Buchholz et al., 2006; Flamant et al., 2006; Harvey and Williams, 2002; Yen, 2001). Two distinct genetic loci encode TRs, denoted α and β, each of which is alternatively spliced to generate additional receptor diversity. TRα1 and TRβ1 are among the most abundant of these receptor isoforms, and exert distinct, if partially overlapping, biological roles (Harvey and Williams, 2002; Lazar, 1993; Murata, 1998; Yen, 2001). Both TRα1 and TRβ1 bind to specific DNA sequences (response elements), and regulate the expression of adjacent target genes up or down (Katz and Koenig, 1993; Lazar, 1993; Lazar, 2003; Yen, 2001). On "positively-regulated" target genes, TRs recruit corepressors and repress gene transcription in the absence of T3, but release corepressors, recruit coactivators, and activate gene transcription in the presence of T3 (Cheng, 2000; Harvey and Williams, 2002; Tsai and Fondell, 2004; Zhang and Lazar, 2000). Less is understood about "negatively-regulated" target genes that are activated by TRs in the absence, but repressed in the presence, of hormone; this inverted T3 response presumably arises due to alterations in coregulator recruitment or function, perhaps reflecting combinatorial interactions with other transcription factors also arrayed on the same promoter (Berghagen et al., 2002; Matsushita et al., 2007; Meyer et al., 1997; Nygard et al., 2003; Tagami et al., 1997).
Disruptions of TR function lead to disease (Cheng, 2005; Yen and Cheng, 2003). Inherited mutations in the TRβ locus result in Human Resistance to Thyroid Hormone (RTH) Syndrome, an endocrine disorder (DeGroot, 1996; Kopp et al., 1996; Refetoff, 1993; Refetoff et al., 1993; Yen, 2003). A virally transduced mutant form of TRα contributes to leukemogenesis by the avian erythroblastosis retrovirus (Graf and Beug, 1983; Privalsky, 1992; Sap et al., 1986; Weinberger et al., 1986). Spontaneous mutations in TRα and TRβ are found at high frequency in human hepatocellular carcinomas (HCCs), renal clear cell carcinomas, and certain thyroid malignancies and are believed to participate in the initiation or progression of these malignancies (Chan and Privalsky, 2006; Cheng, 2003; Gonzalez-Sancho et al., 2003; Kamiya et al., 2002; Lin et al., 1999; Lin et al., 1996; Puzianowska-Kuznicka et al., 2002). Virtually all of these TR mutants are impaired in the ability to exchange coactivator for corepressor in response to physiological concentrations of T3. As a result, the mutant receptors retain corepressor inappropriately and can function as dominant-negative inhibitors of wild-type receptor function (Chan and Privalsky, 2006; Lin et al., 1997; Lin et al., 1996; Privalsky, 2008).
Given this commonality, why do certain dominant-negative TRs produce primarily endocrine disease, whereas others are closely associated with neoplasia? Notably the mutant TRs found in RTH-Syndrome represent single mutational events, whereas the mutant TRs found in neoplasia are typically aggregates of multiple genetic lesions, often including mutations in the DNA recognition domain of these receptors (Kamiya et al., 2002; Lin et al., 1999; Puzianowska-Kuznicka et al., 2002; Refetoff, 1993; Yen and Cheng, 2003). We have proposed that single TR mutations give rise to simple dominant-negative receptors that produce endocrine disorders, whereas additional mutations further modify the dominant-negative phenotype, at least in part by altering target gene recognition, to generate the neoplastic phenotype (Chan and Privalsky, 2006; Privalsky, 2008). To test this hypothesis, we employed a microarray analysis to compare the target gene profiles of wild-type and HCC-mutant TRs. We report that the HCC TR mutants tested displayed widespread alterations in their target gene repertoire compared to the corresponding wild-type controls, and we discuss how these changes may contribute to oncogenesis.
MATERIALS AND METHODS
Isolation of Stable Transformants, Microarray Analysis, and Reporter Assay
HepG2 transformants bearing pCIneo constructs of human wild-type TRα1, wild-type TRβ1, TRα1-I, TRβ1-N or an empty plasmid control were generated using a G418 co-selection methodology (Chan and Privalsky, 2006). Individual cell clones were propagated and screened for integration and expression of the TRs by PCR/reverse-transcriptase PCR. Six to twelve independent cell clones positive for the expression of each receptor were pooled and were treated with 100 nM T3 or with ethanol carrier alone for 6h in DMEM containing 10% hormone-stripped FBS (this time period was chosen to focus primarily on direct response genes). The cells were harvested and RNA was isolated using an RNeasy kit (Qiagen Inc, Valencia CA). The purified RNA was submitted to the University of California, Davis, Cancer Center Gene Expression Resource for subsequent cDNA probe generation, hybridization to and scanning of Affymetrix GeneChip Human Gene 1.0 ST microarrays (Affymetrix Incorporated, Santa Clara, CA, USA). Raw microarray data was normalized by the Robust Multichip Array (RMA) method and p-value adjustments were performed by the Benjamini-Hochberg method using R software and the affylmGUI package (Irizarry et al., 2003; Smyth, 2004). Heat maps were generated with GenePattern (Reich et al., 2006). Three independent biological repeats were analyzed for each of the five transformant pools. Luciferase reporter assays were performed by transient transfection of these stable transformants (Chan and Privalsky, 2006).
Reverse transcriptase PCR
RNA was isolated from cells using Qiagen’s RNeasy kit following the manufacturer’s protocols. Complementary DNA was synthesized using Qiagen’s QuantiTect kit and 1 µg RNA. Semi-quantitative reverse-transcriptase PCR was performed using GoTaq DNA Polymerase (Promega, Madison WI) and the primer sequences listed in supplementary Table S1. PCR products were analyzed on 8% Tris-acetate-EDTA polyacrylamide gels, stained with SYBR Green, and quantified with a Flurochem8900 Imager (Alpha Innotech, San Leandro, CA, USA).
RESULTS
HCC TR mutants are impaired in transcriptional activation and act as dominant negative inhibitors of wild-type TRs in reporter assays
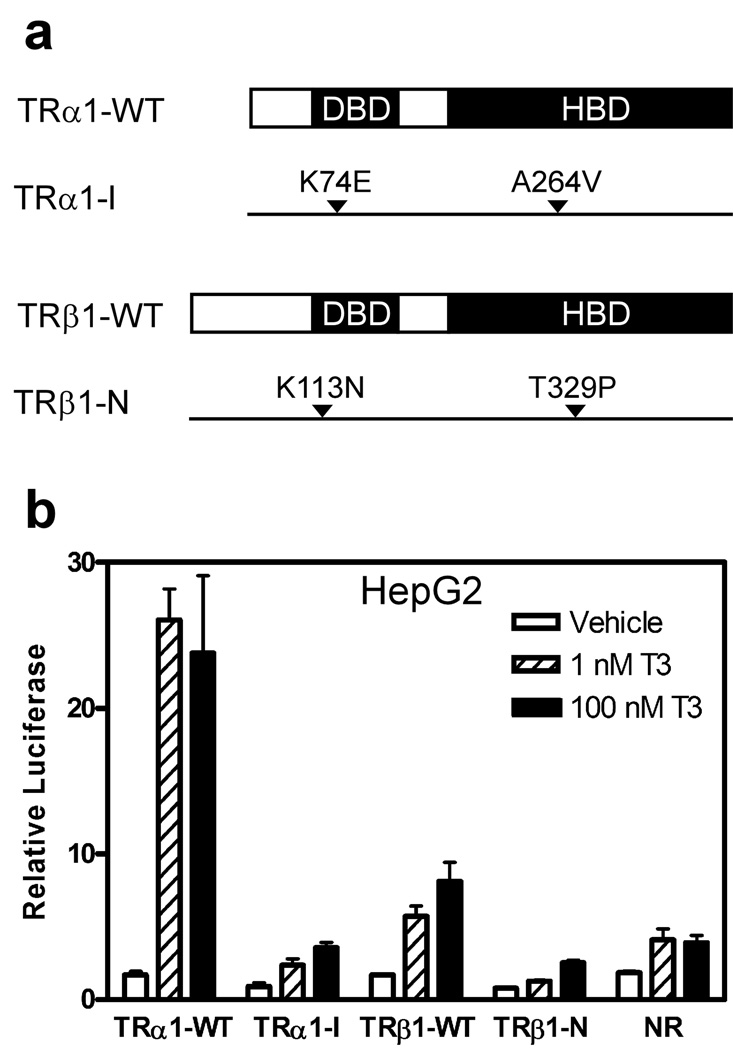
HepG2 are a well-characterized human HCC cell line that lacks known TR mutations and expresses modest levels of wild-type TRα1 and TRβ1 (Chamba et al., 1996; Lin et al., 1996). We generated stable HepG2 transformants expressing a representative HCC-TRα1 mutant (denoted TRα1-I), a representative HCC-TRβ1 mutant (denoted TR-β1-N), wild-type TRα1 (TRα1-WT), wild-type TRβ1 (TRβ1-WT), or an empty expression vector control (Fig. 1a). We first examined the overall T3 response in these cells using a luciferase reporter containing an archetypal DR4 positive-response element (Fig. 1b). The HepG2 transformants containing the empty expression plasmid exhibited a low level T3-mediated regulation of this reporter mediated by the endogenous TRs present in these cells (Darby et al., 1991; Theriault et al., 1992). Transformants bearing the TRα1-WT constructs displayed a significantly enhanced T3 response (Fig. 1b); a similar, though more modest T3 response was also observed for TRβ1-WT (Fig. 1b). In contrast, HepG2 transformants expressing the TRα1-I or TRβ1-N mutants displayed reduced levels of reporter expression in the absence of T3 compared to the empty vector controls, and a severely attenuated reporter gene activation in response to T3. These results support prior findings that these HCC TR mutants are impaired for T3-induced transcriptional activation (Chan and Privalsky, 2006).
Figure 1. HCC TR mutants are impaired in transcriptional activation.
(a) Schematic of wild-type and mutant TRs. The different TRs are depicted with the locations of the DNA binding (DBD), hormone binding domain (HBD), and HCC mutations indicated. (b) Impaired reporter activation in HCC mutant TR transformants. HepG2 cells stably integrated with TRα1-WT, TRβ1-WT, TRα1-I, TRβ1-N or an empty plasmid control (no-receptor, NR) were transiently transfected with a DR4-TK-luciferase reporter and pCH110 as an internal control. The cells were treated 24 hrs later with T3 or vehicle alone, harvested 48 hrs after transfection, and the luciferase activity was determined relative to β-galactosidase activity (mean + S.E.M., n = 3).
The HCC TR mutants repress only a subset of the genes repressed by wild-type TRs in the absence of T3, and have gained a novel ability to repress additional targets not recognized by wild-type TRs
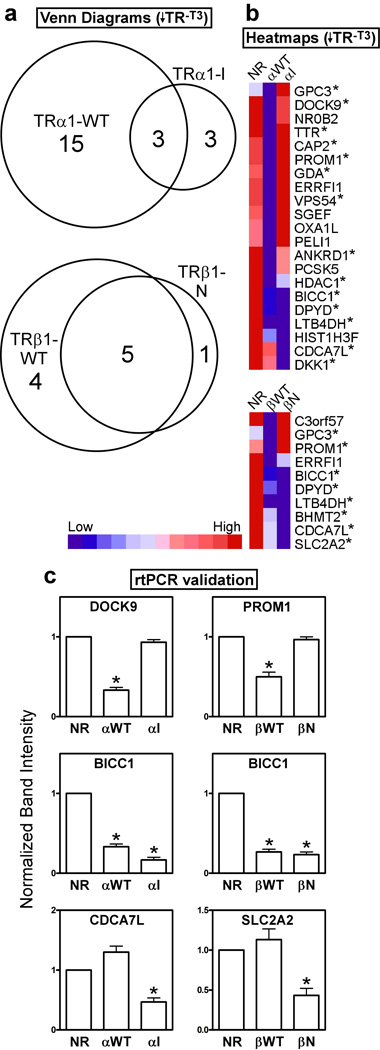
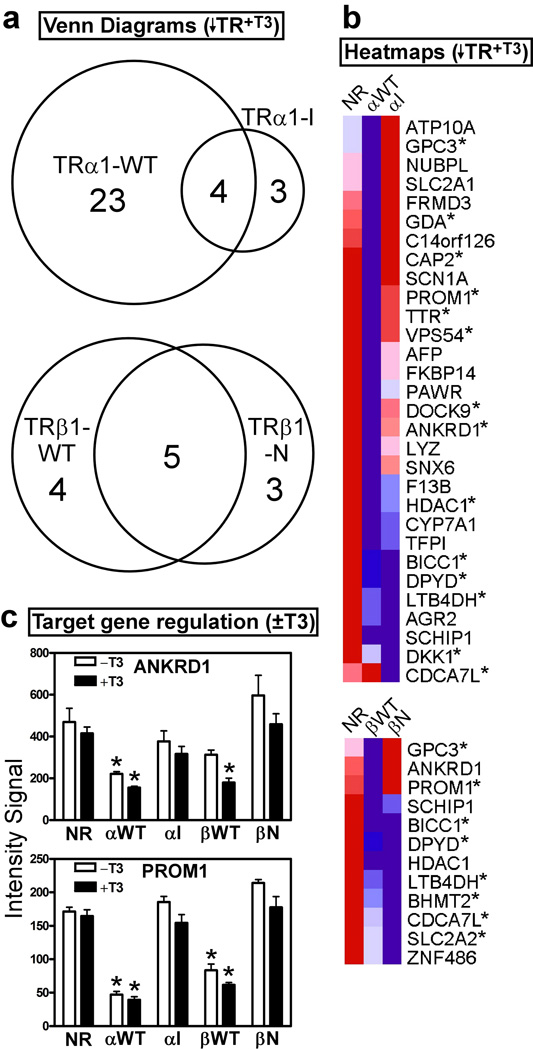
We next compared the patterns of cellular gene expression in our HepG2 transformants by use of a microarray analysis. The TRα1-I and TRβ1-N mutants have been proposed to operate in cancer by constitutively mimicking the repressive functions mediated by the wild-type receptors in the absence of T3 (Chan and Privalsky, 2006). Therefore we first identified the cellular genes repressed by the mutant TRs relative to the empty-vector transformants, and compared these genes to those repressed by the wild-type TRs relative to the empty vector transformants, all in the absence of T3 (symbolized as ↓TR−T3) (Fig. 2a and 2b). Representative target genes were confirmed by rt-PCR (Fig. 2c and data not shown). Notably, several genes, such as BICC1, were strongly repressed by all TRs tested (Fig. 2b and 2c). However, other genes were repressed by the wild-type TR, but not by the corresponding mutant under these conditions. For example, DOCK9 was repressed by TRα1-WT but not TRα1-I, and PROM1 was repressed by both TRα1-WT and TRβ1-WT, but not by either mutant (Figs. 2b, 2c). Reciprocally, certain genes were selectively repressed by the mutant version of a given receptor: for example CDCA7L was repressed only by the TRα1-I mutant, and not by TRα1-WT, whereas SLC2A2 was preferentially repressed by the TRβ1-N mutant compared to TRβ1-WT (Fig. 2b and 2c). A more comprehensive list of all genes described in this study is provided in supplementary Table S2.
Figure 2. In the absence of T3, the HCC TR mutants repress a distinct set of genes compared to wild-type TRs.
(a) Venn diagram of gene transcripts down-regulated in each TR transformant compared to the empty vector (NR) control, all in the absence of T3. HepG2 cells transformed with the TR alleles indicated, or with the empty plasmid control, were incubated in the absence of T3 (vehicle only) for 6 hrs.; RNA was isolated, and used to probe the arrays. Transcripts down-regulated in each TR transformant compared to the empty vector control (NR) were identified using a Benjamini-Hochberg adjusted p-value of <0.05. (b) Heat map clustering of the genes from panel (a). For each gene, dark blue indicates lowest expression, dark red indicates highest expression, with intermediate values represented by lighter shades. These comparisons were minus T3; asterisks indicate genes that were also down-regulated in the presence of T3 (see Figure 6). (c) rtPCR on representative genes identified in panel (b), using gene-specific primers. The level of expression of each gene product was defined as “1” for the empty vector control in the absence of T3 (mean + S.E.M., n = 3). An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.
These results support our hypothesis that the mutant receptors are selective dominant-negatives able to repress some, but not all the genes repressed by their wild-type progenitors. Significantly, our data also demonstrated an additional, if limited, capability of the mutant receptors to repress target genes that were not repressed by the corresponding wild-type receptors, indicating that the mutations in these receptors did not simply narrow, but actually redirected their target gene repertoire.
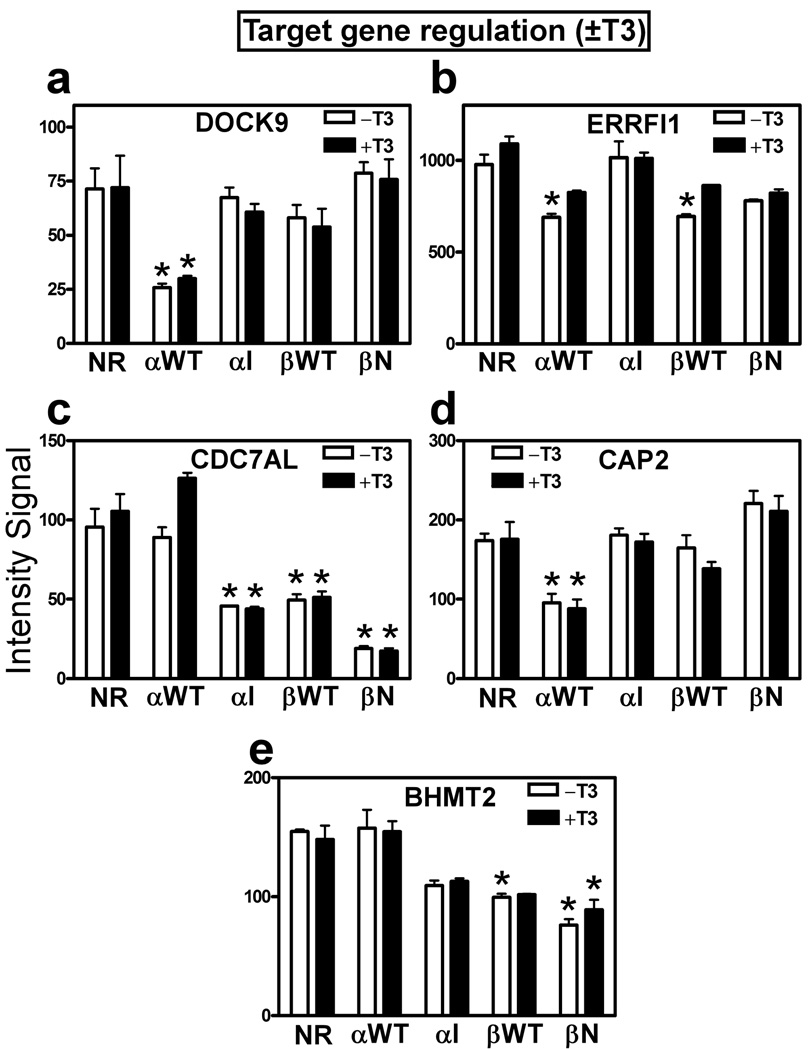
We next examined the effect of T3 on the same panel of genes that were repressed by TRs in the absence of this hormone. As anticipated from our reporter gene assays, the TRα1-I and TRβ1-N mutants behaved as hormone-independent, constitutive repressors (↓TR−T3 remained ↓TR+T3) on most of their cellular target genes (e.g. CDC7AL for TRα1-I and TRβ1-N, or BHMT2 for TRα1-I) (Fig. 3c, 3d, 3e, Table S2). In contrast, repression by the wild-type receptors was at least partially reversed by T3 for certain target genes (e.g. ERRFI1 for both TRα1-WT and TRβ1-WT, and, to a lesser extent, DOCK9 for TRα1-WT; Fig. 3a, 3b, Table S2). However other wild-type TR target genes behaved constitutively and were equally repressed by the wild-type receptor in both the absence and presence of T3 (e.g. CAP2 for TRα1-WT or BHMT2 for TRβ1-WT; Fig. 3d and 3e). These results were confirmed for representative genes by rt-PCR (data not shown). Target genes that are repressed more strongly in response to T3 than in its absence (e.g. negatively regulated genes; ↓TR−T3 changes to ↓↓TR+T3) are discussed separately, below. We conclude that wild-type TRs possess a mix of hormone-independent and hormone-reversible repressive properties, with the latter largely absent for the HCC-mutants.
Figure 3. Certain target genes that are down-regulated by TRs in the absence of T3 are derepressed by T3; others are not.
(a-e) Expression levels, minus or plus T3 treatment, of representative gene transcripts from the panel of genes identified in Figure 2. Microarray intensity signal values (mean + S.D., n = 3) are presented for HepG2 transformants bearing each of the TR expression vectors indicated. An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.
The HCC mutant TRs are not exclusively transcriptional repressors, but are also able to activate a limited set of target genes in the presence of T3
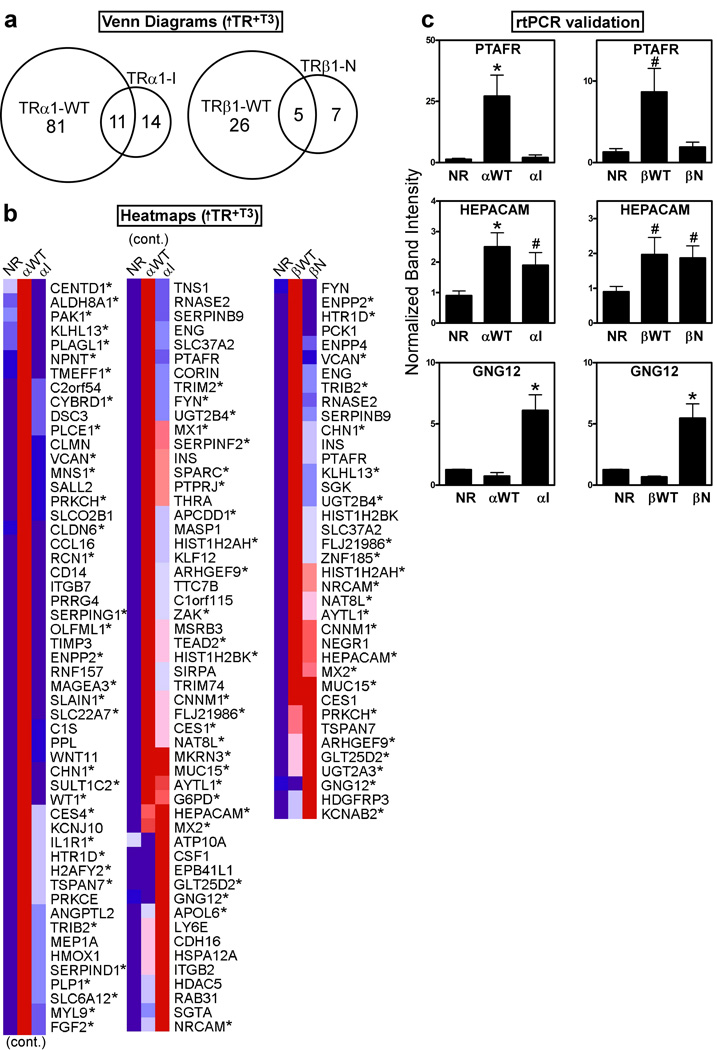
Although several of the genes characterized above were derepressed by T3, most were not activated by T3 above the empty vector controls. We therefore next searched for TR gene targets that were up-regulated above basal levels by T3 (i.e. ↑TR+T3) (Fig. 4a, 4b, and validated by rt-PCR, Fig. 4c). As expected, TRα1-I and TRβ1-N failed to induce many of the cellular genes induced by the corresponding wild-type receptors under these conditions (e.g. PTAFR, Fig. 4b and 4c). However, these mutants retained an unanticipated ability to activate several of the same genes activated by the wild-type TRs (e.g. HEPACAM, Fig. 4b, and by rt-PCR, Fig. 4c), and actually gained an ability to activate a novel set of target genes not regulated by the wild-type TRs (e.g. GNG12; Fig. 4b, and by rt-PCR, Fig. 4c).
Figure 4. The HCC mutant TRs are able to up-regulate a distinct subset of target genes in the presence of T3.
(a) Venn diagram of gene transcripts up-regulated in each TR transformant compared to the empty vector (NR) control, all in the presence of T3. HepG2 cells transformed with the TR alleles indicated, or with the empty plasmid control, were incubated with T3 for 6 hrs.; RNA was isolated, and used to probe the arrays. Transcripts up-regulated in each TR transformant compared to the empty vector control (NR) were identified using a Benjamini-Hochberg adjusted p-value of <0.05. (b) Heat map clustering of the genes from panel (a). For each gene, dark blue indicates lowest expression, dark red indicates highest expression, with intermediate values represented by lighter shades. These comparisons were plus T3; asterisks indicate genes that were also up-regulated in the absence of T3 (see Figure 7). (c) rtPCR on representative genes identified in panel (b), using gene-specific primers. The level of expression of each gene product was defined as “1” for the empty vector control in the absence of T3 (mean + S.E.M., n = 3). An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05; "#" indicates a P value ≤ 0.1.
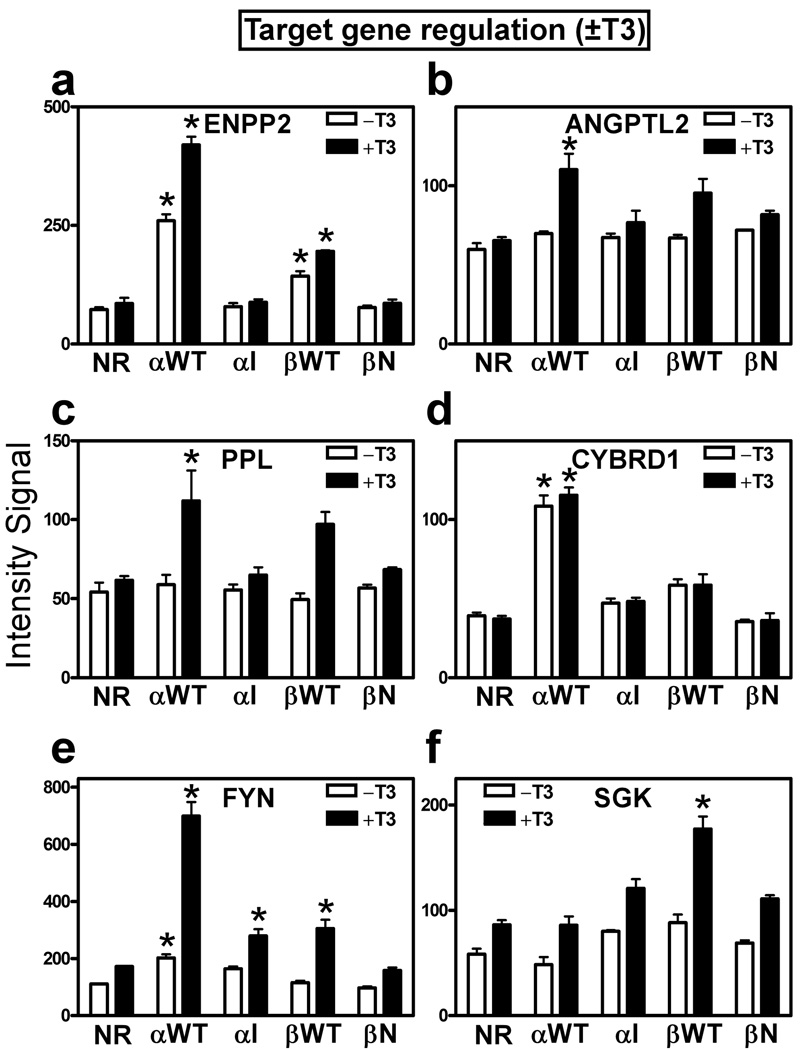
Further comparison of this same panel of genes in the absence as well as in the presence of T3 revealed more information (Fig. 5). As expected, many of the genes regulated by the wild-type receptors were expressed at higher levels in the presence of T3 than in its absence. This grouping included genes activated by receptor in the absence of T3, but expressed at still higher levels in the presence of T3 (i.e. ↑TR−T3 changes to ↑↑TR+T3), such as ENPP2 (Fig. 5a). It also included genes expressed at basal levels (i.e. equal to empty vector controls) in the absence of T3, and induced above basal only in the presence of T3 (i.e. basalTR−T3 changes to ↑TR+T3), such as ANGPTL2 and PPL (Fig. 5b, 5c). Yet other target genes were activated by the wild-type TRs essentially independent of T3 status (i.e. ↑TR−T3 remains ↑TR+T3), such as CYBD1 (Fig. 5d). Although most of the targets activated by the HCC-TR mutants fell into this constitutively activated category, a subset of HCC-TR target genes retained a residual hormone response and were more strongly activated plus T3 versus minus T3 (↑TR−T3 became ↑↑TR+T3; e.g. FYN and SGK, Fig. 5e, 5f). Representative genes were confirmed by rt-PCR (data not shown). Genes that were more strongly repressed in the presence of T3 (e.g. negatively regulated genes) are described below. We conclude that the HCC mutant TRs unexpectedly retain an ability to activate a subset of the genes induced by the corresponding wild-type receptor, and have also gained an ability to activate a novel, if limited, repertoire of additional genes not activated by the wild-type TRs.
Figure 5. Certain target genes are constitutively up-regulated by TRs independent of hormone status; others are up-regulated only in response to T3.
(a-f) Expression levels, minus or plus T3 treatment, of representative gene transcripts from the panel of genes identified in Figure 4. Microarray intensity signal values are presented (mean + S.D., n = 3) are presented for HepG2 transformants bearing each of the TR expression vectors indicated. An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.
HCC TR mutants regulate a narrow subset of the T3-repressed, negative-response genes that are targeted by the wild-type TRs
Genes possessing "negative response elements" operate reciprocally to the positive response genes described above: they are preferentially repressed in the presence of T3, or preferentially activated in the absence of T3. To capture these additional TR targets, our next step was to identify the genes repressed by mutant or wild-type receptors in the presence of T3 (↓TR+T3). This parsing found both overlap and divergence between the genes targeted by the wild-type versus the mutant TRs (Fig. 6). Notably most of the target genes repressed by TRs in the presence of T3 were previously identified as repressed by TRs in the absence of T3 (identified by asterisks in Fig. 2b and 6b); the majority of these are hormone non-responsive genes (↓TR−T3 remains ↓TR+T3), rather than negative-response genes per se (Figure 6 and Table S1) and have already been discussed. However, several genes (such as ANKRD1 and PROM1, Fig. 6c) were reproducibly more strongly repressed in the presence than in the absence of T3 (↓TR−T3 changed to ↓↓TR+T3). The mutant TRs exhibited an attenuated, although often still detectable, negative response to T3 on several of these same genes (Fig. 6c, Table S1).
Figure 6. The mutant and wild-type TRs negatively regulate distinct sets of target genes in response to T3.
(a) Venn diagram of gene transcripts down-regulated in each TR transformant compared to the empty vector (NR) control, all in the presence of T3. HepG2 cells transformed with the TR alleles indicated, or with the empty plasmid control, were incubated with T3 for 6 hrs.; RNA was isolated, and used to probe the arrays. Transcripts down-regulated in each TR transformant compared to the empty vector control (NR) were identified using a Benjamini-Hochberg adjusted p-value of <0.05. (b) Heat map clustering of the genes from panel (a). For each gene, dark blue indicates lowest expression, dark red indicates highest expression, with intermediate values represented by lighter shades. These comparisons were plus T3; asterisks indicate genes that were also down-regulated in the absence of T3 (see Figure 2). (c) Expression levels, minus or plus T3 treatment, of representative gene transcripts from the genes identified in panel (b). Microarray intensity signal values are presented (mean + S.D., n = 3). An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.
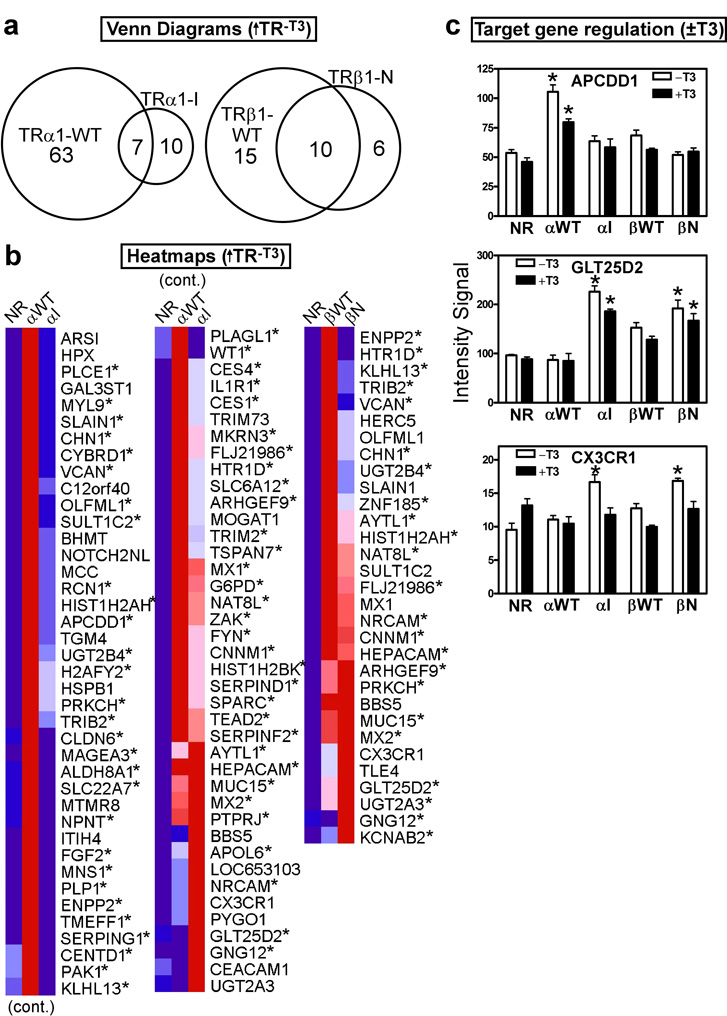
Our final comparison was to identify the genes that are selectively activated by receptors in the absence of T3 (↑TR−T3) (Fig. 7). A significant number of the targets previously identified as induced by the mutant or wild-type receptors in the presence of T3 were also induced in its absence (Figure 7 and Table S1). Many of these targets are not negative response genes per se, but rather fall into the two subclasses of positive response genes already discussed: those that are constitutively up-regulated in a hormone-independent fashion (↑TR−T3 remain ↑TR+T3), and those that are up-regulated in the absence of hormone, and further up-regulated in the presence of T3 (↑TR−T3 change to ↑↑TR+T3); these are indicated by asterisks in Fig. 4b and Fig. 7b. Although much more rare, genes that do fit our definition of negative regulation, preferentially up-regulated in the absence, relative to the presence, of T3 (↑↑TR−T3 changes to ↑TR+T3 or ↑TR−T3 changes to basalTR+T3), were also identified. These included genes regulated preferentially by the wild-type receptors (e.g. APCDD1, Fig. 7c) or preferentially by the mutant receptors (e.g. GLT25D2 and CX3CR1, Fig. 7c; TRα1-WT exhibited little or no regulation of these genes, whereas TRβ1-WT displayed an attenuated response compared to the two TR mutants).
Figure 7. The mutant and wild-type TRs positively regulate distinct sets of target genes in the absence of T3.
(a) Venn diagram of gene transcripts up-regulated in each TR transformant compared to the empty vector (NR) control, all in the absence of T3. HepG2 cells transformed with the TR alleles indicated, or with the empty plasmid control, were incubated in the absence of T3 (vehicle only) for 6 hrs.; RNA was isolated, and used to probe the arrays. Transcripts up-regulated in each TR transformant compared to the empty vector control (NR) were identified using a Benjamini-Hochberg adjusted p-value of <0.05. (b) Heat map clustering of the genes from panel (a). For each gene, dark blue indicates lowest expression, dark red indicates highest expression, with intermediate values represented by lighter shades. These comparisons were minus T3; asterisks indicate genes that were also up-regulated in the presence of T3 (see Figure 4). (c) Expression levels, minus or plus T3 treatment, of representative gene transcripts from the genes identified in panel (b). Microarray intensity signal values are presented (mean + S.D., n = 3). An "*" indicates that the difference between the TR transformant and the empty vector control was significant at a P value ≤ 0.05.
We conclude that the HCC TRα1-I and TRβ1-N mutants are generally restricted to a narrower panel of target genes and a narrower dynamic range of transcriptional output than are the wild-type TRs. Nonetheless, both TRα1-I and TRβ1-N have also acquired an ability to target novel genes not regulated by the wild-type receptors, and can regulate a subset of their target genes up or down at least as strongly as can their wild-type TR progenitors.
The perturbed gene expression mediated by the HCC-TR mutants parallel known alterations of the transcriptosome found in primary HCC tumors
At least seven of the genes we identify as expressed at higher levels in the HCC TR mutant transformants compared to wild-type (i.e. either activated by the mutant TRs but not by the wild-type TRs, or repressed by the wild-type TRs but not by the mutant TRs) have been previously identified as upregulated in primary HCC tumors (Table 1). At least eight of the genes we identify as expressed at lower levels in the TR HCC mutants compared to wild-type TRs (i.e. genes activated by wild-type TRs but not by the mutants) have been previously identified as down-regulated in primary HCC tumors (Table 1). A yet-additional panel of the target genes aberrantly regulated by the TR mutants studied here, although not previously associated with HCC, have been implicated in other forms of neoplasia (Table 2 and supplementary Table S3). These comparisons support the relevance of our cell culture analysis to the tumor transcriptosome in vivo, and suggest that the altered gene repertoire of the TR mutants likely contributes to the overall changes in gene expression characteristic of HCC.
Table 1.
Aberrantly expressed genes previously identified in HCC that are also aberrantly regulated by the HCC TR mutants.
| Gene Symbol |
Gene | Exp. in HCC |
Misreg. By |
|---|---|---|---|
| Repressed by wt, not mutant TR, or activated by mutant, not wt TR | |||
| CAP2 | CAP, adenylate cyclase-associated protein, 2 | ↑ | αI |
| CSF1 | colony stimulating factor 1 (macrophage) | ↑ | αI |
| GPC3 | glypican 3 | ↑ | αI, βN |
| HDAC1 | histone deacetylase 1 | ↑ | αI |
| HDAC5 | histone deacetylase 5 | ↑ | αI |
| PROM1 | prominin 1 | ↑ | αI, βN |
| TTR | transthyretin (prealbumin, amyloidosis type I) | ↑ | αI |
| Activated by wt, not mutant TR | |||
| C1S | complement component 1, s subcomponent | ↓ | αI |
| CES1 | carboxylesterase 1 (monocyte/macrophage serine esterase 1) | ↓ | αI |
| HMOX1 | heme oxygenase (decycling) 1 | ↓ | αI |
| PCK1 | phosphoenolpyruvate carboxykinase 1 | ↓ | βN |
| PLAGL1 | pleiomorphic adenoma gene-like 1 | ↓ | αI |
| PTAFR | platelet-activating factor receptor | ↓ | αI, βN |
| SERPINF2 | serpin peptidase inhibitor, clade F, member 2 | ↓ | αI |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | ↓ | αI |
Table 2.
Partial list of cancer implicated genes aberrantly regulated by the HCC TR mutants.
| Gene Symbol |
Gene | Implicated Role |
|---|---|---|
| AGR2 | anterior gradient 2 homolog | cancer cell survival |
| APOL6 | apolipoprotein l, 6 | cancer cell apoptosis |
| CEACAM1 | carcinoembryonic antigen-related cell adhesion molecule 1 | tumor suppressor |
| CSF1 | colony stimulating factor 1 | cancer progression |
| CX3CR1 | chemokine (c-x3-c motif) receptor 1 | cell migration/survival |
| DKK1 | dickkopf homolog 1 | tumor suppressor |
| ERRFI1 | erbb receptor feedback inhibitor 1 | tumor suppressor |
| FGF2 | fibroblast growth factor 2 | cell survival/metastasis |
| FYN | fyn oncogene related to src, fgr, yes | oncogene |
| GPC3 | glypican 3 | tumor suppressor |
| MCC | mutated in colorectal cancers | tumor suppressor |
| NR0B2 | nuclear receptor subfamily 0, group b, member 2 | tumor suppressor |
| NRCAM | neuronal cell adhesion molecule | cancer progression |
| PAK1 | p21/cdc42/rac1-activated kinase 1 | oncogene |
| PAWR | prkc, apoptosis, wt1, regulator | tumor suppressor |
| PLAGL1 | pleiomorphic adenoma gene-like 1 | tumor suppressor |
| PTPRJ | protein tyrosine phosphatase, receptor type, j | tumor suppressor |
| SGK | serum/glucocorticoid regulated kinase 1 | oncogene |
| TIMP3 | timp metallopeptidase inhibitor 3 | tumor suppressor |
| TMEFF1 | transmembrane protein w/ egf-like and two follistatin-like domains 1 | tumor suppressor |
| VCAN | chondroitin sulfate proteoglycan 2 (versican) | metastasis |
| WT1 | wilms tumor 1 | tumor suppressor |
DISCUSSION
TR mutants implicated in human HCC are selective antimorphs, repressing only a subset of the genes targeted by wild-type TRs
Dominant-negative mutant TRs play roles in RTH-Syndrome, avian erythroleukemia, HCC, RCCC and thyroid cancers (Chan and Privalsky, 2006; Graf and Beug, 1983; Kamiya et al., 2002; Lin et al., 1999; Puzianowska-Kuznicka et al., 2002; Rietveld et al., 2001; Yen and Cheng, 2003; Zenke et al., 1990). What accounts for the ability of certain of these TR mutants to induce neoplasia, whereas others are exclusively associated with endocrine disorders? Notably the mutant TRs isolated from human HCCs function not only as dominant-negative inhibitors on certain reporter genes but often have sustained additional mutations that alter their DNA recognition properties (Chan and Privalsky, 2006). A similar phenomenon was observed for the oncogenic v-Erb A allele and for certain RCCC-TR mutants (Bonde and Privalsky, 1990; Chen et al., 1993; Sharif and Privalsky, 1991; Ventura-Holman et al., 2008; Rosen and Privalsky, 2009). We hypothesized that these changes might confer an altered target gene repertoire that contributes to the ability of these receptors to induce neoplasia. Supporting this commonality, v-Erb A, when expressed in transgenic mice, induces HCC (Barlow et al., 1994).
We report here an analysis of target gene expression in HepG2 cells expressing wild-type or mutant TRs. This approach confirmed a key aspect of our working hypothesis: the TRα1-I and TRβ1-N mutants each repressed only a subset of the cell genes repressed by the corresponding wild-type receptors, and repression by the mutants was typically independent of hormone. This result is consistent with our reporter experiments and with in vitro data that the TRα1-I mutant binds to a more narrow set of natural and artificial DNA binding elements than did wt-TRα1 (supplementary Fig. S1, and Chan and Privalsky, 2006). Notably, our approach also identified an additional set of target genes repressed by the TRα1-I or TRβ1-N mutants but not by the wild-type receptors. TRα1-I binds better to at least one artificial DNA sequence than does TRα1-WT (supplementary Fig. S1 and Chan and Privalsky, 2006), and it is likely these HCC-TR cellular target genes possess related, mutant-specific response elements.
Therefore the mutations in the HCC-TR mutants have not simply narrowed their gene recognition properties, but have also shifted them to encompass novel targets. We favor the model that this altered target gene repertoire arises primarily from the altered DNA sequence recognition properties of these HCC-TR mutants; however we cannot exclude the possibility that alterations in transcriptional regulation after DNA binding may also contribute. For example, a coactivator required for activation of a specific subset of target genes may be recruited by the wild-type but not by the mutant TRs.
Unexpectedly, the HCC-TR mutants were able to activate transcription of a subset of the target genes induced by the wild-type receptors, plus an additional set of mutant-specific target genes
Our study also identified genes whose expression was increased by the introduction of a given TR. A subset of these genes were induced by the wild-type TRs more strongly in the presence than in the absence of T3, presumably reflecting the actions of the T3-dependent "AF-2" activation domain within the receptor hormone-binding domain (Yen, 2001). Interestingly, a second panel of target genes were constitutively up-regulated by the wild-type TRs independent of T3 status; this category may represent the actions of the TR N-terminal "AF-1" domain, which is known to mediate hormone-independent transcriptional activation (Yen, 2001). Our results support prior studies indicating that wild-type TRs exert a spectrum of possible responses to hormone ranging from derepression to activation (Yen, 2001).
Unexpectedly, a panel of genes were also induced above basal levels by introduction of the HCC mutant receptors. These mutant-activated genes were generally a subset of those activated by the wild-type TRs, although a few were unique to a given mutant receptor. Most were activated by the mutant receptors in a T3-independent fashion, perhaps through the actions of the AF-1 domain, which is untouched by the mutations in TRα1-I and TRβ1-N. However, a narrow subset of these target genes were activated more strongly by the HCC mutant receptors in the presence versus the absence of hormone. This is a particularly unexpected result for the TRα1-I mutant, which is largely incapable of activation in artificial reporter gene assays (Chan and Privalsky, 2006). However the TRα1-I mutant retains the ability to bind hormone in vitro, and it is possible that the remnants of the AF2 domain in this mutant are sufficient to regenerate a T3 response on certain promoters. Alternatively, endogenous TRs are present at low levels in the HepG2 cells (Chamba et al., 1996); it is possible that dimmers may form between mutant and endogenous TRs, with the endogenous TR partner conferring a T3-response on target genes determined by the mutant TR partner.
We conclude that the HCC TR mutants can behave both as constitutive repressors on certain target genes, and as constitutive activators on others. In much more restricted circumstances, these same mutants also display a residual T3 response on a specific subset of their target gene repertoire. This opens the possibility that certain aspects of the HCC neoplastic phenotype may be responsive to treatment with TR agonists or antagonists, an issue that should be explored further in future studies.
The altered gene repertoire of the HCC-TR mutants extends to negative response genes and reveals divergent modes of target gene recognition
An additional category of TR targets, denoted negative response genes, are expressed at lower levels in the presence versus the absence of T3, and have been implicated in the ability of wild-type TRs to suppress cell proliferation in the presence of T3. Although relatively few of these negative-response genes were detected by our array approach, we did find examples in all three possible categories: targets negatively regulated by T3 only in the wild-type TR transformants, only in the mutant TR transformants, or in both types of transformants. These results extent our prior observations using a prototypic negative response reporter derived from the collagenase promoter (Chan and Privalsky, 2006).
TRs are believed to be recruited to many negative response genes through protein-protein interactions with other transcription factors, rather than by direct DNA binding (Desbois et al., 1991; Sharif and Privalsky, 1992). Notably TRα1-I has a substitution in a highly conserved lysine (codon 74). K74 is located in the receptor DNA recognition helix and makes base specific contacts in the major groove of the DNA binding site (Rastinejad et al., 1995). As a result, it is not surprising that the TRα1-I mutant displays an altered recognition of genes regulated through direct receptor/DNA contacts (presumably the bulk of the positive-response genes identified here). However, K74 has been shown to also exert unusual functions on negative response genes. An artificial lysine to alanine substitution in TRα1 (K74A) converts the mutant TR from a negative response regulator of the collagenase promoter into a positive response regulator (Uht et al., 2004). It has been suggested that the wild-type lysine at codon 74 may contribute to the direction of the hormone response by sensing whether nuclear receptors are bound to DNA (a positive response gene) or not (a negative response gene) (Meyer et al., 1997; Starr et al., 1996). At least some of the alterations in cellular gene regulation observed in our current study may reflect the dual roles of this lysine both as a sensor and as a mediator of DNA binding.
The TRα1-I and TRβ1-N mutants have both convergent and divergent target repertories
Mutations in TRα1 and in TRβ1 occur separately in certain HCC tumors, indicating that aberrant regulation by either isoform may contribute to neoplasia (Lin et al., 1999). In fact, certain genes identified here, such as PROM1, were regulated by both wild-type receptor isoforms, but not by either mutant; conversely other genes that were targeted by both mutants, but by neither wild-type isoform, such as GNG12. These targets define a common mode of gene expression through which either mutant might independently contribute to establishment of the oncogenic phenotype. Nonetheless, approximately 47% of HCCs bear mutations in both TR isoforms, suggesting that mutations in TRα1 and TRβ1 can also work together to play complementary roles in a single tumor (Lin et al., 1999). In fact, other sets of the target genes characterized here were regulated by one or the other mutant but not by both, perhaps helping to explain how the two mutant TRs might be better than one in tumorigenesis.
What roles do these gene play in cancer initiation or progression? Wild-type TRs can act as inhibitors of tumor invasiveness, metastasis, and proliferation (Garcia-Silva and Aranda, 2004; Martinez-Iglesias et al., 2009); one role of these TR mutants, therefore, may be to interfere in a dominant-negative fashion with the antiproliferative properties of the wild-type TRs. Consistent with this proposal, our TRα1-WT transformants were inhibited for anchorage independent growth, whereas the TRα1-I transformants were not (Chan and Privalsky, 2006). Perhaps also reflecting the same phenomenon, we noted a slight bias against the wild-type TRs when generating our stable HepG2 transformants: 31% of the G418 clones were TRα1-WT positive and 34% were TRβ1-WT positive, whereas 45% were TRα1-I positive and 42% were TRβ1-N positive.
Studies on thyroid and pituitary neoplasia associated with mutant TR expression have implicated several specific pathways that contribute to tumor suppression by wild-type TRs and to tumor promotion by TR mutants, including alterations in Wnt signaling, cyclin D1 regulation, and phosphatidylinositol 3-kinase (PI3K) activity (Furumoto et al., 2005; Furuya et al., 2006; Miller et al., 2001). Our current study is consistent with this concept that the HCC-TR mutants function, in part, by counteracting the growth suppressive properties of the wild-type TRs. However, we also demonstrate that the HCC-TR mutants can regulate novel genes not recognized by the wild-type receptors, and may also operate through novel, pro-oncogenic pathways. A partial list of the genes we identify here as aberrantly regulated by the HCC-TR mutants, and which have been implicated previously in various aspects of oncogenic signaling, is provided in Table 1 and Table 2. Among these are tumor suppressors in the Wnt signaling pathway (DKK1 and WT1), known oncogenes (PAK1 and SGK) and genes involved in other features of cancer such as metastasis (VCAN) and survival (AGR2) (Lee et al., 2004; Mikheev et al., 2008; Ramachandran et al., 2008; Ricciardelli et al., 2009; Vadlamudi and Kumar, 2003; Vogelstein and Kinzler, 2004; Zhang et al., 2005). More analysis will be required to determine the precise role of these targets in HCC, their relationship to other cancers associated with mutant TRs, and their significance relative to the growth regulatory properties of the wild-type TRs.
Supplementary Material
ACKNOWLEDGMENTS
We are extremely grateful to Liming Liu for excellent technical assistance, Michael L. Goodson for expert assistance in analysis of the array data, and Elsie L. Campbell for helpful discussions.
Research Support: This work was supported by Public Health Service/National Cancer Institute award R37-CA53394 and by the UC Davis Cancer Center Gene Expression Resource (NCI P30-CA93373). I. H. C. was supported in part by a PHS pre-doctoral training award, T32-GM007377, from the National Institute of General Medical Sciences.
Footnotes
Supplementary information is available at Oncogene’s website.
REFERENCES
- Barlow C, Meister B, Lardelli M, Lendahl U, Vennstrom B. Thyroid abnormalities and hepatocellular carcinoma in mice transgenic for v-erbA. Embo J. 1994;13:4241–4250. doi: 10.1002/j.1460-2075.1994.tb06744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghagen H, Ragnhildstveit E, Krogsrud K, Thuestad G, Apriletti J, Saatcioglu F. Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element. J Biol Chem. 2002;277:49517–49522. doi: 10.1074/jbc.M209546200. [DOI] [PubMed] [Google Scholar]
- Bonde BG, Privalsky ML. Sequence-specific DNA binding by the v-erbA oncogene protein of avian erythroblastosis virus. J Virol. 1990;64:1314–1320. doi: 10.1128/jvi.64.3.1314-1320.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent GA. Tissue-specific actions of thyroid hormone: insights from animal models. Rev Endocr Metab Disord. 2000;1:27–33. doi: 10.1023/a:1010056202122. [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Paul BD, Fu L, Shi YB. Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol. 2006;145:1–19. doi: 10.1016/j.ygcen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Chamba A, Neuberger J, Strain A, Hopkins J, Sheppard MC, Franklyn JA. Expression and function of thyroid hormone receptor variants in normal and chronically diseased human liver. J Clin Endocrinol Metab. 1996;81:360–367. doi: 10.1210/jcem.81.1.8550778. [DOI] [PubMed] [Google Scholar]
- Chan IH, Privalsky ML. Thyroid hormone receptors mutated in liver cancer function as distorted antimorphs. Oncogene. 2006;25:3576–3588. doi: 10.1038/sj.onc.1209389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smit-McBride Z, Lewis S, Sharif M, Privalsky ML. Nuclear hormone receptors involved in neoplasia: erb A exhibits a novel DNA sequence specificity determined by amino acids outside of the zinc-finger domain. Mol Cell Biol. 1993;13:2366–2376. doi: 10.1128/mcb.13.4.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY. Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev Endocr Metab Disord. 2000;1:9–18. doi: 10.1023/a:1010052101214. [DOI] [PubMed] [Google Scholar]
- Cheng SY. Thyroid hormone receptor mutations in cancer. Mol Cell Endocrinol. 2003;213:23–30. doi: 10.1016/j.mce.2003.10.051. [DOI] [PubMed] [Google Scholar]
- Cheng SY. Thyroid hormone receptor mutations and disease: beyond thyroid hormone resistance. Trends Endocrinol Metab. 2005;16:176–182. doi: 10.1016/j.tem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Damm K, Thompson CC, Evans RM. Protein encoded by v-erbA functions as a thyroid-hormone receptor antagonist. Nature. 1989;339:593–597. doi: 10.1038/339593a0. [DOI] [PubMed] [Google Scholar]
- Darby IA, Bouhnik J, Coezy ED, Corvol P. Thyroid hormone receptors and stimulation of angiotensinogen production in HepG2 cells. In Vitro Cell Dev Biol. 1991;27:21–24. doi: 10.1007/BF02630890. [DOI] [PubMed] [Google Scholar]
- DeGroot LJ. Resistance to thyroid hormone. Ann Intern Med. 1996;125:623. doi: 10.7326/0003-4819-125-7-199610010-00022. [DOI] [PubMed] [Google Scholar]
- Desbois C, Aubert D, Legrand C, Pain B, Samarut J. A novel mechanism of action for v- ErbA: abrogation of the inactivation of transcription factor AP-1 by retinoic acid and thyroid hormone receptors. Cell. 1991;67:731–740. doi: 10.1016/0092-8674(91)90068-a. [DOI] [PubMed] [Google Scholar]
- Flamant F, Baxter JD, Forrest D, Refetoff S, Samuels H, Scanlan TS, et al. International Union of Pharmacology. LIX. The pharmacology and classification of the nuclear receptor superfamily: thyroid hormone receptors. Pharmacol Rev. 2006;58:705–711. doi: 10.1124/pr.58.4.3. [DOI] [PubMed] [Google Scholar]
- Furumoto H, Ying H, Chandramouli GV, Zhao L, Walker RL, Meltzer PS, et al. An unliganded thyroid hormone beta receptor activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol Cell Biol. 2005;25:124–135. doi: 10.1128/MCB.25.1.124-135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya F, Hanover JA, Cheng SY. Activation of phosphatidylinositol 3-kinase signaling by a mutant thyroid hormone beta receptor. Proc Natl Acad Sci U S A. 2006;103:1780–1785. doi: 10.1073/pnas.0510849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Silva S, Aranda A. The thyroid hormone receptor is a suppressor of ras-mediated transcription, proliferation, and transformation. Mol Cell Biol. 2004;24:7514–7523. doi: 10.1128/MCB.24.17.7514-7523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sancho JM, Garcia V, Bonilla F, Munoz A. Thyroid hormone receptors/THR genes in human cancer. Cancer Lett. 2003;192:121–132. doi: 10.1016/s0304-3835(02)00614-6. [DOI] [PubMed] [Google Scholar]
- Graf T, Beug H. Role of the v-erbA and v-erbB oncogenes of avian erythroblastosis virus in erythroid cell transformation. Cell. 1983;34:7–9. doi: 10.1016/0092-8674(83)90130-7. [DOI] [PubMed] [Google Scholar]
- Harvey CB, Williams GR. Mechanism of thyroid hormone action. Thyroid. 2002;12:441–446. doi: 10.1089/105072502760143791. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Puzianowska-Kuznicka M, McPhie P, Nauman J, Cheng SY, Nauman A. Expression of mutant thyroid hormone nuclear receptors is associated with human renal clear cell carcinoma. Carcinogenesis. 2002;23:25–33. doi: 10.1093/carcin/23.1.25. [DOI] [PubMed] [Google Scholar]
- Katz RW, Koenig RJ. Nonbiased identification of DNA sequences that bind thyroid hormone receptor alpha 1 with high affinity. J Biol Chem. 1993;268:19392–19397. [PubMed] [Google Scholar]
- Kopp P, Kitajima K, Jameson JL. Syndrome of resistance to thyroid hormone: insights into thyroid hormone action. Proc Soc Exp Biol Med. 1996;211:49–61. doi: 10.3181/00379727-211-43951. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Thyroid hormone action: a binding contract. J Clin Invest. 2003;112:497–499. doi: 10.1172/JCI19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, He B, You L, Xu Z, Mazieres J, Reguart N, et al. Dickkopf-1 antagonizes Wnt signaling independent of beta-catenin in human mesothelioma. Biochem Biophys Res Commun. 2004;323:1246–1250. doi: 10.1016/j.bbrc.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Lin KH, Shieh HY, Chen SL, Hsu HC. Expression of mutant thyroid hormone nuclear receptors in human hepatocellular carcinoma cells. Mol Carcinog. 1999;26:53–61. doi: 10.1002/(sici)1098-2744(199909)26:1<53::aid-mc7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Lin KH, Zhu XG, Hsu HC, Chen SL, Shieh HY, Chen ST, et al. Dominant negative activity of mutant thyroid hormone alpha1 receptors from patients with hepatocellular carcinoma. Endocrinology. 1997;138:5308–5315. doi: 10.1210/endo.138.12.5625. [DOI] [PubMed] [Google Scholar]
- Lin KH, Zhu XG, Shieh HY, Hsu HC, Chen ST, McPhie P, et al. Identification of naturally occurring dominant negative mutants of thyroid hormone alpha 1 and beta 1 receptors in a human hepatocellular carcinoma cell line. Endocrinology. 1996;137:4073–4081. doi: 10.1210/endo.137.10.8828459. [DOI] [PubMed] [Google Scholar]
- Martinez-Iglesias O, Garcia-Silva S, Tenbaum SP, Regadera J, Larcher F, Paramio JM, et al. Thyroid hormone receptor beta1 acts as a potent suppressor of tumor invasiveness and metastasis. Cancer Res. 2009;69:501–509. doi: 10.1158/0008-5472.CAN-08-2198. [DOI] [PubMed] [Google Scholar]
- Matsushita A, Sasaki S, Kashiwabara Y, Nagayama K, Ohba K, Iwaki H, et al. Essential role of GATA2 in the negative regulation of thyrotropin beta gene by thyroid hormone and its receptors. Mol Endocrinol. 2007;21:865–884. doi: 10.1210/me.2006-0208. [DOI] [PubMed] [Google Scholar]
- Meyer T, Starr DB, Carlstedt-Duke J. The rat glucocorticoid receptor mutant K461A differentiates between two different mechanisms of transrepression. J Biol Chem. 1997;272:21090–21095. doi: 10.1074/jbc.272.34.21090. [DOI] [PubMed] [Google Scholar]
- Mikheev AM, Mikheeva SA, Maxwell JP, Rivo JV, Rostomily R, Swisshelm K, et al. Dickkopf-1 mediated tumor suppression in human breast carcinoma cells. Breast Cancer Res Treat. 2008;112:263–273. doi: 10.1007/s10549-007-9867-2. [DOI] [PubMed] [Google Scholar]
- Miller LD, Park KS, Guo QM, Alkharouf NW, Malek RL, Lee NH, et al. Silencing of Wnt signaling and activation of multiple metabolic pathways in response to thyroid hormone-stimulated cell proliferation. Mol Cell Biol. 2001;21:6626–6639. doi: 10.1128/MCB.21.19.6626-6639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y. Multiple isoforms of thyroid hormone receptor: an analysis of their relative contribution in mediating thyroid hormone action. Nagoya J Med Sci. 1998;61:103–115. [PubMed] [Google Scholar]
- Nygard M, Wahlstrom GM, Gustafsson MV, Tokumoto YM, Bondesson M. Hormone-dependent repression of the E2F-1 gene by thyroid hormone receptors. Mol Endocrinol. 2003;17:79–92. doi: 10.1210/me.2002-0107. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. v-erb A, nuclear hormone receptors, and oncogenesis. Biochim Biophys Acta. 1992;1114:51–62. doi: 10.1016/0304-419x(92)90006-k. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. Thryoid hormone receptors, coregulators, and disease. London: World Scientific Publishing, Ltd.; 2008. [Google Scholar]
- Puzianowska-Kuznicka M, Krystyniak A, Madej A, Cheng SY, Nauman J. Functionally impaired TR mutants are present in thyroid papillary cancer. J Clin Endocrinol Metab. 2002;87:1120–1128. doi: 10.1210/jcem.87.3.8296. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Arumugam T, Wang H, Logsdon CD. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 2008;68:7811–7818. doi: 10.1158/0008-5472.CAN-08-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- Refetoff S. Resistance to thyroid hormone. Clin Lab Med. 1993;13:563–581. [PubMed] [Google Scholar]
- Refetoff S, Weiss RE, Usala SJ. The syndromes of resistance to thyroid hormone. Endocr Rev. 1993;14:348–399. doi: 10.1210/edrv-14-3-348. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev. 2009;28:233–245. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- Rietveld LE, Caldenhoven E, Stunnenberg HG. Avian erythroleukemia: a model for corepressor function in cancer. Oncogene. 2001;20:3100–3109. doi: 10.1038/sj.onc.1204335. [DOI] [PubMed] [Google Scholar]
- Rosen MD, Privalsky ML. Thyroid hormone receptors involved in renal clear cell carcinoma alter corepressor release and reveal helix 12 as a key determinant of corepressor specificity. Mol Endocrinol. 2009 doi: 10.1210/me.2009-0126. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, et al. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature. 1986;324:635–640. doi: 10.1038/324635a0. [DOI] [PubMed] [Google Scholar]
- Sharif M, Privalsky ML. v-erbA oncogene function in neoplasia correlates with its ability to repress retinoic acid receptor action. Cell. 1991;66:885–893. doi: 10.1016/0092-8674(91)90435-2. [DOI] [PubMed] [Google Scholar]
- Sharif M, Privalsky ML. V-erbA and c-erbA proteins enhance transcriptional activation by c-jun. Oncogene. 1992;7:953–960. [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Starr DB, Matsui W, Thomas JR, Yamamoto KR. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- Tagami T, Madison LD, Nagaya T, Jameson JL. Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol. 1997;17:2642–2648. doi: 10.1128/mcb.17.5.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault A, Ogbonna G, Adeli K. Thyroid hormone modulates apolipoprotein B gene expression in HepG2 cells. Biochem Biophys Res Commun. 1992;186:617–623. doi: 10.1016/0006-291x(92)90791-i. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Fondell JD. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm. 2004;68:93–122. doi: 10.1016/S0083-6729(04)68003-4. [DOI] [PubMed] [Google Scholar]
- Uht RM, Webb P, Nguyen P, Price RH, Jr, Valentine C, Favre H, et al. A conserved lysine in the estrogen receptor DNA binding domain regulates ligand activation profiles at AP-1 sites, possibly by controlling interactions with a modulating repressor. Nucl Recept. 2004;2:2. doi: 10.1186/1478-1336-2-2. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Kumar R. P21-activated kinases in human cancer. Cancer Metastasis Rev. 2003;22:385–393. doi: 10.1023/a:1023729130497. [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Mamoon A, Subauste JS. Modulation of expression of RA-regulated genes by the oncoprotein v-erbA. Gene. 2008;425:23–27. doi: 10.1016/j.gene.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM. The c-erb-A gene encodes a thyroid hormone receptor. Nature. 1986;324:641–646. doi: 10.1038/324641a0. [DOI] [PubMed] [Google Scholar]
- Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- Yen PM. Molecular basis of resistance to thyroid hormone. Trends Endocrinol Metab. 2003;14:327–333. doi: 10.1016/s1043-2760(03)00114-0. [DOI] [PubMed] [Google Scholar]
- Yen PM, Cheng SY. Germline and somatic thyroid hormone receptor mutations in man. J Endocrinol Invest. 2003;26:780–787. doi: 10.1007/BF03347365. [DOI] [PubMed] [Google Scholar]
- Zenke M, Munoz A, Sap J, Vennstrom B, Beug H. v-erbA oncogene activation entails the loss of hormone-dependent regulator activity of c-erbA. Cell. 1990;61:1035–1049. doi: 10.1016/0092-8674(90)90068-p. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui R, Cheng X, Du J. Antiapoptotic effect of serum and glucocorticoid-inducible protein kinase is mediated by novel mechanism activating I{kappa}B kinase. Cancer Res. 2005;65:457–464. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.