Abstract
Gastrin-17 (G17) processing intermediates bind to non-CCK receptors which mediate growth of the colonic mucosa but also the formation and development of colonic cancers. In previous studies, we removed the C-terminal region of G17 to form G17(1-12) and considerably shorter C-terminally amidated and non-amidated analogs. Peptides as short as G17(1-4) continued to bind to a single site on DLD-1 human colonic carcinoma cells, while only the G17(1-6)-NH2 and G17(1-12) peptides retained the ability to activate the receptor and stimulate cell proliferation in vitro. In this report, we studied the structure of these analogs, using a combination of ECD and VCD spectroscopy and replica exchange Molecular Dynamics (REMD) simulations in water, TFE, and membrane-mimicking environments, in order to determine preferred conformations that may have importance in promoting the biological activities. Mostly random meander structures, punctuated by a β-turn at residues 1–4, were found in most peptides by REMD simulations. G17(1-3)-NH2, which cannot form a β-turn, failed to bind the non-CCK receptor, suggesting the importance of this feature for binding. Additionally, the β-turn appeared more frequently longer sequences, perhaps explaining the higher affinity of the non-CCK receptor for these peptides seen previously. Finally, C-terminally amidated peptides generally showed greater formation of turn structure than their non-amidated counterparts as shown ECD spectra, suggesting the importance of peptide length in stabilizing turn structure in N-terminal sequences, and perhaps explaining the ability of G17(1-6)-NH2 to activate the non-CCK receptor where as the non-amidated G17(1-6) and shorter peptides do not.
1. Introduction
The mature, amidated peptide, gastrin-17 (G17; pGlu-Gly-Pro-Trp-Leu-Glu-Glu-Glu-Glu-Glu-Ala-Tyr-Gly-Trp-Met-Asp-Phe-NH2) is considered to have a role in the development and growth of gastric cancers. Strong evidence suggests that processing intermediates of G17, progastrin and G17-Gly, are more likely to be responsible for the growth and development of colonic cancers. G17 produced in the stomach does not arrive in the circulation in the colon in sufficient concentration to promote cancer growth in the colon [9, 22, 23, 43, 48, 54], and CCK2 receptors, which mediate the growth effects of G17 in the stomach, are largely not present in the colon [4, 17, 18, 19, 50, 52]. G17 intermediates are secreted far more frequently in the colon than G17 [11,29,40] and have been shown to stimulate growth of the colonic mucosa of rats in vitro and in vivo [5, 12, 30, 44, 54, 56] as well as proliferation of normal and neoplastic colonic cells which do not express CCK2 receptors [6, 26, 31, 55, 58].
Several groups have demonstrated the existence of previously unknown receptors with nanomolar and micromolar affinity for G17-Gly in primary tissues and cultured cell lines, which may mediate the effects of this peptide [25, 27, 38, 55, 58, 64, 68]. In previous work, this group confirmed the existence of nanomolar and micromolar affinity receptors on the DLD-1 and HT-29 colon cancer cell lines [2]. It was also demonstrated that the N-terminal region of G17, G17(1-12), was able to stimulate the proliferation of HT-29 cells, showing that the C-terminal tetrapeptide (Trp-Met-Asp-Phe-NH2) of G17 which is essential for binding and activation of the CCK2 receptor is not necessary for binding and activation of the putative G17-Gly growth promoting receptor [3]. Furthermore, we have shown [13] that 1, G17(1-12) binds to high and low affinity receptors on DLD-1 cells as does [Leu15]G17-Gly and promotes the proliferation of such cells; 2, further excision of the sequence Glu7-Tyr12 still gives a peptide, G17(1-6)-NH2, which binds, albeit with low affinity, and activates the putative growth promoting receptor on DLD-1 cells; 3, shorter N-terminal analogs with and without C-terminal amidation, including G17(1-6), bind with decreasing affinity than that of G17(1-6)-NH2 [15]. Sequences shorter than G17(1-4) fail to stimulate proliferation of DLD-1 cells.
The secondary structure of the N-terminal region of G17 has not been well studied, as most groups have focused on the C-terminal region of the peptide [1, 21, 34, 35, 36, 37, 39, 45, 46, 49, 59, 60, 62, 63, 67]. In this study the structure of N-terminal analogs, as well as the N-terminal to central G17(1-12) was examined using a combination of electronic and vibrational circular dichroism spectroscopy (ECD and VCD) and Molecular Dynamics simulations, in order to determine which structural features may play a role in binding and/or activation of the putative G17-Gly growth-promoting receptor.
2. Experimental Procedures
2.1 Materials
2.1.1 Solid phase peptide synthesis resins and amino acids
Rink Amide AM, Fmoc-Trp(Boc)-Wang, and Fmoc-Tyr(OtBu)-Wang resins were from NovaBioChem (San Diego, CA, USA). Fmoc-Glu(OtBu)-Wang and Fmoc-Leu-Wang resins were from Advanced ChemTech (Louisville, KY, USA). H-Tyr(OtBu)-HMPB-ChemMatrix resin was from Matrix Innovations (Montreal, Quebec, Canada). Fmoc-Gly-OH, Fmoc-Leu-OH, Fmoc-Trp(Boc)-OH, Fmoc-Pro-OH, Fmoc-Ala-OH, and pGlu-OH were from Advanced ChemTech. Fmoc-Glu(OtBu)-OH and Fmoc-Tyr(OtBu)-OH were from NovaBioChem.
2.1.2 Peptide synthesis, cleavage, and purification reagents
DMF, DMSO, NMP, DCM, acetone, methanol, ammonium bicarbonate, acetonitrile, sodium hydroxide, and sodium chloride were from Fisher (Fair Lawn, NJ, USA). DIEA and HBTU were from Chem-Impex (Wood Dale, IL). TIS, thioanisole, EDT, piperidine, and diethyl ether were from Sigma-Aldrich (Milwaukee, WI, USA). TFA was from Sigma-Aldrich (peptide synthesis grade), Chem-Impex (peptide synthesis grade), and Pierce (Rockford, IL) (HPLC grade).
2.1.3 ECD and VCD spectroscopy reagents
Na2HPO4 was from Fisher. D2O, DCl, DMSO-d6, TFE-d, TFE, and SDS were from Sigma-Aldrich.
2.2 Methods
2.2.1 N-terminal G17 analog synthesis, cleavage and purification
Synthesis, cleavage, and purification of all analogs was conducted as described previously [15]. The identities of all purified peptides were verified by mass spectrometry using a Perkin Elmer ESI-MS quadrupole mass spectrometer.
2.2.2 ECD spectroscopy
ECD spectra were measured using a Jasco J-810 spectropolarimeter at 20 °C, at a scanning speed of 100 nm/min and a pitch of 0.1 nm. Peptides (200 mM – 800 mM) were dissolved in either nanopure water, 15 mM Na2HPO4 solution containing 100 mM NaCl (PBS), TFE, TFE and H2O mixtures, or 80 mM SDS and their spectra were recorded using either 0.5 mm or 0.1 mm path length cells. Spectra in the far UV region were obtained by averaging data from 20 scans.
Peptide concentrations were determined by comparing of the HPLC peak size with a standard curve obtained using a 20 residue random sequence peptide containing all of the natural amino acids and having a molecular weight of 2339 Da [61].
Secondary structure content was calculated by analysis of ECD spectra the CDSSTR program [57] available on the DICHROWEB web-site [32].
2.2.3 VCD spectroscopy
All measurements were carried out using a 75 μm path length CaF2 cell on a BOMEM-Biotools Chiralir Fourier Transform VCD spectrometer (Elmhurst, IL, USA). Purified peptides were first lyophilized in 0.1 mM DCl/D2O to remove TFA salts from cleavage, as TFA absorbs strongly in the amide I and amide I′ region of the IR spectra at 1650 cm−1. These peptides were then dissolved at a concentration of 10–20 mg/mL in either 80 mM SDS in D2O, TFE-d1/DMSO-d6 (95:5, v/v) mixture, neat DMSO-d6, or 15 mM Na2HPO4 in D2O containing 100 mM NaCl. All spectra were measured at 20 °C with a resolution of 8 cm−1. Data were collected in 8 one-hour-long blocks with 2250 VCD scans and 113 IR scans per block. Spectra were corrected by subtracting the background spectra obtained for each of the respective solvents. VCD and IR spectra were normalized by dividing each value by the highest peak absorbance in the amide I′ region of the IR spectrum, giving an Amax of 1.0 in the amide I′ region. The VCD spectrum, thus, is directly expressed as ΔA/A at Amax [14]. In plotting VCD and IR data, all spectra were set to zero A and A, respectively, at 1800 cm−1.
2.2.4 REMD simulations
MD simulations of structures of peptides were performed in the presence of either H2O, DMSO or TFE. To begin, simulated annealing calculations, using an in-house perl script [33], were used to generate energy minimized starting conformations of each peptide using the OPLS-AA/L [20] force field as implemented in the TINKER v4.2 program package. Each peptide structure, starting from a fully extended conformation, was rapidly heated to 1050 K for 2 ps and then exponentially cooled to 50 K in 2 ps. This process was repeated 1000 times. The starting conformation for each cycle was the annealed conformation from the previous cycle. The 1000 annealed conformations were then energy-minimized to a final gradient of 0.0001 kcal/mol/Å2 [20]. The lowest energy conformation for each peptide was then modified by removal of aliphatic hydrogens for use with the united-atom GROMOS96 force field to provide the starting conformation for MD simulation.
REMD simulations were performed with the GROMACS package [65] using the GROMOS96 force field with the 53a6 parameter set [41, 42]. Each starting structure was converted to GROMACS structure format and then placed in a 125 nm3 cubic box. The peptide was then immersed in explicit solvent molecules (either SPC water, DMSO or TFE). The systems were energy-minimized using the steepest descent method. To neutralize negative charges, solvent molecules at the largest negative electrostatic potential were replaced with Na+ ions and the system was energy-minimized again. A 1 ns 300 K NVT (constant number of molecules, volume and temperature) simulation was performed with the peptide positionally restrained but the solvent free to move, in order to both homogenize solvent density and generate initial velocities for solvent molecules.
An NPT (constant number of molecules, pressure and temperature) REMD simulation was performed for each peptide. Run-input files were generated for replicas at 20 temperatures (K): 277.7, 283.1, 288.6, 294.3, 300, 305.6, 311.8, 317.9, 324.1, 330.6, 336.9, 343.4, 350.1, 356.9, 363.9, 371, 378.2, 385.6, 393.1, and 400.8. A 2 fs time step was used for integration; a non-bonded interactions list was updated every 10 steps; to avoid the effect of cutoff on long range electrostatic interactions, non-bonded interactions were treated by the twin-range method. 0.8 nm was used for the short range cutoff, 1.4 nm was used for the long range electrostatic cutoff, and 1.4 nm was used for the long range van der Waals cutoff. A reaction-field correction was used for long range electrostatic interactions, and an energy dispersion correction was used for energy and pressure. The LINCS constraint algorithm, as a quantum harmonic oscillator, was used to maintain correct bond lengths [24]. In cases where the LINCS algorithm failed, the SHAKE algorithm was used [51]. The peptide, solvent, and ions were coupled to separate temperature baths with a relaxation constant of 0.1 ps, and the peptide and solvent were coupled to a pressure bath with a relaxation constant of 1 ps. The dielectric constants and isothermal compressibility values of each of the solvents were included in the respective simulation parameters. The simulations were 200.1 ns long (the first 0.1 ns being an equilibration period) under the same conditions as for the respective standard simulation, except for G17(1-12), when simulations were 100.1 ns. Replica exchange was attempted every 1000 steps. When the simulation was complete, the 20 replica trajectories were demultiplexed using a perl script provided in the GROMACS package to generate a single trajectory file. Following completion of the simulation, calculation of secondary structures from the peptide trajectory was performed using the DSSP method [28], and a cluster analysis of the peptide trajectory, based on the RMSD of the peptide backbone, was performed using the gromos method as described by Daura and associates [16].
3. Results
3.1 Far UV ECD spectroscopy
3.1.1 G17(1-12)
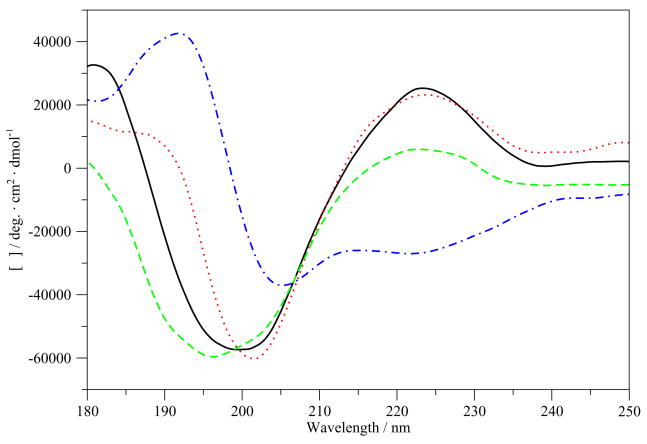
CDSSTR secondary structure analyses of ECD spectra of 200 μM G17(1-12) obtained using 0.05 cm pathlength cell are summarized in Table 1. Figure 1 shows the averaged ECD scans of G17(1-12) in various of solvents. CDSSTR analysis showed that the structure of the peptide is about 35% helical (or type I/III β-turns, which give similar spectra to those of 310-helices) or other turn structure in water and about 46% helical/turn in PBS, with a large percentage of unordered and β-sheet structures. Helical/turn propensity is substantially increased in SDS micelles or an aqueous solution of TFE. The peptide is 82% helix/turn in a 100% TFE solution.
Table 1.
CDSSTR analysis of CD spectra of G17(1-12) in various solvents.
| Peptide | Solvent | Helix/turn | β-strand/bridge | Unordered | Total |
|---|---|---|---|---|---|
| G17(1-12) | H2O | 0.39 | 0.33 | 0.28 | 1 |
| 15 mM Na2HPO4/100 mM NaCl | 0.46 | 0.20 | 0.34 | 1 | |
| 80 mM SDS | 0.53 | 0.25 | 0.22 | 1 | |
| TFE | 0.82 | 0.09 | 0.09 | 1 |
Figure 1.
ECD spectra of G17(1-12) in various solvents. –, H2O; …, Na2HPO4/NaCl; - - -, SDS;; -.-.-, TFE.
3.1.2 G17(1-6)-NH2 and G17(1-6)
CDSSTR secondary structure analyses of ECD spectra of 300 μM G17(1-6)-NH2 and 1.5 mM G17(1-6) obtained using 0.05 cm and 0.01 cm, respectively, pathlength cells are shown in Table 2. Figures 1S and 2S (see supplementary material) show the averaged ECD scans in various of solvents. In the case of the G17(1-6)-NH2, helical/turn structure increases in SDS and TFE, although not as much as does for G17(1-12). For G17(1-6), helix/turn formation also increases upon dissolution in TFE and in SDS, however, the increase is smaller than for the amidated peptide.
Table 2.
CDSSTR analysis of CD spectra of G17(1-6)-NH2 and G17(1-6) in various solvents.
| Peptide | Solvent | Helix/turn | β-strand/bridge | Unordered | Total |
|---|---|---|---|---|---|
| G17(1-6)-NH2 | H2O | 0.38 | 0.28 | 0.34 | 1 |
| 15 mM Na2HPO4/100 mM Nacl | 0.32 | 0.35 | 0.33 | 1 | |
| 80 mM SDS | 0.59 | 0.19 | 0.24 | 1 | |
| TFE | 0.57 | 0.23 | 0.20 | 1 | |
| G17(1-6) | H2O | 0.32 | 0.37 | 0.31 | 1 |
| 15 mM Na2HPO4/100 mM NaCl | 0.39 | 0.35 | 0.26 | 1 | |
| 80 mM SDS | 0.48 | 0.24 | 0.28 | 1 | |
| TFE | 0.44 | 0.29 | 0.27 | 1 | |
3.5.3 G17(1-5)-NH2 and G17(1-5)
CDSSTR secondary structure analyses of ECD spectra of 400 μM G17(1-5)-NH2 and G17(1-5) obtained using 0.05 cm pathlength cells are shown in Table 3. Figures 3S and 4S (see supplementary material) show the averaged ECD scans. G17(1-5)-NH2 shows similar behavior to G17(1-6)-NH2, in that helix/turn structure increases in SDS and in TFE, although these increases are not nearly as substantial as with the G17(1-6) analogs or G17(1-12). The non-amidated peptide, unlike G17(1-6), shows no increase in helix/turn propensity in any tested solvent.
Table 3.
CDSSTR analysis of CD spectra of G17(1-5)-NH2 and G17(1-5) in various solvents.
| Peptide | Solvent | Helix/turn | β-strand/bridge | Unordered | Total |
|---|---|---|---|---|---|
| G17(1-5)-NH2 | H2O | 0.30 | 0.33 | 0.37 | 1 |
| 15 mM Na2HPO4/100 mM NaCl | 0.30 | 0.35 | 0.35 | 1 | |
| 80 mM SDS | 0.56 | 0.19 | 0.25 | 1 | |
| TFE | 0.40 | 0.32 | 0.28 | 1 | |
| G17(1-5) | H2O | 0.28 | 0.44 | 0.28 | 1 |
| 15 mM Na2HPO4/100 mM NaCl | 0.30 | 0.42 | 0.28 | 1 | |
| 80 mM SDS | 0.31 | 0.39 | 0.30 | 1 | |
| 100% TFE | 0.28 | 0.34 | 0.38 | 1 | |
3.1.4 G17(1-4)-NH2 and G17(1-4)
CDSSTR secondary structure analyses of ECD spectra of 500 μM G17(1-4)-NH2 and G17(1-4) obtained using 0.05 cm pathlength cells are shown in Table 4. Figures 5S and 6S (see supplementary material) show the averaged ECD scans. Only the amidated peptide shows a slightly increasing tendency towards helix/turn structure in SDS and in TFE.
Table 4.
CDSSTR analysis of CD spectra of G17(1-4)-NH2 and G17(1-4) in various solvents.
| Peptide | Solvent | Helix/turn | β-strand/bridge | Unordered | Total |
|---|---|---|---|---|---|
| G17(1-4)-NH2 | H2O | 0.25 | 0.41 | 0.33 | 0.99 |
| 15 mM Na2HPO4/100 mM NaCl | 0.23 | 0.46 | 0.31 | 1 | |
| 80 mM SDS | 0.33 | 0.44 | 0.33 | 1 | |
| TFE | 0.30 | 0.35 | 0.35 | 1 | |
| G17(1-4) | H2O | 0.22 | 0.36 | 0.39 | 0.97 |
| 15 mM Na2HPO4/100 mM NaCl | 0.24 | 0.47 | 0.29 | 1 | |
| 80 mM SDS | 0.23 | 0.51 | 0.26 | 1 | |
| TFE | 0.24 | 0.44 | 0.32 | 1 | |
3.2 VCD spectroscopy
3.2.1 G17(1-12)
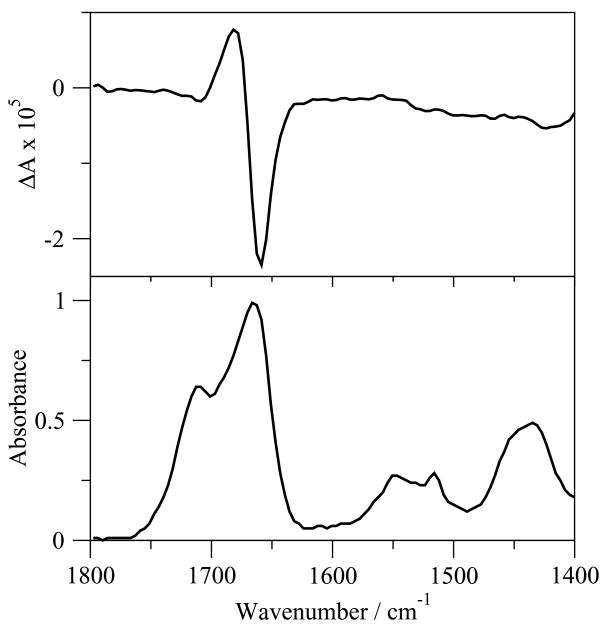
G17(1-12) was insufficiently soluble for VCD and IR measurements in D2O, PBS in D2O, TFE-d1/DMSO-d6 (95:5, v/v), and 80 mM SDS in D2O at concentrations as low as 5 mg/mL. The VCD spectrum of G17(1-12) in DMSO-d6 (Figure 2.) shows a negatively biased negative couplet in the amide I′ region, with a maximum at 1682 cm−1 and a minimum at 1658 cm−1. No clear maximum/minimum is seen in the amide II/II′ regions except at 1420 cm−1. In the IR spectrum, two overlapping peaks occur in the amide I′ region, with the larger peak occurring very close (1667 cm−1) to the midpoint of the couplet in the VCD spectrum (1670 cm−1), and the smaller peak occurring upfrequency at 1719 cm−1. Smaller, broader bands are also present in the amide II and II′ regions, with peaks at 1551 and 1516 cm−1 in the amide II region and a peak at 1433 cm−1 in the amide II′ region.
Figure 2.
VCD (top) and IR(bottom) spectra of G17(1-12) in DMSO-d6.
3.2.2 G17(1-6)-NH2 and G17(1-6)
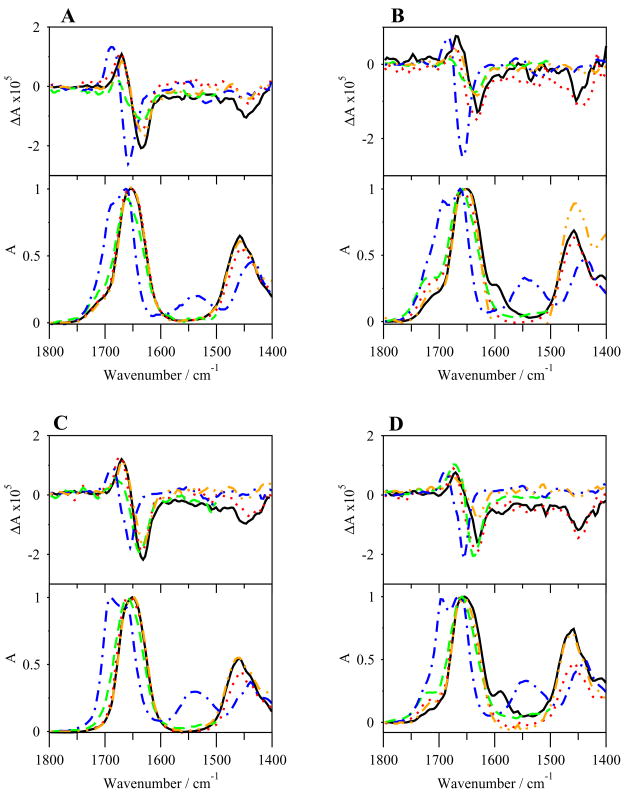
The VCD and IR spectra of G17(1-6)-NH2 in various solvents are shown in Figure 3A. The TFE-d1/DMSO-d6 (95:5, v/v) spectra are omitted below 1500 cm−1 because of strong solvent absorbance in that region. The VCD spectra in all solvents, except DMSO-d6, display negative couplets with negative bias of varying magnitudes in the amide I′ region, with maxima near 1670 cm−1 and minima near 1633 cm−1. The amide II/II′ regions are mostly devoid of bands, though a negative band appears near 1446 cm-1 in the amide II′ region of spectra in D2O, PBS and SDS. The VCD spectrum in DMSO-d6 also shows a negatively biased negative couplet in the amide I′ region, though the couplet is more strongly biased and shifted upfrequency, with a maximum at 1686 cm−1 and a minimum at 1654 cm−1. The IR spectra (Figure 3A) in the amide I′ region show a series of extensively overlapped peaks in all solvents. A maximum occurs in D2O, PBS, and SDS solutions at 1652 cm−1, but strong intensity shoulders are also apparent at 1640 cm−1 and 1664 cm−1. These are more apparent in TFE-d1/DMSO-d6 (95:5, v/v). High frequency shoulders exist near 1714 cm−1 in all solvents. The spectra are flat in the amide II region, but large peaks exist near 1460 cm−1 in the amide II′ region. The IR spectrum of the peptide in DMSO-d6 is different, showing a peak at 1663 cm−1 with a large shoulder at 1695 cm−1, and small broad bands in the amide II/II′ regions.
Figure 3.
VCD (top) and IR (bottom) spectra of G17(1-6)-NH2 (A), G17(1-6) (B), G17(1-5)-NH2 (C) and G17(1-5) (D) in various solvents. –, Na2HPO4/NaCl in D2O; …, SDS in D2O; - - -, TFE-d1/DMSO-d6; -.-.-, DMSO-d6; -..-, D2O.
The VCD spectra of G17(1-6) (Figure 3B) are similar to that of G17(1-6)-NH2. Negatively biased negative couplets are present in the amide I′ region, with maxima at 1670 cm−1 and minima at 1632 cm−1 (though the maximum in TFE-d1/DMSO-d6 (95:5, v/v) is shifted to 1681 cm−1), while the spectrum in DMSO-d6 has a shifted, highly biased couplet similar to that of G17(1-6)-NH2, with a maximum at 1684 cm−1 and minimum at 1656 cm−1. Negative peaks in the amide II′ region exist in the D2O, PBS and SDS solutions near to 1450 cm−1. The IR spectra (Figure 3B) in the amide I′ region closely resembles those of G17(1-6)-NH2 in all solvents, with a maximum at 1650 cm−1 and strongly overlapping shoulders at 1664 and 1641 cm−1 in D2O and PBS solutions. The spectrum for G17(1-6) in SDS is more similar to that in TFE-d1/DMSO-d6 (95:5, v/v) than for G17(1-6)-NH2, showing one peak near 1660 cm−1 and a shoulder near 1641 cm−1. High frequency shoulders are again seen, this time at 1718 cm−1 (with that in TFE-d1/DMSO-d6 (95:5, v/v) shifted to 1730 cm−1), and a low frequency shoulder at 1597 cm−1 in PBS. Spectra are flat in the amide II region, but large peaks are seen in the amide II′ region around 1458 cm−1. The IR spectrum of G17(1-6) in DMSO-d6 displays a double peak in the amide I′ region at 1663 and 1694 cm−1, and a small shoulder with broad bands in both the amide II and II′ regions, around 1544 and 1440 cm−1, respectively.
3.2.3 G17(1-5)-NH2 and G17(1-5)
The VCD and IR spectra of G17(1-5)-NH2 are shown in Figure 3C. The VCD spectra again show a negatively biased negative couplet in all solvents, with maxima at 1670 cm−1 and minima at 1633 cm−1. The spectrum in TFE-d1/DMSO-d6 (95:5, v/v) is slightly shifted upfrequency, with a maximum and minimum at 1679 and 1639 cm−1, respectively. The couplet in DMSO-d6 is shifted to a maximum and minimum of 1687 cm−1 and 1655 cm−1. Negative peaks exist in the amide II′ region at ~1447 cm−1 in D2O, PBS and SDS solutions, while a negative peak also exists in amide II region for the spectrum in DMSO-d6 solution. The IR spectra (Figure 3C) show a maximum at 1652 cm−1 in D2O and PBS solutions. High frequency shoulders are not as apparent here as for the G17(1-6) and G17(1-6)-NH2, but a shoulder appears at 1660 cm−1 and possibly also at 1641 cm−1 as well in these solvents. The peaks of the spectra in SDS and in TFE-d1/DMSO-d6 (95:5, v/v) are slightly shifted to 1659 cm−1, with noticeable shoulders at 1641 cm−1. No high frequency, low intensity shoulders are seen as in spectra of G17(1-6) and G17(1-6)-NH2. The amide II region is flat, while the amide II′ region contains a peak at 1460 cm−1. The spectrum in DMSO-d6, as before, shows a double peak with a well defined, high intensity shoulder at 1692 and 1663 cm−1. Again, broad positive bands appear in the amide II/II′ regions at 1540 cm−1 and 1440 cm−1.
The VCD and IR spectra of G17(1-5) are shown in Figure 3D. The VCD spectra in all solvent show a negatively biased negative couplet in the amide I′ region, with maxima near 1670 cm−1 and minima near 1631 cm−1, while in TFE-d1/DMSO-d6 (95:5, v/v) solution the minimum is shifted to 1638 cm−1. Negative peaks are seen in the amide II′ region in the SDS and PBS solutions. The amide I′ couplet in DMSO-d6 is once again shifted upfrequency, with a maximum at 1690 cm−1 and a minimum at 1656 cm−1. In the IR spectra (Figure 3D), maxima in the amide I′ region are seen at 1655 cm−1 in D2O and PBS solutions, with weak shoulders at 1639 cm−1. Peaks in SDS and TFE-d1/DMSO-d6 (95:5, v/v) solutions are shifted upfrequency to 1660 cm−1, with similar 1639 cm−1 shoulders. All spectra show high frequency shoulders at 1719 cm−1 (1729 cm−1 for TFE-d1/DMSO-d6 (95:5, v/v) solution), and the PBS solution spectrum has a low frequency shoulder at 1590 cm−1. The amide II region is flat in all solvents except DMSO-d6, but the amide II′ region has peaks at 1458 cm−1. The spectrum in DMSO-d6 again shows a double peak in the amide I′ region, with maxima at 1697 and 1664 cm−1. Additionally, the spectrum in DMSO-d6 shows broad bands in the amide II/II′ regions at 1546 cm−1 and 1439 cm−1, respectively.
3.2.4 G17(1-4)-NH2 and G17(1-4)
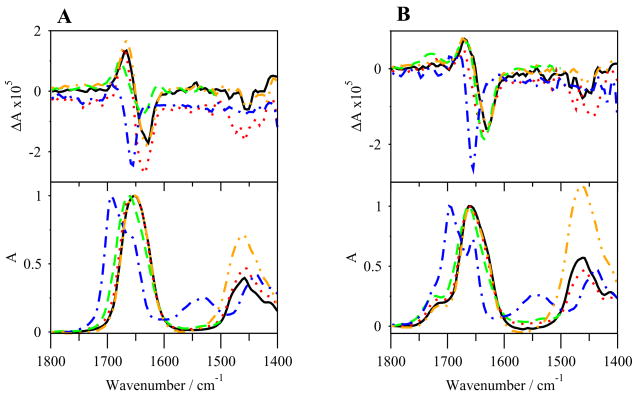
The VCD and IR spectra of G17(1-4)-NH2 are shown in Figure 4A. Negatively biased negative couplets are seen in the amide I′ region, with maxima at 1667 cm−1 and minima at 1629 cm−1 in D2O and PBS solutions. The couplets in SDS and TFE-d1/DMSO-d6 (95:5, v/v) are shifted slightly upfrequency, with the couplet in SDS having a maximum at 1673 cm−1 and a minimum of 1635 cm−1 and in the TFE-d1/DMSO-d6 (95:5, v/v) a maximum at 1679 cm−1 and a minimum at 1638 cm−1. The couplet in DMSO-d6 amide I′ is also shifted upfrequency, with a maximum at 1697 cm−1 and a minimum at 1656 cm−1. Small negative bands are also seen in the amide II′ region near 1458 cm−1 in all solvents except DMSO-d6. The IR spectra (Figure 4A) show amide I′ region maxima at 1652 cm−1 in D2O and in PBS solutions. Shoulders are extremely overlapped and only barely discerned. The peaks are shifted upfrequency in SDS and in TFE-d1/DMSO-d6 (95:5, v/v) to 1657 and 1663 cm−1, respectively, but only in the latter can a 1641 cm−1 shoulder be discerned. The non-DMSO-d6 spectra are once again flat in the amide II region, but positive bands are seen at 1461 cm−1 in the amide II′ region. The spectrum in DMSO-d6 once again shows a double peak in the amide I′ region, with maxima at 1695 cm−1 and 1656 cm−1, and bands in the amide II/II′ regions at 1533 cm−1 and 1439 cm−1, respectively.
Figure 4.
VCD (top) and IR (bottom) spectra of G17(1-4)-NH2 (A) and G17(1-4) (B) in various solvents. –, Na2HPO4/NaCl in D2O; …, SDS in D2O; - - -, TFE-d1/DMSO-d6; -.-.-, DMSO-d6; -..-, D2O.
The VCD and IR spectra of G17(1-4) are shown in Figure 4B. Negatively biased negative couplets are seen in the amide I′ region, with maxima at 1670 cm−1 and minima of 1629 cm−1 in D2O and PBS solutions. The couplets are shifted upfrequency in SDS and in TFE-d1/DMSO-d6 (95:5, v/v) with spectrum in SDS having a maximum at 1672 cm−1 and a minimum at 1634 cm−1, and in TFE-d1/DMSO-d6 (95:5, v/v) having a maximum at 1671 cm−1 and a minimum at 1636 cm−1. The spectrum in DMSO-d6 also has a strongly biased couplet shifted upfrequency with a maximum at 1679 cm−1 and a minimum at 1656 cm−1. Negative peaks are seen at different wavenumbers in the amide II′ region in all non DMSO-d6 solvents.
In the IR spectra (Figure 4B), the maximum for all solvents except DMSO-d6 is seen at 1661 cm−1, with a shoulder in all solvents at 1641 cm−1. High frequency, low intensity shoulders are seen at 1718 cm−1 for all solvents except TFE-d1/DMSO-d6 (95:5, v/v), which has a peak at 1727 cm−1, and peaks are seen in the amide II′ region near 1464 cm−1. In DMSO-d6, a double peak is again prominent, this one more separated than in the previous cases, with maxima at 1697 cm−1 and 1656 cm−1. Positive bands are also seen in the amide II/II′ regions at 1543 and 1440 cm−1, respectively.
3.3 REMD simulations
3.3.1 G17(1-12)
The DSSP analysis of the trajectory of G17(1-12) simulations in various solvents is shown in Figure 7S (see supplementary material). The simulations indicate a predominance of random meander structure throughout the peptide in all solvents. β-bend structure, defined as a region of high curvature in which the peptide backbone bends at least 70° (but lacks hydrogen bonding), is also prevalent from Pro3 to Glu10 in all solvents. The region containing the most ordered structure is pGlu1-Trp4, in which a turn structure is formed throughout the H2O and TFE simulations. Some β-bridge structure (a short region containing hydrogen bonds characteristic of β-sheet structure) is also seen at Gly2 and Ala11 in the standard water simulation.
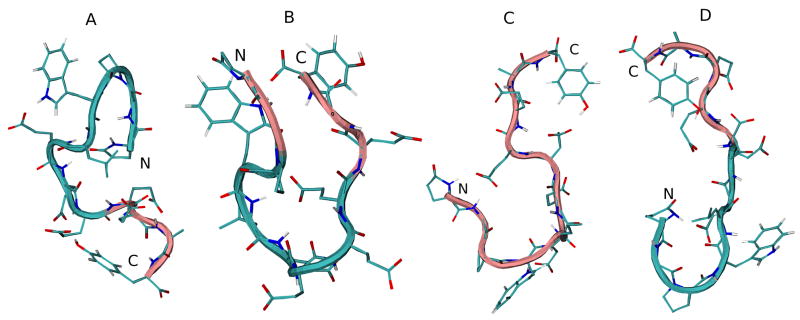
A cluster analysis was also performed to determine the most prevalent conformations of the peptide backbone over the duration of the simulation. Output trajectories of peptide backbones that differed in RMSD by less than 0.1 nm were grouped into clusters. In addition to the energy minimized initial structure, the backbone of the average middle structure for the most populated cluster for each REMD simulation is shown in Figure 5. The initial N-terminal β-turn structure migrates to the central part of the backbone in water, is lost completely in DMSO, and stays at the N-terminal region (pGlu1-Glu7) in TFE. Table 5 additionally shows the backbone dihedral angles of the middle structure of the most populated cluster for G17(1-12) in each solvent.
Figure 5.
Initial energy minimized structure and middle structures of the most populated cluster of G17(1-12) in various solvents: A, initial structure; B, H2O; C, DMSO; D, TFE. Backbone structure is color coded, with pink representing random meander structure and cyan representing β-turn structure. N and C indicate the N- and C-terminus, respectively.
Table 5.
Backbone dihedral angles of the representative structures of G17(1-12) of cluster 1 in various solvents.
| G17(1-12) | |||||||
|---|---|---|---|---|---|---|---|
| Residue | Solvent | Angle/degrees |
Residue | Solvent | Angle/degrees |
||
| φ | ψ | φ | ψ | ||||
| pGlu1 | H2O | 90.2 | 41.2 | Glu7 | H2O | −68.9 | −55.4 |
| DMSO | 110.8 | 72.8 | DMSO | −141.7 | 162.2 | ||
| TFE | 169.4 | 144.8 | TFE | −70.4 | 112.4 | ||
| Gly2 | H2O | 150.4 | −160.7 | Glu8 | H2O | −100.5 | 93.4 |
| DMSO | −76 | 125 | DMSO | −78.4 | 126.6 | ||
| TFE | 63.3 | −128.2 | TFE | −149.1 | 30.1 | ||
| Pro3 | H2O | −67.3 | 142.5 | Glu9 | H2O | −125.8 | 140.1 |
| DMSO | −62 | −45.1 | DMSO | −87.1 | −38 | ||
| TFE | −52.9 | 103.7 | TFE | −119.5 | 103.7 | ||
| Trp4 | H2O | −110.8 | −39.6 | Glu10 | H2O | −37.2 | 112.4 |
| DMSO | −127.4 | 162.2 | DMSO | −87.1 | 155.9 | ||
| TFE | 55.4 | 156.7 | TFE | −61.7 | −56.2 | ||
| Leu5 | H2O | −111.6 | −80.7 | Ala11 | H2O | −72 | 98.1 |
| DMSO | −68.9 | 87.9 | DMSO | −95.8 | 106 | ||
| TFE | −118.9 | 84.7 | TFE | −113.2 | 141.7 | ||
| Glu6 | H2O | −75.2 | 169.4 | Tyr12 | H2O | −101.3 | −56.2 |
| DMSO | −116.3 | −52.2 | DMSO | −148 | −47.5 | ||
| TFE | −85.5 | 81.5 | TFE | −117.9 | 147.2 | ||
3.3.2 G17(1-6)-NH2 and G17(1-6)
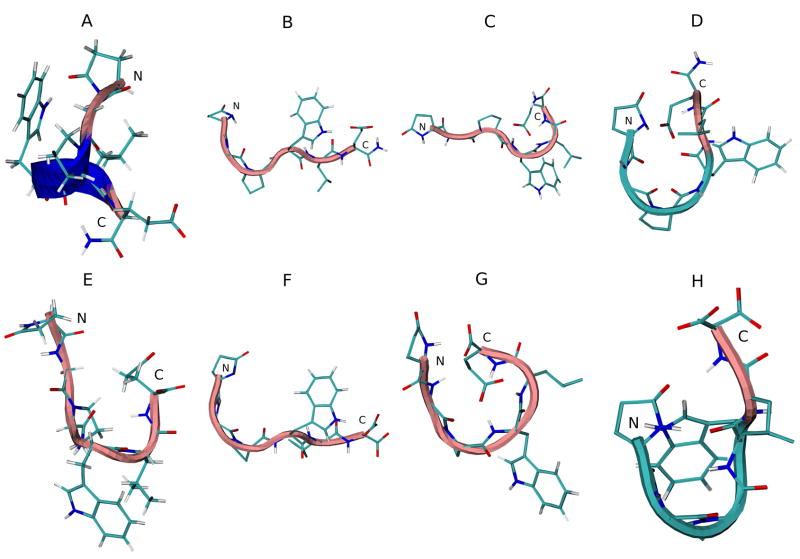
The DSSP analysis of the trajectories of the G17(1-6)-NH2 simulations in various solvents is seen in Figure 8S (see supplementary material). Random meander structure is seen in the outlying residues, while bend and turn structure is seen frequently in pGlu1-Trp4. α-helical and some 310-helical structure is momentarily seen from pGlu1-Glu6 early in both the standard and REMD water simulations. All three (β-turn, α-helix, and 310-helix) structures are again seen in the TFE simulations, where they last longer. The DMSO simulations are again mostly devoid of non-β-bend (hydrogen bonded) structures. Figure 6A–D shows the initial and average middle structures of the most populated cluster in each REMD simulation. The initial energy minimized structure shows 310-helical structure. No secondary structure is seen in water and DMSO, but the peptide in TFE shows β-turn structure at the N-terminal end.
Figure 6.
Initial energy minimized structure and middle structures of the most populated cluster of G17(1-6)-NH2 (A–D) and G17(1-6) (E–F) in various solvents: A and E, initial structure; B and F, H2O; C and G, DMSO; D and H, TFE. Backbone structure is color coded: blue, representing 310 helix; pink, random meander structure; cyan, representing β-turn structure. N and C indicate the N- and C-terminus, respectively.
Trajectories of MD simulations of G17(1-6) (Figure 9S, see supplementary material) do not vary greatly from those of G17(1-6)-NH2. Turn structure is prevalent from pGlu1-Trp4 in water and TFE, Some α-helical and 310-helical structure is present, though marginally less than in G17(1-6)-NH2. DMSO simulations contain more turn structure than for the amidated peptide, especially in Pro3-Glu6, and β-bend structure is more pronounced. However, in water and TFE, turn structure is diminished. Figure 6E–H shows the initial and average middle structures of the most populated cluster in each REMD simulation. Once again, no secondary structure is seen in water and DMSO, but β-turn structure is evident at the N-terminal end in the TFE simulation. Table 6 shows the backbone dihedral angles of the middle structure of the most populated cluster for both peptides in each solvent.
Table 6.
Backbone dihedral angles of the representative structures of G17(1-6)-NH2 and G17(1-6) of cluster 1 in various solvents.
| G17(1-6)-NH2 | G17(1-6) | ||||||
|---|---|---|---|---|---|---|---|
| Residue | Solvent | Angle/degrees |
Residue | Solvent | Angle/degrees |
||
| φ | ψ | φ | ψ | ||||
| pGlu1 | H2O | −171.7 | 139.3 | pGlu1 | H2O | 79.9 | 79.9 |
| DMSO | 65.7 | 73.6 | DMSO | 84.7 | 63.3 | ||
| TFE | −134.5 | 148 | TFE | −176.5 | 97.3 | ||
| Gly2 | H2O | 125.8 | −136.9 | Gly2 | H2O | 95 | −105.3 |
| DMSO | −109.2 | 140.9 | DMSO | −122.7 | −154.3 | ||
| TFE | 57.8 | −131.4 | TFE | 76.8 | −126.6 | ||
| Pro3 | H2O | −67.6 | 128.2 | Pro3 | H2O | −73.9 | 140.9 |
| DMSO | −65.2 | 147.2 | DMSO | −51.3 | −54.6 | ||
| TFE | −73.5 | 124.3 | TFE | −59 | −46.7 | ||
| Trp4 | H2O | −64.9 | 122.7 | Trp4 | H2O | −109.2 | 124.3 |
| DMSO | −104.5 | −77.6 | DMSO | −115.6 | −76 | ||
| TFE | 73.6 | 140.9 | TFE | −100.5 | 139.3 | ||
| Leu5 | H2O | −121.1 | 118.7 | Leu5 | H2O | −116.3 | 113.2 |
| DMSO | −112.4 | −21.4 | DMSO | −125.8 | 24.5 | ||
| TFE | −118.7 | 88.6 | TFE | −104.5 | 88.6 | ||
| Glu6 | H2O | −127.4 | 132.2 | Glu6 | H2O | −125.8 | 146.4 |
| DMSO | −63.3 | −51.4 | DMSO | −106.8 | 128.2 | ||
| TFE | −85.5 | 100.5 | TFE | −107.6 | 110 | ||
3.3.3 G17(1-5)-NH2 and G17(1-5)
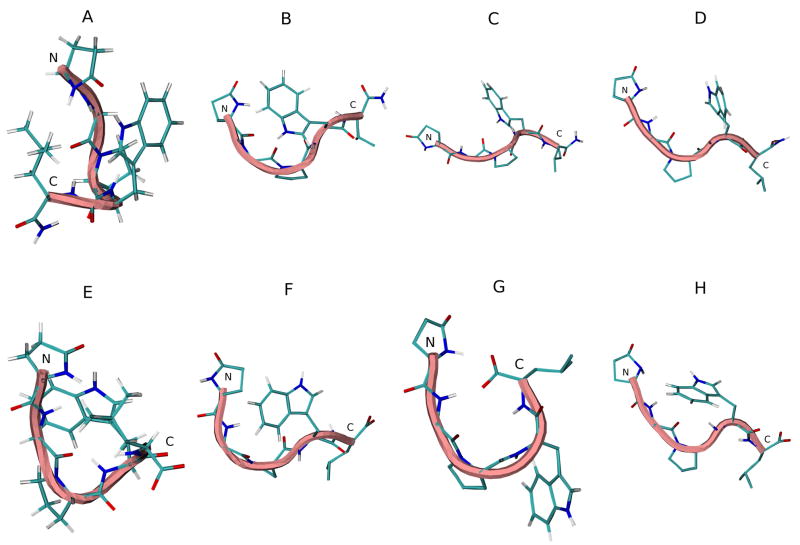
The DSSP analysis of the trajectories of G17(1-5)-NH2 simulations in various solvents is seen in Figure 10S (see supplementary material). Turn structure is prevalent in pGlu1-Trp4. No α-helical and very little 310-helical structure is seen in water or TFE as for the longer analog. Bend structure is restricted to Pro3. Simulation in DMSO again showed predominant random coil and bend structure, with greatly diminished turn structure by comparison with results of the water and TFE simulations. Figure 7A–D shows the initial and average middle structures of the most populated cluster in each REMD simulation. No turn structure is seen in any of the solvents.
Figure 7.
Initial energy minimized structure and middle structures of the most populated cluster of G17(1-5)-NH2 (A–D) and G17(1-5) (E–F) in various solvents: A and E, initial structure; B and F, H2O; C and G, DMSO; D and H, TFE. Backbone structure is color coded: blue, representing 310 helix; pink, random meander structure; cyan, representing β-turn structure. N and C indicate the N- and C-terminus, respectively.
The DSSP analysis of the trajectories of G17(1-5) simulations in various solvents (Figure 11S; see supplementary material) resemble those of the peptide amide, though the frequency of turn structure is somewhat diminished, as for G17(1-6)-NH2 and G17(1-6). Figure 7E–H shows the initial and average middle structures of the most populated cluster in each REMD simulation. Again, no turn structure is seen in any solvent. Table 7 shows the backbone dihedral angles of the middle structure of the most populated cluster of both peptides in each solvent.
Table 7.
Backbone dihedral angles of the representative structures of G17(1-5)-NH2 and G17(1-5) of cluster 1 in various solvents.
| G17(1-5)-NH2 | G17(1-5) | ||||||
|---|---|---|---|---|---|---|---|
| Residue | Solvent | Angle/degrees | Residue | Solvent | Angle/degrees | ||
| φ | ψ | φ | ψ | ||||
| pGlu1 | H2O | −167 | 110 | pGlu1 | H2O | −170.2 | −170.2 |
| DMSO | 98.1 | 128.2 | DMSO | 87.9 | 45.9 | ||
| TFE | 86.3 | 53 | TFE | −155.9 | 114 | ||
| Gly2 | H2O | 73.6 | −134.5 | Gly2 | H2O | −151.2 | −83.1 |
| DMSO | −126.6 | 171 | DMSO | −124.3 | −137.7 | ||
| TFE | −172.5 | −163.8 | TFE | 151.2 | −171 | ||
| Pro3 | H2O | −55.3 | 114 | Pro3 | H2O | −60.4 | 129 |
| DMSO | −75.5 | 125 | DMSO | −43.8 | −89.4 | ||
| TFE | −73.5 | 136.1 | TFE | −66 | 104.5 | ||
| Trp4 | H2O | −67.6 | 113.2 | Trp4 | H2O | −109.2 | 101.3 |
| DMSO | −79.1 | 112.4 | DMSO | −84.7 | −37.2 | ||
| TFE | −91.8 | 115.6 | TFE | −74.4 | 87.1 | ||
| Leu5 | H2O | −108.4 | 104.5 | Leu5 | H2O | −104.5 | −134.5 |
| DMSO | −127.4 | 118.7 | DMSO | −129.8 | −37.2 | ||
| TFE | −115.6 | 97.3 | TFE | −72.8 | 110.8 | ||
3.3.4 G17(1-4)-NH2 and G17(1-4)
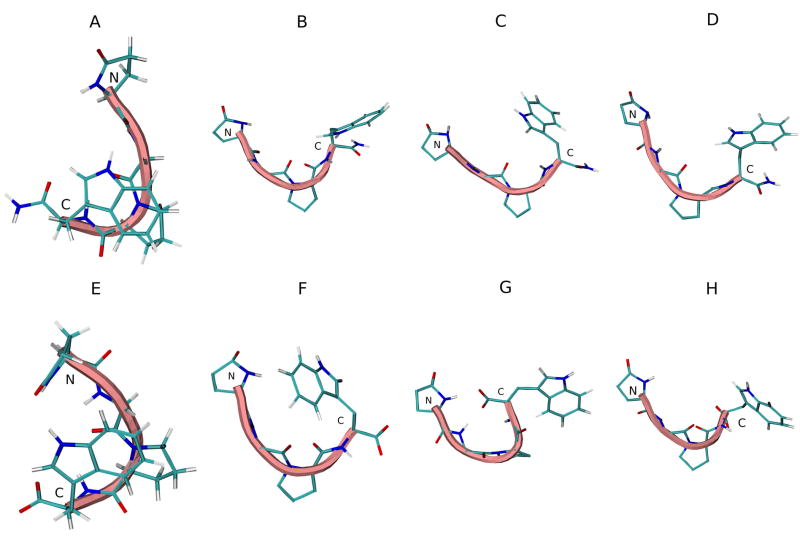
The DSSP analysis of the trajectories of G17(1-4)-NH2 simulations in various solvents are shown in Figure 12S (see supplementary material). β-bend noticeably vanishes from Pro3 in all solvents. Turn structure again exists from pGlu1-Trp4, though in diminished frequency. Figure 8A–D shows the initial and average middle structures of the most populated cluster in each REMD simulation. No turn structure is seen in any of the solvents.
Figure 8.
Initial energy minimized structure and middle structures of the most populated cluster of G17(1-4)-NH2 (A–D) and G17(1-4) (E–F) in various solvents: A and E, initial structure; B and F, H2O; C and G, DMSO; D and H, TFE. Backbone structure is color coded: blue, representing 310 helix; pink, random meander structure; cyan, representing β-turn structure. N and C indicate the N- and C-terminus, respectively.
In G17(1-4) (Figure 13S; see supplementary material), as in the longer analogs, turn structure is less frequent than in the peptide amide. Figure 8H-E shows the initial and average middle structures of the most populated cluster in each REMD simulation. Again, no turn structure is seen in any of the solvents. Table 8 shows the backbone dihedral angles of the middle structure of the most populated cluster of both peptides in each solvent.
Table 8.
Backbone dihedral angles of the representative structures of G17(1-4)-NH2 and G17(1-4) of cluster 1 in various solvents.
| G17(1-4)-NH2 | G17(1-4) | ||||||
|---|---|---|---|---|---|---|---|
| Residue | Solvent | Angle/degrees | Residue | Solvent | Angle/degrees |
||
| φ | ψ | φ | ψ | ||||
| pGlu1 | H2O | 174.9 | 156.7 | pGlu1 | H2O | 161.5 | 133 |
| DMSO | 48.3 | 146.4 | DMSO | 98.9 | 40.4 | ||
| TFE | −176.5 | 121.1 | TFE | 66.5 | 125 | ||
| Gly2 | H2O | 68.1 | −144.8 | Gly2 | H2O | 110.8 | −150.4 |
| DMSO | 133.8 | 139.3 | DMSO | −137.7 | −100.5 | ||
| TFE | 159.1 | −178.1 | TFE | 117.9 | −168.6 | ||
| Pro3 | H2O | −60 | 130.6 | Pro3 | H2O | −72.3 | 140.1 |
| DMSO | −76.7 | 107.6 | DMSO | −67.2 | −41.2 | ||
| TFE | −58.1 | 131.4 | TFE | −76.7 | 148 | ||
| Trp4 | H2O | −121.1 | 148 | Trp4 | H2O | −136.9 | 179.7 |
| DMSO | −83.9 | 116.3 | DMSO | −126.6 | −35.6 | ||
| TFE | −130.6 | 159.1 | TFE | −69.6 | −51.4 | ||
4. Discussion
G17(1-12)
Four classes of ECD spectra were defined by Woody [47]. The ECD spectra of G17(1-12) in all non-TFE solutions are obviously reminiscent of random coil spectra, with some hints of class C spectra (particularly the positive band near 180 nm), signifying type I/III β-turn structure (Figure 1). The spectrum of the peptide in TFE, however, is more classically class C, signifying helical or type I/III β-turn structure, in agreement with the CDSSTR results (Table 1). This is quite similar to spectrum of G17, which was seen in several studies to have minimal structure in aqueous solution or in DMSO [35, 45, 46, 62]. VCD spectra in solvents other than DMSO-d6 could not be obtained due to solubility problems in the comparable deuterated solvents. In DMSO-d6, the dual peaks seen in the IR spectrum (Figure 2) in the amide I′ region are attributed to the effects of DMSO as a proton acceptor and breaker of intramolecular hydrogen bonds, destabilizing secondary structure and freeing carbonyl groups, which do not form hydrogen bonds with DMSO, to shift their vibrations upfrequency [7, 53]. The small size of the peak shifted upfrequency to 1718 cm−1 compared to the peak for other analogs indicate less free carbonyl groups and thus relatively less unfolded structure in DMSO-d6. The shifted couplet in the VCD spectrum, as discussed below, is representative of mixed β-turn/random meander structure [16, 47].
The results of the REMD simulations in H2O echo those of the ECD spectra, showing largely random and β-bend structures throughout the peptide, with a β-turn structure near the N-terminus (Figure 7S) from residues 1–4 featuring hydrogen bond between the amide groups of pGlu1 and Trp4. A cluster analysis of the G17(1-12) REMD simulations (Figure 5) reveals that the middle structures of the most populated cluster (“cluster 1”) in water and TFE show β-turn structure in the middle portion and on the N-terminal end. Further examination of the cluster 1 dihedral angles of the Gly2 and Pro3 residues in H2O and TFE (Table 5) indicates that they do not match type I/III β-turn or other helical or turn structures, implying either a broader range type IV β-turn or random meander structure. Similarly, measurement of the distance between the alpha carbons of pGlu1 and Trp4 of the cluster 1 structure, which is another indicator of β-turn type, gives a distance of about 0.81 nm in water and 0.63 nm in TFE. This is much greater than the distance for type I/III turns but similar to that of type IV turns measured previously [10]. Immersing the peptide in TFE, however, does not significantly increase the β-turn structure content of the peptide in the REMD simulation from that of the peptide conformation in H2O (Figure 7S-C), in contrast to the ECD spectra and CDSSTR results (Figure 1; Table 1). It is possible that the peptide does not adopt the ordered structure seen in the ECD spectra during the simulations. With the exchange of the trajectories of neighboring replicas at different temperatures, REMD simulations are expected to increase the rate of sampling of the various conformations available to the peptide and therefore find the minimum energy conformation as quickly as possible. It is possible to use a variety of sets of replicas at different temperatures, which may increase or decrease the probability of the exchange of replicas over the course of the simulation and thus, either increase or decrease the sampling rate. An ensemble of replicas with a higher probability of exchange may be required in order to see the conformational change seen in the ECD spectra.
G17(1-6)-NH2 and G17(1-6)
Both G17(1-6)-NH2 and G17(1-6) show ECD spectra (Figure 1S and 2S) which display a mixture of unordered and class C features, and a tendency towards greater helix/turn structure in SDS and in TFE. The CDSSTR results, however, show that G17(1-6)-NH2 has a greater percentage of helix/turn structure in these solvents than does G17(1-6) (Table 2). In lieu of other structural and binding differences, if indeed these conformational features represent the bioactive conformation of G17(1-6)-NH2, then they may be why G17(1-6)-NH2 stimulates proliferation while G17(1-6) does not [13, 15].
The VCD spectra of the two peptides are quite similar and reflect the structural features revealed by ECD spectra. The negatively biased negative couplet in the amide I′ region, as well as the major peak in the IR spectra, indicates the presence of random meander structure of the peptide backbone in D2O, PBS, SDS micelles, and in TFE-d1/DMSO-d6 (95:5, v/v) (Figures 3A–B). The high and low frequency shoulders seen in the amide I′ region of the IR spectra seen near 1660 cm−1 and 1640 cm−1 are indicative of either extended (β-sheet) or mixed β-turn structures [16, 47]. While both meander and turn structures are indicated, the lack of an overlapping positive couplet in the amide I′ region VCD spectra, as observed by Borics and coworkers when studying disulfide bridged cyclic tetrapeptides, seems to rule out the possibility of type II β-turn structures [8]. β-turn structures are probably more frequent than β-sheet, as indicated by the intensity of the 1660 cm−1 shoulder/peak relative to the 1640 cm−1 shoulder, especially in TFE-d1/DMSO-d (95:5, v/v), DMSO-d6, and SDS micelles in both analogs, where the 1660 cm−1 peak/shoulder is prominent. The extremely high frequency shoulders seen in the region 1730–1710 cm−1 are difficult to interpret. They might be a complement to the 1640 cm−1 shoulders and represent β-sheet formation, though they are at higher frequency than the typical (1690–1670 cm−1).
The spectra of the peptides in DMSO-d6 are unique in showing shifted, negatively biased negative couplets in the VCD amide I′ region, and a double peak in the IR amide I′ region. The shifted, negatively biased couplet with extrema at ~1685 cm−1 and ~1655 cm−1 is represents β-turn/random meander structure [7, 53]. The double peak in the IR spectra is attributable to the nature of DMSO as a hydrogen bond acceptor; the solvent could break hydrogen bonds formed between the amide groups of the peptide backbone, and the resultant change in the stretching of backbone carbonyl groups would cause their vibration to be shifted upfrequency [53, 66]. The double peak in the spectra of both peptides probably represents a mixture of β-turn structures, some with broken hydrogen bonds and others with those still intact. This was the conclusion of Borics and coworkers, who saw similar VCD and IR spectra of tetrapeptides dissolved in DMSO-d6 [7].
DSSP analysis of REMD simulations (Figure 8S and 9S) reveals greater β-turn structure in residues pGlu1-Trp4 in TFE than in H2O simulations. The peptides in DMSO are largely devoid of this structure, probably because of the destabilization of backbone hydrogen bonding. Though the DSSP plots might not suggest much difference between the conformations shown by the water and TFE simulations, the cluster 1 structures for both peptides shows β-turn structure at residues 1–4 in TFE, but not in water, suggesting a more stable and frequent β-turn conformation in that solvent (Figure 6). However, an examination of the cluster 1 dihedral angles does not indicate type I/III turn or other defined turn structures, implying a type IV β-turn or random meander structure (Table 6). The distance between the alpha carbons of pGlu1 and Trp4 1 in the representative structure of cluster 1 is about 0.81 nm in water and 0.74 nm in TFE for G17(1-6)-NH2 and 0.77 nm in water and 0.58 nm in TFE for G17(1-6); again more indicative of type IV turn or random meander structure [10]. These data suggest that the β-turn structure in the N-terminal region represents the predominant conformation for binding the G17-Gly receptor. However, this does not explain the inability of the G17(1-6) analog to activate the receptor. Results from studies both of biological activity and structure of the two peptides are similar aside from the results of proliferation assays and ECD studies [13, 15]. The slightly greater tendency of G17(1-6)-NH2 to form helix/turn structure (as seen in the ECD studies) may be due to stabilization of the peptide structure by the C-terminal amide, and this structural difference, though not detectable by qualitative review of the VCD/IR spectra or REMD simulations, may account for the difference in activity.
G17(1-5)-NH2 and G17(1-5)
The ECD spectra of the G17(1-5)-NH2 and G17(1-5) show similar structural features (Figures 3S and 4S). The spectra of both peptides in water and in PBS indicate mostly unordered, random coil structure with hints of class C structure. Placing the peptides in the membrane-mimicking solvents, SDS and TFE, however, causes G17(1-5)-NH2 to exhibit stronger class C characteristics (particularly, a negative band at 220 nm), while G17(1-5) retains mostly unordered structure. This is seen in the CDSSTR results. The tendency towards helix/turn structure in the pentapeptide amide, however, is clearly not as strong as in G17(1-6)-NH2. The VCD spectra of the pentapeptides (Figures 3C–D) are once again indicative of unordered structure mixed with β-turn structure and possibly some extended (β-sheet) structure. Upon dissolution of the peptide in DMSO a shifted VCD couplet and IR double peak in the amide I′ region is observed, signifying H-bonded and non-H-bonded β-turn structures and unordered structure. These spectra for the two peptides are not appreciably different. The REMD simulations and cluster analyses (Figures 7, 10S and 11S), similarly, do not show major structural differences between the two peptides in structure, nor do they show a major increase in helix/turn structure when the peptides are placed in TFE. In both cases, β-turn structure is located in pGlu1-Trp4. The representative structure of cluster 1 does not demonstrate turn structure in any solvent, showing only random meander structure. The dihedral angles of Gly2 and Pro3 in the representative structure of cluster 1 indicate type IV β-turn or random meander structure in this region (Table 7). The distance between the alpha carbons of pGlu1 and Trp4 is 0.89 nm in water and 0.90 nm in TFE for G17(1-5)-NH2 and 0.77 nm in water and 0.91 nm in TFE for G17(1-5), which is not indicative of type I/III β-turn structure [10]. These peptides seem to form turn structures infrequently.
G17(1-4)-NH2 and G17(1-4)
G17(1-4)-NH2 and G17(1-4) display almost no more in turn structure in SDS or TFE-d1/DMSO-d6 (95:5, v/v) than in water or PBS in ECD studies (Figures 5S and 6S). Turn structure exists in residues pGlu1-Trp4, as indicated by the 1660 cm−1 peaks of the IR spectra and the shifted couplet/double peak in the DMSO-d6 VCD and IR spectra (Figures 4A–B), as well as by the MD simulations (Figures 12S and 13S) and the ECD spectra. However, turn structure only marginally increases in G17(1-4)-NH2 and not at all in G17(1-4), as shown by the ECD spectra and the results of the REMD simulations. The representative structure of cluster 1 does not indicate turn in the peptides (Table 8). The distance between the alpha carbons of pGlu1 and Trp4 of the representative structure of cluster 1 is 0.77 nm in water and 0.87 nm in TFE for G17(1-4)-NH2 and 0.8 nm in water and 0.84 nm in TFE for G17(1-4), again do not indicate type I/III β-turn [10]. These tetrapeptides, as well as the pentapeptides, can bind the receptor, most likely because they form some N-terminal β-turn structure, but they cannot activate the receptor, perhaps due to the lack of the glutamyl residue and C-terminal amide [15]. Also, the lesser tendency to form turn structures, as seen in the DSSP plots and the cluster 1 structures (Figure 8), probably accounts for the lower affinity of these short peptides for the G17-Gly receptor.
5. Conclusions
The failure of G17(1-3)-NH2 to bind the G17-Gly receptor indicates that the sequence pGlu1-Trp4 probably in a β-turn structure is required, though this is still inadequate for activation of the receptor. The hexapeptide G17(1-6)-NH2 but not the pentapeptides meets the structural requirement for stimulation of proliferation of DLD-1 cells. C-terminal capping with an amide group appears to provide some measure of structural stabilization. Significantly, binding and activation does not require the C-terminus of G17 or the central pentaglutamyl sequence of gastrin. The N-terminal tetra- and pentapeptides, however, may have sufficient binding activity function as core compounds for the development of antagonists acting on the G17-Gly receptors in colonic cancer.
Supplementary Material
Acknowledgments
This work was supported by NIH-INBRE grant (1 P20 RR16469) and the Carpenter Endowed Chair in Biochemistry, Creighton University.
Abbreviations
- Boc
butyloxycarbonyl
- CCK
cholecystokinin
- DCM
dichloromethane
- DIEA
N,N-diisopropylethylamine, DMF, N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- DSSP
Dictionary of Secondary Structure Prediction
- ECD
electronic circular dichroism
- EDT
1,2-ethanedithiol
- ESI-MS
electrospray ionization mass spectrometry
- Fmoc
9H-fluoren-9-yloxycarbonyl
- G17
gastrin-17
- G17-Gly
gastrin-17-Gly
- HBTU
O-benzotriazol-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- HPLC
high performance liquid chromatography
- IR
infrared
- NMP
N-methylpyrrolidinone
- OtBu
O-t-butyl
- pGlu
pyroglutamic acid
- REMD
replica exchange molecular dynamics
- RMSD
root mean square deviation
- SDS
sodium dodecyl sulfate
- TFA
trifluoroacetic acid
- TFE
2,2,2-trifluoroethanol
- TIS
triisopropylsilane
- VCD
vibrational circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abillon E, Chuong PPV, Fromageot P. Conformational calculations on gastrin C-terminal tetrapeptide. Int J Peptide Protein Res. 1981;17:480–485. doi: 10.1111/j.1399-3011.1981.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed S, Budai B, Heredi-Szabo K, Farkas J, Toth G, Murphy RF, et al. High and low affinity receptors mediate growth effects of gastrin and gastrin-Gly on DLD-1 human colonic carcinoma cells. FEBS Lett. 2004;556:199–203. doi: 10.1016/s0014-5793(03)01408-x. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed S, Murphy RF, Lovas S. Importance of N- and C-terminal regions of gastrin-Gly for preferential binding to high and low affinity gastrin-Gly receptors. Peptides. 2005;26:1207–1212. doi: 10.1016/j.peptides.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Aly A, Shulkes A, Baldwin GS. Gastrins, cholecystokinins and gastronintestinal cancer. Biochimica et Biophysica Acta. 2004;1704:1–10. doi: 10.1016/j.bbcan.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Aly A, Shulkes A, Baldwin GS. Short term infusion of glycine-extended (17) stimulates both proliferation and formation of aberrant crypt foci in rat colonic mucosa. Int J Cancer. 2001;94:307–313. doi: 10.1002/ijc.1483. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin GS. The role of gastrin and cholecystokinin in normal and neoplastic gastrointestinal growth. J Gastroenterol Hepatol. 1995;10:215–232. doi: 10.1111/j.1440-1746.1995.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 7.Borics A, Murphy RF, Lovas S. Conformational analysis of Ac-NPGQ-NH(2) by vibrational circular dichroism spectroscopy combined with molecular dynamics and quantum chemical calculations. Protein Pept Lett. 2007;14:353–359. doi: 10.2174/092986607780363907. [DOI] [PubMed] [Google Scholar]
- 8.Borics A, Murphy RF, Lovas S. Optical spectroscopic elucidation of β-turns in disulfide bridged cyclic tetrapeptides. Biopolymers. 2007;85:1–11. doi: 10.1002/bip.20593. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Destree M, Hakanson R, Willems G. Endogenous hypergastrinemia does not promote growth of colonic mucosa or of transplanted colon adenocarcinoma in rats. Eur J Gastroenterol Hepatol. 1998;10:293–299. doi: 10.1097/00042737-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Chou K-C. Prediction of tight turns and their types in proteins. Anal Biochem. 2000;286:1–16. doi: 10.1006/abio.2000.4757. [DOI] [PubMed] [Google Scholar]
- 11.Ciccotosto G, McLeish A, Hardy K, Shulkes A. Expression processing and secretion of gastrin in patients with colorectal carcinoma. Gastroenterology. 1995;109:1142–1153. doi: 10.1016/0016-5085(95)90572-3. [DOI] [PubMed] [Google Scholar]
- 12.Cobb S, Wood T, Ceci J, Varro A, Velasco M, Singh P. Intestinal expression of mutant and wild-type progastrin significantly increases colon carcinogenesis in response to azooxymethane in transgenic mice. Cancer. 2004;100:1311–1323. doi: 10.1002/cncr.20094. [DOI] [PubMed] [Google Scholar]
- 13.Copps J, Ahmed S, Murphy RF, Lovas S. Gastrin 1-6 promotes growth of colon cancer cells through non-CCK receptors. Peptides. 2007;28:632–635. doi: 10.1016/j.peptides.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Copps J, Murphy RF, Lovas S. Avian pancreatic polypeptide fragments refold to native aPP conformation when combined in solution: A CD and VCD study. Biopolymers. 2006;83:32–38. doi: 10.1002/bip.20524. [DOI] [PubMed] [Google Scholar]
- 15.Copps J, Murphy RF, Lovas S. Bioactivity of analogs of the N-terminal region of gastrin-17. Peptides. 2009 doi: 10.1016/j.peptides.2009.09.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: when simulation meets experiment. Angew Chem Int Ed. 1999;38:236–240. [Google Scholar]
- 17.Dufresne M, Seva C, Fourmy D. Cholecystokinin and gastrin receptors. Physiol Rev. 2006;86:805–847. doi: 10.1152/physrev.00014.2005. [DOI] [PubMed] [Google Scholar]
- 18.Fontana MG, Donato F, Villanacci V, Ghirardi M, Moneghini D, Di Betta E, et al. Inhibitory effect of a gastrin receptor antagonist, CR 2495, on 1,2-dimethylhydrazine-induced colorectal cancer in mice. Eur Surg Res. 1999;31:406–411. doi: 10.1159/000008719. [DOI] [PubMed] [Google Scholar]
- 19.Fontana MG, Moneghini D, Villanacci V, Donato F, Rindi G. Effect of cholecystokinin-B gastrin receptor blockade on chemically induced colon carcinogenesis in mice: follow up at 52 weeks. Digestion. 2002;65:35–40. doi: 10.1159/000051929. [DOI] [PubMed] [Google Scholar]
- 20.Kaminski GA, Friesner RA. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 21.Gohring W, Moroder L, Borin GF, Lobbia A, Bali JP, Wunsch E. Synthesis of peptides with gastrinlike activity. Studies on the structure-activity relationship of the natural hormone human little gastrin I. Hoppe Seylers Z Physiol Chem. 1984;365:83–94. doi: 10.1515/bchm2.1984.365.1.83. [DOI] [PubMed] [Google Scholar]
- 22.Hakanson R, Axelson J, Ekman R, Sundler F. Hypergastrinemia evoked by omprazole stimulates growth of gastric mucosa but not of pancreas or intestines in hamster, guinea pig and chicken. Regul Pept. 1988;23:105–115. doi: 10.1016/0167-0115(88)90426-0. [DOI] [PubMed] [Google Scholar]
- 23.Hakanson R, Blom H, Carlsson E, Larsson H, Ryberg B, Sundler F. Hypergastrinemia produces trophic effects in stomach but not in pancreas and intestine. Regul Pept. 1986;13:225–233. doi: 10.1016/0167-0115(86)90041-8. [DOI] [PubMed] [Google Scholar]
- 24.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 25.Higashide S, Gomez G, Greeley GH, Jr, Townsend CM, Jr, Thompson JC. Glycine-extended gastrin potentiates gastrin-stimulated gastric acid secretion in rats. Am J Physiol Gastrointest Liver Physiol. 1996;270:G220–224. doi: 10.1152/ajpgi.1996.270.1.G220. [DOI] [PubMed] [Google Scholar]
- 26.Hollande F, Imdahl A, Mantamadiotis T, Ciccotosto GD, Shulkes A, Baldwin GS. Glycine-extended gastrin acts as an autocrine growth factor in a nontransformed colon cell line. Gastroenterology. 1997;113:1576–1588. doi: 10.1053/gast.1997.v113.pm9352860. [DOI] [PubMed] [Google Scholar]
- 27.Imdahl A, Mantamadiotis T, Eggstein S, Farthmann EH, Baldwin GS. Expression of gastrin, gastrin/CCK-B and gastrin/CCK-C receptors in human colorectal carcinomas. J Cancer Res Clin Oncol. 1995;121:661–666. doi: 10.1007/BF01218524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 29.Kochman ML, Del Valle J, Dickinson CJ, Borland CR. Post-translational processing of gastrin in neoplastic human colonic tissues. Biochem Biophys Res Commun. 1992;189:1165–1169. doi: 10.1016/0006-291x(92)92326-s. [DOI] [PubMed] [Google Scholar]
- 30.Koh TJ, Dockray GJ, Varro A, Cahill RJ, Dangler CA, Fox JG, et al. Overexpression of glycine-extended gastrin in transgenic mice results in increased colonic proliferation. J Clin Invest. 1999;103:1119–1126. doi: 10.1172/JCI4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litvak DA, Hellmich MR, Iwase K, Evers BM, Martinez J, Amblard M, et al. JMV1155: a novel inhibitor of glycine-extended progastrin-mediated growth of a human colon cancer in vivo. Anticancer Res. 1999;19:45–49. [PubMed] [Google Scholar]
- 32.Lobley A, Wallace BA. DICHROWEB: a website for the analysis of protein secondary structure from circular dichroism spectra. Biophysical J. 2001;80:373a. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 33.Lovas S, Murphy RF. Molecular modeling of neuropeptides in methods in molecular biology. In: Irvine GB, Williams CH, editors. Neuropeptide Protocols. Vol. 73. Humana Press; Totowa: 1997. pp. 209–217. [DOI] [PubMed] [Google Scholar]
- 34.Mammi S, Foffani MT, Peggion E, Galleyrand JC, Bali JP, Simonetti M, et al. Conformational and biological properties of the Ala10 analogue of human des-Trp1, Nle12-minigastrin. Biochemistry. 1989;28:7182–7188. doi: 10.1021/bi00444a008. [DOI] [PubMed] [Google Scholar]
- 35.Mammi S, Goodman M, Peggion E, Foffani MT, Moroder L, Wuensch E. Conformational studies on gastrin related peptides by high resolution 1H-NMR. Int J Peptide Protein Res. 1986;27:145–152. doi: 10.1111/j.1399-3011.1986.tb01804.x. [DOI] [PubMed] [Google Scholar]
- 36.Mammi S, Mammi NJ, Foffani MT, Peggion E, Moroder L, Wunsch E. Conformation of human little gastrin and minigastrin analogs in surfactant solution. Biopolymers. 1987;26:S1–S10. doi: 10.1002/bip.360260005. [DOI] [PubMed] [Google Scholar]
- 37.Mammi S, Mammi NJ, Peggion E. Conformational studies of human des-Trp1, Nle12-minigastrin in water-trifluoroethanol mixtures by 1H NMR and circular dichroism. Biochemistry. 1988;27:1374–1379. doi: 10.1021/bi00404a043. [DOI] [PubMed] [Google Scholar]
- 38.Mauss S, Niederau C, Hengels KJ. Effects of gastrin, proglumide, loxiglumide and L-365,260 on growth of human colon carcinoma cells. Anticancer Res. 1994;14:215–220. [PubMed] [Google Scholar]
- 39.Morley JS, Tracy HJ, Gregory RA. Structure-function relationships in the active C-terminal tetrapeptide sequence of gastrin. Nature (London) 1965;207:1356–1359. doi: 10.1038/2071356a0. [DOI] [PubMed] [Google Scholar]
- 40.Nemeth J, Taylor B, Pauwels S, Varro A, Dockray GJ. Identification of progastrin derived peptides in colorectal carcinoma extracts. Gut. 1993;34:90–95. doi: 10.1136/gut.34.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oostenbrink C, Soares TA. Validation of the 53a6 GROMOS force field. Eur Biophys J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 42.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53a5 and 53a6. J Comp Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 43.Oscarson J, Veen H, Ross J, Malt R. Dimethylhydrazine induced colonic neoplasia: dissociation from endogenous gastrin levels. Surgery. 1982;91:525–530. [PubMed] [Google Scholar]
- 44.Ottewell PD, Varro A, Dockray GJ, Kirton CM, Watson AJ, Wang TC, et al. COOH-terminal 26-amino acid residues of progastrin are sufficient for stimulation of mitosis in murine colonic epithelium in vivo. Am J Physiol Gastrointest Liver Physiol. 2005;288:G541–G549. doi: 10.1152/ajpgi.00268.2004. [DOI] [PubMed] [Google Scholar]
- 45.Peggion E, Foffani MT, Wunsch E, Moroder L, Borin G, Goodman M, et al. Conformational Properties of Gastrin Fragments of Increasing Chain Length. Biopolymers. 1985;24:647–666. [Google Scholar]
- 46.Peggion E, Jaeger E, Knof S, Moroder L, Wuensch E. Conformational aspects of gastrin-related peptides: a circular dichroism study. Biopolymers. 1981;20:633–652. doi: 10.1002/bip.1981.360200402. [DOI] [PubMed] [Google Scholar]
- 47.Perczel A, Turns Hollosi M. In: Circular dichroism and the conformational analysis of biomolecules. Fasman GD, editor. Plenum Press; 1996. pp. 285–380. [Google Scholar]
- 48.Pinson DM, Havu N, Sztern MI, Mattsson H, Looney GA, Kimler BF, et al. Drug-induced hypergastrinemia: absence of trophic effects on colonic carcinoma in rats. Gastroenterology. 1995;108:1068–1074. doi: 10.1016/0016-5085(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 49.Previero A, Mourier G, Bali JP, Lignon MT, Moroder L. N alpha-glycosylgastrin-related peptides. Synthesis, characterization and biological activity. Hoppe Seylers Z Physiol Chem. 1982;363:813–818. doi: 10.1515/bchm2.1982.363.2.813. [DOI] [PubMed] [Google Scholar]
- 50.Reubi JC, Waser B, Schmassmann A, Laissue JA. Receptor autoradiographic evaluation of cholecystokinin, neurotensin, somatostatin and vasoactive intestinal peptide receptors in gastrointestinal adenocarcinoma samples: where are they really located? Int J Cancer. 1999;81:376–386. doi: 10.1002/(sici)1097-0215(19990505)81:3<376::aid-ijc11>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 51.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints; molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 52.Schmitz F, Otte JM, Stechele HU, Reimann B, Banasiewicz T, Folsch UR, et al. CCK-2/gastrin receptors in human colorectal cancer. Eur J Clin Investig. 2001;31:812–820. doi: 10.1046/j.1365-2362.2001.00870.x. [DOI] [PubMed] [Google Scholar]
- 53.Shanmugam G, Polavarapu PL, Gopinath D, Jayakumar R. The structure of antimicrobial pexiganan peptide in solution probed by Fourier transform infrared absorption, vibrational circular dichroism, and electronic circular dichroism spectroscopy. Biopolymers (Peptide Science) 2005;80:636–642. doi: 10.1002/bip.20132. [DOI] [PubMed] [Google Scholar]
- 54.Singh P, Velasco M, Given R, Wargovich M, Varro A, Wang TC. Mice overexpressing progastrin are predisposed for developing aberrant colonic crypt foci in response to AOM. Am J Physiol Gastroenterol Liver Physiol. 2000;278:G390–G399. doi: 10.1152/ajpgi.2000.278.3.G390. [DOI] [PubMed] [Google Scholar]
- 55.Singh P, Owlia A, Espeijo R, Dai B. Novel gastrin receptors mediate mitogenic effects of gastrin and processing intermediates of gastrin on Swiss 3T3 fibroblasts. Absence of detectable cholecystokinin CCK-A and CCK-B receptors. J Biol Chem. 1995;270:8429–8438. doi: 10.1074/jbc.270.15.8429. [DOI] [PubMed] [Google Scholar]
- 56.Singh P, Velasco M, Given R, Varro A, Wang TC. Progastrin expression predisposes mice to colon carcinomas and adenomas in response to a chemical carcinogen. Gastroenterology. 2000;119:162–171. doi: 10.1053/gast.2000.8527. [DOI] [PubMed] [Google Scholar]
- 57.Sreerama N, Woody RW. Estimation of protein secondary structure from CD spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 58.Stepan VM, Sawada M, Todisco A, Dickinson CJ. Glycine-extended gastrin exerts growth-promoting effects on human colon cancer cells. Mol Med. 1999;5:147–159. [PMC free article] [PubMed] [Google Scholar]
- 59.Stone SR, Giragossian C, Mierke DF, Jackson GE. Further evidence for a C-terminal structural motif in CCK2 receptor active peptide hormones. Peptides. 2007;28:2211–2222. doi: 10.1016/j.peptides.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Stone SR, Mierke DF, Jackson GE. Evidence for a C-terminal structural motif in gastrin and its bioactive fragments in membrane mimetic media. Peptides. 2007;28:1561–1571. doi: 10.1016/j.peptides.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Szendrei GI, Fabian H, Mantsch HH, Lovas S, Nyeki O, Schon I, et al. Aspartate-bond isomerization affects the major conformations of synthetic peptides. Eur J Biochem. 1994;226:917–924. doi: 10.1111/j.1432-1033.1994.t01-1-00917.x. [DOI] [PubMed] [Google Scholar]
- 62.Torda HJ, Baldwin GS, Norton RS. High-resolution proton nuclear magnetic resonance studies of human gastrin. Biochemistry. 1985;24:1720–1727. doi: 10.1021/bi00328a023. [DOI] [PubMed] [Google Scholar]
- 63.Tracy HJ, Gregory RA. Physiological properties of a series of synthetic peptides structurally related to gastrin I. Nature (London) 1964;204:935–938. doi: 10.1038/204935a0. [DOI] [PubMed] [Google Scholar]
- 64.Upp JR, Singh P, Townsend CM, Jr, Thompson JC. Clinical significance of gastrin receptors in human colon cancers. Cancer Res. 1989;49:488–492. [PubMed] [Google Scholar]
- 65.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 66.Vass E, Hollosi M, Besson F, Buchet R. Vibrational spectroscopic detection of beta- and gamma-turns in synthetic and natural peptides and proteins. Chem Rev. 2003;103:1917–1954. doi: 10.1021/cr000100n. [DOI] [PubMed] [Google Scholar]
- 67.Wunsch E, Scharf R, Peggion E, Foffani MT, Bali JP. Conformational properties of gastrin peptides: synthesis, biological activity, and preliminary CD characterization of the (Asp)5 analog of des-Trp1, Leu12-minigastrin. Biopolymers. 1986;25:229–234. doi: 10.1002/bip.360250204. [DOI] [PubMed] [Google Scholar]
- 68.Yang C-H, Ford J, Karelina Y, Shulkes A, Xiao S-D, Baldwin GS. Identification of a 70-kDa gastrin-binding protein on DLD-1 human colorectal carcinoma cells. Int J Biochem Cell Biol. 2001;33:1071–1079. doi: 10.1016/s1357-2725(01)00077-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.