Abstract
GDNF signaling through the Ret receptor tyrosine kinase is critical for ureteric bud branching morphogenesis during kidney development, yet few of the downstream genes are currently known. We find that the ETS transcription factors Etv4 and Etv5 are positively regulated by Ret signaling in the ureteric bud tips. Etv4−/−, Etv5+/− mice display either renal agenesis or severe hypodysplasia, while kidney development fails completely in double homozygotes. We identify several genes whose expression in the ureteric bud depends on Etv4 and Etv5, including Cxcr4, Myb, Met, Mmp14. Thus, Etv4 and Etv5 are key components of a gene network downstream of Ret that promotes and controls renal branching morphogenesis.
Introduction
During kidney development, the ureteric bud (UB), a tubular outgrowth of the Wolffian duct (WD), develops into the epithelium of the urinary collecting system through a complex process of branching morphogenesis. These events are controlled largely by signals to the UB cells from an adjacent population of mesenchymal cells, the “metanephric mesenchyme” (MM), and several of the responsible growth factors and their receptors have been identified1–3. In a reciprocal inductive process, factors secreted by the UB induce progenitor cells in the MM to condense, epithelialize and differentiate into nephrons, the functional units of the kidney2–4. Defects in UB branching morphogenesis can lead to reduced nephron number, which has been linked to the progression of renal diseases and hypertension5.
One secreted protein that plays a crucial role in UB growth and branching is glial cell line-derived neurotrophic factor (GDNF), which is produced by the MM cells and signals via the receptor tyrosine kinase Ret and the co-receptor Gfrα16, which are co-expressed in the Wolffian duct and later at the tips of the UB. The ablation of any of these genes in mice results in absence of the kidney (agenesis), due to failure of UB outgrowth, or severe malformation (hypodysplasia) due to very limited UB branching7. In humans, RET mutations are associated with renal agenesis as well as Hirschsprung disease and cancer6,8. While the consequences of GDNF signaling include UB cell proliferation, cell survival, and branching of the epithelium, the precise mechanisms by which GDNF exerts its effects on UB cells remain to be elucidated7.
Some of the genes regulated by GDNF are likely to promote cell proliferation, movement, adhesion, or shape changes, and thus contribute to morphogenesis of the epithelium, while others may encode inductive proteins secreted by the UB, or promote collecting duct cell differentiation. Only a few genes regulated by GDNF have been identified, and none of them can account for the profound effects of GDNF on UB morphogenesis. For this reason, we performed a genome-wide analysis of mRNA expression in the isolated UB cultured with or without GDNF. Among the genes identified were two members of the Pea3 family of ETS transcription factors9, Etv4 (Pea3) and Etv5 (Erm), and here we focus on their roles in kidney development. Etv4 and Etv5 are known to be important in processes including neuronal, spermatogonial and limb development10–12, but a role in renal development had not been investigated. We show that the overlapping expression of Etv4 and Etv5 in UB tip cells is positively regulated by GDNF/Ret signaling, and that these transcription factors are jointly required for kidney development. Thus, mice lacking both Etv4 alleles and one Etv5 allele have a high frequency of renal agenesis or hypodysplasia, due to branching defects, while in mice completely deficient in Etv4 and Etv5, kidney development fails entirely. We next identify several genes whose expression in the UB tips depends on Etv4 and Etv5, and which together may contribute to the defects seen in Etv4/Etv5 as well as Gdnf, Ret or Gfra1 mutant kidneys. These studies begin to elucidate a gene regulatory network activated by GDNF that controls ureteric bud branching morphogenesis.
Results
A screen for genes whose expression in the ureteric bud is altered by GDNF
To identify genes whose expression is altered by GDNF/Ret signaling, we took advantage of the ability of isolated UBs to grow and branch in the absence of mesenchyme, when cultured in GDNF-supplemented Matrigel13. The mesenchyme was removed from E11.5 metanephroi, and the ureteric buds were cultured overnight +/−GDNF (Fig. 1a). Without GDNF, the UBs do not grow or branch appreciably13, but after 16 hours remain viable and capable of resuming growth if GDNF is added.
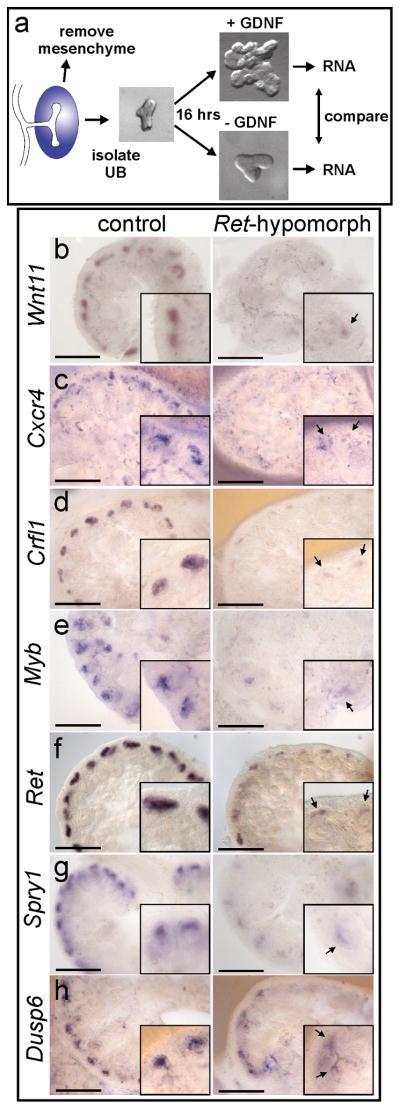
Figure 1. A screen for genes upregulated by GDNF/Ret signaling in the ureteric bud.
a, E11.5 UBs isolated from mesenchyme were cultured with or without GDNF. RNA was analyzed with Affymetrix U74Av2 and 430A arrays (Table 1 and Supplementary Tables 1 and 2). b–h, in situ hybridization to control or Rettm2(RET)Vpa/Rettm2(RET)Vpa kidneys (E14.5–E15.5). Unlike Ret−/− mice, which usually have renal agenesis, Rettm2(RET)Vpa homozygotes (expressing only the Ret51 isoform) have a milder phenotype with reduced branching. Thus, the Rettm2(RET)Vpa allele behaves as a hypomorph due to decreased Ret signaling activity15. Scale bars 100 μM. Insets, 2x enlargements of UB tips (arrows).
We surveyed Affymetrix murine gene chips U74Av2 and 430A using RNA from three pools of UBs grown with, and two without, GDNF. Of the ~12,000 transcripts represented on the U74Av2 array, 89 were upregulated by ≥ 2-fold with GDNF, 14 by ≥ 3-fold, and four by ≥ 4-fold, while only three were downregulated ≥ 2-fold. Of the 22,626 transcripts represented on the 430A array, 78 were upregulated by >2-fold, six by >3-fold, and three by >4-fold, and four were downregulated by ≥2-fold.
Table 1 lists the genes most highly upregulated by GDNF (more extensive lists are provided in Supplementary Tables 1 and 2). The validity of the screen was supported, first, by the fact that eight of these genes had been independently identified as GDNF/Ret targets in the UB (Wnt11, Ret, Spry1) or in other cell types (Etv4, Etv5, Cxcr4, Ccnd1, Dusp6)10,14–20. A second criterion was that genes upregulated by GDNF should be more highly expressed in the UB tip, the subdomain of epithelium where Ret and Gfrα1 are expressed, than in the trunk. Twelve of the listed genes were examined by in situ hybridization (ISH), and all displayed tip-specific expression in the UB (Fig. 1b–h, Fig. 2i–j, and data not shown).
Table 1.
Genes showing the most elevated expression in UBs cultured with GDNF, including those with an average fold-change of >2.5 in the U74Av2 and 430A arrays, or >3 if represented on only one array (430A * or U74Av2 **).
| Gene symbol | fold-change +/− GDNF |
|---|---|
| Wnt11 | 15 |
| Cxcr4 | 10 |
| Etv5 | 5.5* |
| Crlf1 | 4.7 |
| Myb | 3.8 |
| Arg2 | 3.5 |
| Car2 | 3.5** |
| E2f8 | 3.3** |
| Ret | 3.3 |
| Spry1 | 3.3* |
| Mtm1 | 3.1 |
| Etv4 | 2.9 |
| Dusp6 | 2.9 |
| Ccnd1 | 2.9 |
| Ncaph | 2.7 |
| Ect2 | 2.5 |
| Spred2 | 2.6 |
| Ccnb1 | 2.6 |
| Snrpd3 | 2.5 |
Figure 2. Similar expression patterns of Etv4 and Etv5 during kidney development.
a–f, expression of Etv4-lacZ and Etv5-lacZ at E9.5–11.5 in WD (white arrows) and UB (black arrows). g–h, Etv4 and Etv5 ISH at E11.5. Arrows, UB; dotted lines delineate MM. i–j, Etv4 and Etv5 ISH in E14.5 kidneys. Arrows, UB tips; arrowheads, MM; open arrows, nascent nephrons. k, Etv4 protein expression (red) in UB tips (arrows), MM (arrowheads) and nascent nephrons (open arrows); l, lack of Etv4 staining in Etv4−/− kidney shows antibody specificity. Scale bars 100 μM except for k,l (50 μM).
A third criterion for positive regulation by GDNF was reduced expression in a hypomorphic Ret mutant, in which GDNF/Ret signaling is reduced15. We examined the expression of nine GDNF-upregulated genes in Ret-hypomorphic kidneys, and all were significantly reduced in the mutant UB tips (Fig. 1b–h, Fig. 3c–f).
Figure 3. GDNF/Ret signaling regulates Etv4 and Etv5 expression in UB tips.
a–b, Increased expression of Etv4-lacZ and Etv5-lacZ in the vicinity of GDNF-soaked beads (black asterisks) but not control BSA-soaked beads (red asterisk). E15.5 (a) or E13.5 kidneys (b) were cultured for 48 hrs. c–f, Greatly reduced expression of Etv4 and Etv5 mRNAs in UB tips (arrows) of Ret-hypomorphic E14.5 kidneys (e, f) compared to WT kidneys (c, d). However, expression in nascent nephrons (arrowheads) persists in the mutant, consistent with the lack of Ret expression in the developing nephrons. Insets in c–f, enlargement of UB tips. Scale bars 100 μM.
The genes we found to be upregulated by GDNF encode secreted factors (Wnt11, Crlf1), receptors (Cxcr4, Ret), regulators of signal transduction (Spry1, Dusp6, Spred2), transcription factors (Etv5, Myb, E2f8, Etv4), and enzymes (Arg2, Car2). Several are associated with DNA replication, mitosis and cell cycle regulation (e.g., Myb, Ect2, Ccnd1, Ccnb1, Ncaph), consistent with a role of GDNF in proliferation of UB tip cells14,21. We hypothesize that these genes together mediate the effects of GDNF on branching morphogenesis, and that mutations in some of them may reveal roles in kidney development. Only Ret, Wnt11 and Spry1 are known to be required for normal UB branching morphogenesis16,22,23, while most of the others have been knocked out but caused no reported renal defects, or caused early embryonic lethality (http://www.informatics.jax.org/). Therefore, more complex genetic approaches, such as double or conditional knockouts, may be required to reveal potential functions during kidney development.
Etv4 and Etv5 are regulated primarily by GDNF/Ret signaling in the developing ureteric bud
The observation that two closely related genes, Etv4 and Etv5, were both identified in the GDNF screen, together with their roles in other developing systems, suggested that they might encode key transcription factors that mediate the effects of extracellular cues during kidney development. Both genes are expressed in several organs undergoing branching morphogenesis, including the lung, mammary and salivary glands, and kidney24. We surveyed their expression during kidney development, and found that both Etv4 and Etv5 are expressed in a pattern similar to Ret: in the Wolffian duct at E9.5 and E10.5 (Fig. 2a–d), in the T-shaped UB at E11.5 (Fig. 2e–h) and in the UB tips throughout subsequent development (Fig. 2i–k). Unlike Ret, they are also expressed in the MM and nascent nephrons, but at lower levels than in the UB (Fig. 2g–j). The third Pea3-family member, Etv1, was not detectably expressed in E14.5 and E18.5 kidneys (data not shown).
To further test whether Etv4 and Etv5 are regulated by GDNF, we performed gain- and loss-of-function studies. Culture of kidneys heterozygous for Etv4-lacZ or Etv5-lacZ alleles with GDNF-soaked beads caused elevated lacZ expression in the UB tips close to the beads (Fig. 3a–b), confirming upregulation by GDNF. Conversely, in Ret-hypomorphic kidneys (Fig. 3c–f), or normal kidneys cultured with GDNF-blocking antibody (Supplementary Fig. 1a–d), Etv4 and Etv5 expression was greatly reduced.
Etv4 and Etv5 are regulated by FGFs in several developing organs12,25,26. The two FGFs implicated in UB branching in vivo are FGF7 and FGF10, which are expressed in the MM and stroma27 and signal to the UB, mainly via FGFR2. In Fgf10−/− kidneys, or in kidneys in which Fgfr2 was deleted in the UB (Fgfr2UB−/−), both of which are hypoplastic with reduced UB branching28,29 Etv4 and Etv5 expression remained normal (Supplementary Fig 2a–h). Thus, in the developing UB, the normal expression of Etv4 and Etv5 depends strongly on GDNF/Ret signaling but apparently not on FGF7 or FGF10 signaling via FGFR2, and the defects in Fgf7, Fgf10 and Fgfr2UB−/− mutant kidneys are evidently not due to reduced Etv4/Etv5 expression. However, Etv4 and Etv5 expression in the UB can be upregulated by exogenous FGF10 (but not FGF7), as shown by culturing kidneys with FGF-soaked beads (Supplementary Fig. 1e–f, Supplementary Fig. 2i–n).
Ret signaling activates several intracellular signal transduction pathways, including Erk MAP kinase, PI3-kinase, and PLCγ 6. To investigate the mechanism that lead to expression of Etv4 and Etv5, we employed mutant mice as well as pharmacological inhibitors. Ret tyrosine-1015 (Y1015) is required for PLC-γ activation, and RetY1015F mutants have decreased UB branching30. However, Etv4 and Etv5 were expressed normally in RetY1015F/− kidneys (Supplementary Fig. 3a–d), indicating that PLC-γ signaling is dispensable for their expression. Mutation of Ret tyrosine-1062 (Y1062) reduces (but does not eliminate) both Erk MAP kinase and PI3K signaling, and also causes reduced UB branching in vivo31. RetY1062F mutant kidneys expressed Etv4 and Etv5 at normal levels (Supplementary Fig. 3e–h). As there is not a Ret mutation that specifically eliminates Erk or PI3K signaling, we employed specific chemical inhibitors of these pathways in wild type kidneys cultures. While each inhibitor substantially reduced UB branching, as previously reported32,33, the PI3K inhibitor eliminated Etv4 and Etv5 expression in the UB, while the Erk MAP kinase inhibitor had little or no effect on expression (Supplementary Fig. 3i–x). Thus, the PI3K pathway appears to be critical for expression of Etv4 and Etv5 in the UB tips.
Etv4 and Etv5 are together required for normal kidney development
To test the importance of Etv4 and Etv5 in kidney development, we examined single and compound mutant fetal and newborn mice, initially using Etv4-lacZ10 and Etv5-lacZ knockin/knockout alleles, which delete exons encoding the DNA binding domains (Fig. 4a–h). While Etv4+/− and Etv5+/− heterozygotes were normal, Etv4−/− homozygotes and Etv4+/−;Etv5+/− compound heterozygotes had occasional renal agenesis (2% and 8%, respectively) or hypoplasia (9% and 15%) (Supplementary Table 3a–b). Etv5−/− homozygotes died before kidney development begins, so the most severe compound mutant examined was Etv4−/−;Etv5+/−, and in these newborn mice nearly all of the kidneys were either absent (41%) (Fig. 4d, left side) or hypoplastic (52%) (Fig. 4d, right side) (Supplementary Table 3b), although the mice were grossly normal. The hypoplastic kidneys were not only small but were cystic, lacked a nephrogenic zone, and had greatly reduced tubular elements and glomeruli (Fig. 4f), but the few glomeruli were apparently normal (Fig. 4h). Therefore, Etv4 and Etv5 are functionally redundant, and collectively support kidney development.
Figure 4. Severe renal developmental defects in compound Etv4/Etv5 mutant newborn mice with two different Etv5 knockout alleles.
a–b, Etv4-lacZ and Etv5-lacZ alleles. c–h, renal defects in Etv4−/−;Etv5+/− compound mutants. c, normal kidneys (k) in WT. d, unilateral renal agenesis and unilateral hypodysplasia in Etv4−/−;Etv5+/− mutant. e–f, histology of WT and Etv4−/−;Etv5+/− kidneys. m, medulla; c, cortex; n, nephrogenic zone. g–h, enlargements of glomeruli. Note the small size, absent nephrogenic zone, sparse but apparently normal glomeruli, and multiple cysts in mutant kidney. i, Etv5M allele. Exons 2–5, including initiation codon, are deleted. j–m, renal defects in compound mutants with Etv5M allele. j, Etv4−/−;Etv5M/+ mutant with bilateral hypoplasia. k, Etv4−/−;Etv5M/M double homozygote with one ureter and tiny kidney rudiment. l, histology of normal kidney, with numerous glomeruli (arrows). m, histology of the Etv4−/−;Etv5M/M kidney rudiment in k, with few epithelial elements and no glomeruli. Scale bars:1 mm in c, d, j, k; 25 μM in other panels.
We next examined a second Etv5 allele11, Etv5tm1Kmm (abbreviated Etv5M), in which exons 2–5, including the initiation codon, were deleted (Fig. 4i–m). Etv5M is apparently weaker than Etv5-lacZ, as Etv5M/M homozygotes are viable, and Etv4−/−;Etv5+/M mutants had less frequent renal defects (e.g., Fig. 4j) than Etv4−/−;Etv5+/− (Supplementary Table 3b–c) (the reason for this difference is unclear, but Etv5M could potentially encode a partially functional protein fragment including the DNA binding domain). The viability of Etv5M/M homozygotes allowed us to generate Etv4−/−;Etv5M/M double homozygotes, and in these mutants kidney development failed completely. 6/7 newborn mice had bilateral renal agenesis, and only one had a single, tiny kidney rudiment (Fig. 4k, Supplementary Table 3c), which was disorganized and showed no signs of UB branching or nephrogenesis (Fig. 4m). Despite the differences in severity of the two Etv5 alleles, their qualitatively similar effects (combined with Etv4 mutation) leave no doubt that these two ETS transcription factors play crucial and overlapping roles in kidney development. Thus, our studies identify, for the first time, two genes that are targets of the GDNF/Ret pathway, and when mutated recapitulate the phenotype of Gdnf, Ret or Gfra1 mutants.
Renal agenesis or hypoplasia results from defects in ureteric bud formation and branching
To examine the cause of the renal agenesis/hypoplasia in Etv4−/−;Etv5+/− mutants, we examined UB branching by introducing a Hoxb7/myrVenus transgene expressing a fluorescent protein in the WD and UB34. At E11.5-E12.5, when the UB has normally emerged and started to branch, 16% of UBs were absent, 40% showed no branching, and 19% had retarded branching (Fig. 5a–d, Supplementary Table 4). At later stages, kidneys were often absent (Fig. 5i–j), and when present contained a reduced number and irregular pattern of UB tips (Fig. 5g–h). Thus, defects in UB formation and branching appear to underlie the severe renal defects, consistent with a role for Etv4 and Etv5 downstream of GDNF/Ret signaling.
Figure 5. Ureteric bud branching defects in Etv4−/−;Etv5+/− compoundmutants.
a–d, culture of one control (a) and three representative compound mutant E11.5 kidney primordia, all carrying Hoxb7/myrVenus. Mutant b has a WD (arrowhead) but fails to make a UB, c has a UB (arrow) that fails to branch, d has a normal T-shaped UB at E11.5 but branches less than control when cultured. e–j, Hoxb7/myrVenus fluorescence reveals normal UB branching in a control kidney at E16.5 (e–f), reduced and irregular branching in the hypoplastic kidneys of one mutant (g–h), and a single unbranched ureter with no kidney in another mutant (i–j). Boxed areas are enlarged in f, h, j. k–m, culture of isolated, mesenchyme-free UBs in Matrigel. The Etv4+/− (k) and Etv4−/− (l) UBs grow and branch extensively but the Etv4−/−;Etv5+/− (m) does so only slightly. Scale bars 100 μM (a–d, k–m), 500 μM (e–j).
As Etv4 and Etv5 are expressed in MM as well as UB, a deficiency in one or both lineages might contribute to branching defects. To test their requirement in the UB lineage, we cultured mutant UBs, isolated free of MM, under conditions where wild types grow and branch extensively13. None of the Etv4−/−;Etv5+/− UBs (0/4) branched significantly (Fig. 5m), whereas the majority with only one or two mutant alleles branched extensively (11/17; e.g., Fig. k–l) (p<0.05, t-test). This confirms that Etv4 and Etv5 are together important in the UB lineage for growth and branching.
A survey of candidate genes identifies several that require Etv4/Etv5 for normal expression in the ureteric bud
As Etv4 and Etv5 are transcription factors, we hypothesized that the renal defects in compound mutants result from a failure to regulate their target genes. We therefore examined the expression of a variety of candidate genes by ISH or immunofluorescence, using kidneys from Etv4−/−;Etv5+/− embryos (rather than Etv−/−;Etv5M/M double homozygotes, which had no renal tissue to analyze).
The expression levels of Gdnf, and several other genes with important roles in the metanephric mesenchyme (Wt1, Six1, Six2, Eya1, Pax2), were not significantly altered in the MM of hypoplastic Etv4−/−;Etv5+/− kidneys, nor were markers of nascent nephron epithelia (Lim1, Jag1, Pax2), podocytes (Wt1, Lim1), and stroma (Foxd1) (Supplementary Fig. 4). This is consistent with the morphologically normal glomerular differentiation in these mutant kidneys (Fig. 4h).
Several of the genes positively regulated by GDNF/Ret signaling in the UB tips (Fig. 1 and Table 1), were examined in the hypoplastic Etv−/−;Etv5+/− kidneys. At E15.5, Crlf1, Dusp6, Etv5 and Spry1 were expressed at close to normal levels (Fig. 6, data not shown). Ret remained highly expressed in many mutant UB tips, although it was low in a subset of the tips, while Wnt11 was uniformly reduced. However, at E13.5, expression of Ret (not shown) and Wnt11 (Fig. 6i) was normal, suggesting that their reduced expression at E15.5 is a secondary effect of the progressively abnormal renal development in Etv4−/−;Etv5+/− mutants. In contrast, expression of the chemokine receptor Cxcr4 and the transcription factor Myb were strongly reduced in the mutant kidneys at both E13.5 and E15.5 (Fig. 6f,g,j,k), suggesting they are more likely to be primary targets of Etv4/Etv5 activity.
Figure 6. Gene expression in UB tips of Etv4−/−;Etv5+/− compoundmutant kidneys.
a–d, Expression of Ret, Crlf, Dusp6 and Etv5 mRNAs was similar in the UB tips of mutant and control kidneys at E15.5, although Ret was reduced in some tips. e–h, At E15.5, expression of Wnt11, Cxcr4, Myb and Met was greatly reduced in mutant kidneys. i–l, At E13.5, Wnt11 expression was normal, but Cxcr4, Myb and Met were greatly reduced. Insets show 2x enlargements of UB tips. Black arrows indicate tips with normal expression, white arrows those with reduced expression, and asterisks indicate nascent nephrons. Controls include WT, Etv4+/− or Etv5+/−. Scale bars 100 μM.
We also examined Met and Mmp14, which were not identified in our screen as GDNF targets (perhaps for technical or biological reasons, e.g., effects of Matrigel or culture medium on gene regulation, or low expression at E11.5), but are regulated via Etv4 in other cell types and have been implicated in kidney development35–38. Met, the tyrosine kinase receptor for hepatocyte growth factor (HGF) is regulated by Etv4 in motor neurons39, and is expressed in UB tips. Met mRNA was dramatically reduced in Etv4−/−;Etv5+/− kidneys at both E13.5 and E15.5 (Fig. 6h,l). Consistent with its regulation by Etv4, Met expression was also absent in the UB of Ret-hypomorphic kidneys (not shown). The matrix metalloprotease MMP14 is likely a direct target of Etv4 in tumor cells40, and is implicated in branching morphogenesis in kidney and other organs36,41–43. In wild type kidneys, MMP14 protein was detected in the UB, MM and peripheral stroma (Fig. 7a,e), while in Etv4−/−;Etv5+/−, it was strongly reduced in both the UB and MM (Fig. 7c,g). Similarly, in Ret-hypomorphic kidneys, MMP14 was greatly diminished in both UB and MM (Fig. 7k). Since Ret is only expressed in the UB, the reduced expression of MMP14 in MM is likely an indirect effect of the Ret mutation. Consistent with this interpretation, Mmp14 mRNA is normally expressed in the UB and the stroma, but not the MM (Fig. 7m), and it is downregulated in the UB of Ret-hypomorphic kidneys (Fig. 7n); therefore, the MMP14 protein observed in the MM is probably shed from the UB, a known property of MMP1444. We conclude that Mmp14 is normally expressed in the UB, where it is positively regulated by Ret signaling, via Etv4 and Etv5, and in the stroma where it is independent of these mechanisms.
Figure 7. Reduced MMP14 expression in Etv4−/−;Etv5+/− compoundmutant and Ret-hypomorphic kidneys.
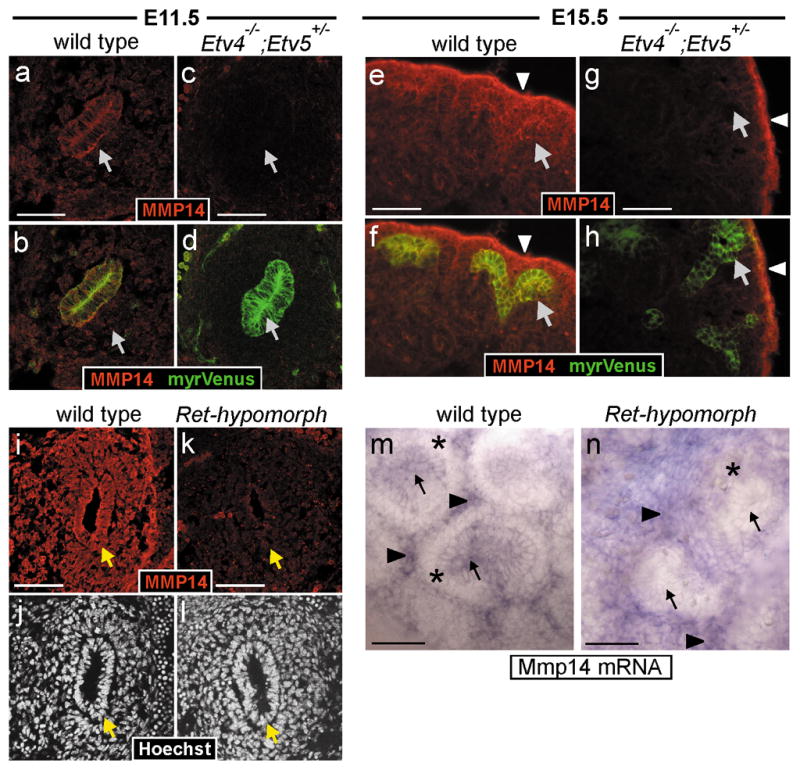
a–h, MMP14 protein in WT and Etv4−/−;Etv5+/− kidneys. At both E11.5 and E15.5, MMP14 is detected in UB epithelium (arrows) and surrounding mesenchyme of WT kidneys (a–b, e–f) but is greatly reduced in UB and MM of Etv4−/−;Etv5+/− kidneys (c–d, g–h). The UB is marked by Hoxb7/myrVenus in b, d, f, h. In the Etv4−/−;Etv5+/− at E15.5 (g–h), strong MMP14 expression persists in the peripheral stroma (arrowheads). i–n, analysis of Ret-hypomorphic kidneys. MMP14 protein (i,k) is reduced in the UB epithelium and the surrounding MM of Ret-hypomorphic kidneys at E11.5. In WT E15.5 kidney (m), Mmp14 mRNA is detected in UB epithelium (arrows) and stroma (arrowheads) but not in cap mesenchyme surrounding UB tips (asterisks). In Ret-hypomorphic kidney (n), Mmp14 mRNA is reduced in UB epithelium but persists in stroma (cap mesenchyme is reduced in quantity in the mutant). Scale bars 50 μM.
Discussion
To identify genes that mediate the important effects of Ret signaling on branching morphogenesis, we conducted a screen for genes whose expression is altered by GDNF in UB cultures. In addition to several previously described GDNF “targets” in the UB (Ret, Wnt11, Spry1) or in other cell types (Etv4, Etv5), we identified several new downstream genes including Cxcr4, Crlf1, Myb and Dusp6. We then focused on the roles of the transcription factors Etv4 and Etv5, and found that they exhibit largely overlapping expression throughout kidney development, and that both are positively regulated by GDNF/Ret signaling in UB tips. Genetic crosses revealed that mice lacking three of the four Etv4/Etv5 alleles have moderate to severe defects in renal development, due to reduced UB branching, while those lacking all four alleles fail to develop kidneys. We next identified four genes whose expression is greatly reduced in the hypoplastic kidneys of Etv4−/−;Etv5+/− compound mutants, and thus represent direct or indirect targets of these transcription factors. Cxcr4, Met, Mmp14, and Myb may participate in different aspects of epithelial morphogenesis, and collectively, their reduced expression likely contributes to the renal defects in Etv4/Etv5 mutants as well as Ret, Gdnf or Gfra1 mutant mice (Supplementary Fig. 5).
Etv4 and Etv5 are transcriptionally regulated by FGFs in several organs12,25,26, and FGFRs activate several signaling pathways common to Ret45. Therefore, it was surprising that renal expression of Etv4 and Etv5 was unaffected by Fgf10 or Fgfr2 mutations. Most likely, the strong GDNF/Ret signaling in the UB overwhelms the effects of FGF signaling, so a reduction in the latter has no noticeable effect on Etv4/Etv5 expression. However, Etv4 and Etv5 expression was increased in kidneys cultured with exogenous FGF10, indicating that they can be regulated by signaling pathways common to FGFRs and Ret. Studies with signaling inhibitors suggested that PI3K (activated by both Ret and FGFRs) is one such pathway. The inability of FGF7 to upregulate Etv4/Etv5 may underlie the distinct effects of FGF7 and FGF10 on branching of isolated UBs; only the latter induces formation of elongated tubules with distinct ampullae 46.
Etv4 and Etv5 are most strongly expressed in the WD/UB lineage, and in the Etv4−/−;Etv5+/− mutants, the UB either failed to emerge from the WD or was defective in later branching. This is consistent with a cell-autonomous role of Etv4/Etv5 within UB cells, downstream of GDNF/Ret signaling. It is possible that Etv4 and Etv5 also function in the MM lineage, indirectly, to promote UB growth and branching. However, the normal expression levels of several important mesenchymally-expressed genes (including Gdnf) in Etv4−/−;Etv5+/− mutant kidneys, as well as the failure of isolated UBs to branch in culture, suggest that the branching defects are intrinsic to the mutant UB epithelium. Etv4−/−;Etv5+/− kidneys contained apparently normal glomeruli (in reduced numbers), while the more severely affected Etv4−/−;Etv5M/M kidneys lacked any sign of nephrogenesis, which could be due either to a role of Etv4/Etv5 in the nephron lineage, or to a failure of the mutant UB to induce nephrogenesis.
To begin to identify both direct and indirect targets of Etv4/Etv5, we examined the expression of a variety of candidate genes in hypoplastic Etv4−/−;Etv5+/− kidneys. Many genes, several of which are regulated by GDNF in the UB tips, showed essentially normal expression. Thus, either a single Etv5 allele is sufficient for their normal expression, or they are regulated by Etv4/Etv5-independent mechanisms (Supplementary Fig. 5). In either case, they are not among the genes responsible for the phenotypic defects in Etv4−/−;Etv5+/− kidneys. However, we identified four genes whose expression in the UB tips was strongly reduced in themutants and may contribute to the renal phenotypes.
CXCR4 and its ligand CXCL12 play important roles in cell migration in many situations47. During kidney development, Cxcr4 is expressed similarly to Etv4/Etv5, in the WD and UB tips, MM and nascent nephrons (Fig. 6, data not shown). In our microarray screen, Cxcr4 was highly upregulated by GDNF, and its expression was barely detectable in either Ret-hypomorphic or Etv4−/−;Etv5+/− kidneys. In a breast cancer cell line, Cxcr4 is regulated by the ETS factor Ets148, which binds the same GGAA/T core motif as Etv4 and Etv5, suggesting that Cxcr4 may be a direct Etv4/Etv5 target in UB cells. CXCR4 might contribute to Ret-dependent epithelial cell movements during renal branching morphogenesis49,50, although the Cxcr4 knockout alone has no major effects on UB branching51.
Myb encodes a transcription factor with documented roles in hematopoiesis, colon crypt maintenance, adult brain neurogenesis, and cancer52. Interestingly, in these organs it seems to promote the proliferation of a stem or progenitor cell population, while dampening cell differentiation52. Given its specific expression in UB tips, a proliferative cell population21 that both self-renews and gives rise to differentiated derivatives in the UB trunks49,53 (P. Riccio and F.C., unpublished data), Myb might function downstream of Ret and Etv4/Etv5 to maintain the proliferation of tip cells. Transcription of Myb, like Cxcr4, can be regulated by Ets154,55, and thus it might be a direct Etv4/Etv5 target in kidney. While kidney defects were not reported in Myb−/− mice, which die at E15.5, or in hypomorphic Myb mutants that survive postnatally, this might be due to redundancy with the closely related gene Mybl252 which is also expressed in the UB (unpublished data).
We examined Met because it is upregulated by Gdnf and Etv4 in motor neurons39, and is a direct Ets1 target in cell lines 56. Its expression in UB tips was greatly reduced in both Etv4−/−;Etv5+/− and Ret-hypomorphic kidneys. HGF, the Met ligand, is expressed in the MM and stroma57, and promotes branching in MDCK cell and renal organ cultures57,58. Furthermore, a UB-specific knockout revealed that Met contributes to UB branching in cooperation with Egfr35. In motor neurons, Gdnf first induces Etv4 expression, which activates Met expression, allowing HGF to further induce Etv439. A similar positive feedback mechanism might occur in the kidney (Supplementary Fig. 5).
Matrix metalloproteases (MMPs) promote branching morphogenesis by degrading components of the extracellular matrix (ECM), thus remodeling the ECM and releasing bound growth factors41,42. Several Mmp genes have been identified as potentially direct Etv4 targets59, and among these, Mmp14 is expressed in the UB37,43 and can be transactivated by Etv440. MMP14 has been implicated in branching in kidney36 and UB cell cultures37, and Mmp14 −/− mice have hypoplastic kidneys, suggesting a branching defect38 (R. Zent and K. Riggins, personal communication). We found that MMP14 expression was considerably reduced in both Etv4−/−;Etv5+/− and Ret-hypomorphic kidneys, indicating that another function of Etv4/Etv5 is to upregulate Mmp14 in the UB.
In conclusion, our studies shows that Etv4 and Etv5 are key components of a gene network downstream of Ret that promotes and controls renal branching morphogenesis. As RET mutations in humans result in renal abnormalities8, these findings are likely to be important for our understanding of the causes of renal birth defects in humans.
Methods
Ureteric bud cultures and Affymetrix array analysis
Ureteric buds were dissected from Swiss Webster mouse embryos at E11.5 and cultured in Matrigel (BD Biosciences) with 2X-concentrated conditioned medium from BSN cells, 10% fetal calf serum, with or without 100 ng/ml GDNF46. They were then removed from the Matrigel, homogenized in TRIzol plus 25 ng/μl glycogen, and stored at −80°C. RNA was isolated from batches of 30–40 pooled UBs, and quantified and checked for integrity using the Agilent 2100 Bioanalyzer. Approximately 8 μg total RNA (from ~200–250 UBs) was used to generate an amount of unamplified probe sufficient for each Affymetrix array. The RNAs were subjected to reverse transcription, second-strand synthesis, and probe generation according to the Expression Analysis Technical Manual (Affymetrix). The same five probe preparations (from three pools of UBs cultured +GDNF and two pools −GDNF) were used for hybridization to U74Av2 and 430A arrays. Hybridization, scanning and low level analysis (generation of raw expression data) were performed by a core facility of the Herbert Irving Comprehensive Cancer Center. Sample comparisons and statistical analysis were performed with dChip 1.3 60, using the Comparison Analysis feature to identify genes differentially expressed between the two groups. The following filtering criteria were used: the “lower limit” for fold-change between the means of the +GDNF and −GDNF samples must exceed 1.2, with 90% confidence limit; and the absolute difference between the two group means must exceed 100.
Mouse strains
Etv4tm1Arbr(also called Etv4-lacZ)10, Etv5tm1Kmm 11, Fgf10 61, Fgfr2 UB-specific knockout29, Rettm2(RET)Vpa 15, RetY1062F 31 and RetY1015F mice 30 have been described. To generate the Etv5-lacZ targeting vector, a partial genomic clone of Etv5 was isolated from a strain129/Sv mouse genomic phage library. A 4Kb PstI-XhoI DNA fragment containing exon 10 and part of exon 11 was cloned 5′ to IRES::lacZ and PGK::neo in pBluescript. A 5.2Kb BamHI-NotI Etv5 genomic fragment containing exon 13 was used for the 3′ homology arm (NotI is from phage sequence). The vector was electroporated into W9.5 ES cells and targeted clones identified by Southern blotting (not shown). The targeted allele thus lacks most of exon 11 and all of exon 12, resulting in loss of the DNA binding domain. Structure of the Etv4lacZ and Etv5lacZ alleles and genotyping is described in Supplementary Fig. 6. Etv5tm1Kmm animals were genotyped by PCR as described11.
Immunofluorescence
Cryosections were washed in PBS, permeabilized in PBS/0.1% Triton, and blocked in blocking buffer (PBS, 0.05% Triton-X 100, and 1% normal donkey serum). Antibodies against Etv4 (1:50)62 or MMP14 (1:200, Sigma) were diluted in blocking buffer. Cy2- or Cy3- conjugated secondary antibodies (Jackson ImmunoResearch) were diluted in PBT. Images were taken with Zeiss LSM510 Meta or Axio Observer microscopes.
In situ hybridization
Embryos or dissected kidneys were fixed in 4% paraformaldehyde overnight at 4°C and cryosections were obtained by standard procedures. For vibratome sections, fixed tissues were washed in PBS and embedded in 0.3% gelatin/20% BSA/13% sucrose, which was polymerized with 1% glutaraldehyde. 75μm sections were cut and fixed for 20 min. in 4% paraformaldehyde before whole-mount ISH, which was carried out using digoxigenin-labeled probes as described63.
Organ cultures and induction with growth factor-soaked beads
Kidneys or lungs were isolated at the indicated stages and cultured on Transwell filters (Fisher) at the air-medium interface in DMEM with 10% fetal calf serum, 1% glutamine, 1% penicillin/streptomycin at 37°C and 5% CO2. For GDNF beads, Affigel blue beads (100–200 mesh, Bio-Rad) were washed with PBS/0.1% BSA before incubating with 50ng/μl recombinant GDNF. FGF7 or FGF10 beads were prepared as described64. Anti-GDNF blocking antibody (R&D Systems) was used at 5 μg/ml.
Supplementary Material
Acknowledgments
We thank Scott Simonet (Amgen, Inc.) for Fgf10 mice, Ken Murphy (Washington University) for Etv5tm1Kmm mice, Linda Williams and Zaiqi Wu for excellent technical assistance, and Natasza Kurpios and Cathy Mendelsohn for critical comments on the manuscript. This work was supported by grant DK55388 to F.C. and J.B., grant DK075578 to F.C., and fellowships from the National Kidney Foundation to C.C., from the American Heart Association to X.C., and from the Sigrid Juselius Foundation and Finnish Cultural Foundation to S.K.
Footnotes
Accession Numbers
Microarray data are available from GEO (accession numbers GSE18267 and GSE18268, approval pending).
Author Contributions
BL, CC, XC, and SK designed, performed and interpreted experiments. RK performed experiments. CB, MH, SJ, NA and MT contributed mutant kidneys. SA, LM and JH generated the Etv5-lacZ and Etv4-lacZ mice. KS-O and JB assisted with microarray analyses. VD’A analyzed renal histopathology. FC conceived and directed the project and wrote the manuscript. All authors edited the manuscript.
Competing Financial Interests
The authors have no competing financial interests.
References
- 1.Costantini F. Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation. 2006;74:402–21. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- 2.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–29. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 3.Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 4.Saxen L. Organogenesis of the Kidney. Cambridge University Press; Cambridge: 1987. [Google Scholar]
- 5.Hoy WE, et al. Nephron number, glomerular volume, renal disease and hypertension. Curr Opin Nephrol Hypertens. 2008;17:258–65. doi: 10.1097/MNH.0b013e3282f9b1a5. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–73. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- 7.Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–27. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- 8.Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ. Renal aplasia in humans is associated with RET mutations. Am J Hum Genet. 2008;82:344–51. doi: 10.1016/j.ajhg.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 10.Livet J, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–92. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–4. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brent AE, Tabin CJ. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development. 2004;131:3885–96. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- 13.Qiao J, Sakurai H, Nigam SK. Branching morphogenesis independent of mesenchymal-epithelial contact in the developing kidney. Proc Natl Acad Sci U S A. 1999;96:7330–5. doi: 10.1073/pnas.96.13.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepicelli CV, Kispert A, Rowitch DH, McMahon AP. GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol. 1997;192:193–8. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- 15.de Graaff E, et al. Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev. 2001;15:2433–44. doi: 10.1101/gad.205001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basson MA, et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–39. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Haase G, et al. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 18.Castellone MD, et al. Functional expression of the CXCR4 chemokine receptor is induced by RET/PTC oncogenes and is a common event in human papillary thyroid carcinomas. Oncogene. 2004;23:5958–67. doi: 10.1038/sj.onc.1207790. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, et al. Characterization of gene expression induced by RET with MEN2A or MEN2B mutation. Am J Pathol. 2002;161:249–56. doi: 10.1016/S0002-9440(10)64176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colucci-D’Amato GL, et al. Abrogation of nerve growth factor-induced terminal differentiation by ret oncogene involves perturbation of nuclear translocation of ERK. J Biol Chem. 2000;275:19306–14. doi: 10.1074/jbc.275.25.19306. [DOI] [PubMed] [Google Scholar]
- 21.Michael L, Davies JA. Pattern and regulation of cell proliferation during murine ureteric bud development. J Anat. 2004;204:241–55. doi: 10.1111/j.0021-8782.2004.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuchardt A, D’Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–3. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 23.Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–85. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- 24.Chotteau-Lelievre A, et al. PEA3 transcription factors are expressed in tissues undergoing branching morphogenesis and promote formation of duct-like structures by mammary epithelial cells in vitro. Dev Biol. 2003;259:241–57. doi: 10.1016/s0012-1606(03)00182-9. [DOI] [PubMed] [Google Scholar]
- 25.Firnberg N, Neubuser A. FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev Biol. 2002;247:237–50. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev Biol. 2003;261:10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 27.Chi L, et al. Sprouty proteins regulate ureteric branching by coordinating reciprocal epithelial Wnt11, mesenchymal Gdnf and stromal Fgf7 signalling during kidney development. Development. 2004;131:3345–56. doi: 10.1242/dev.01200. [DOI] [PubMed] [Google Scholar]
- 28.Ohuchi H, et al. FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–9. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, et al. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–15. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain S, Encinas M, Johnson EM, Jr, Milbrandt J. Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev. 2006;20:321–33. doi: 10.1101/gad.1387206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jijiwa M, et al. A targeting mutation of tyrosine 1062 in Ret causes a marked decrease of enteric neurons and renal hypoplasia. Mol Cell Biol. 2004;24:8026–36. doi: 10.1128/MCB.24.18.8026-8036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher CE, Michael L, Barnett MW, Davies JA. Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development. 2001;128:4329–38. doi: 10.1242/dev.128.21.4329. [DOI] [PubMed] [Google Scholar]
- 33.Tang MJ, Cai Y, Tsai SJ, Wang YK, Dressler GR. Ureteric bud outgrowth in response to RET activation is mediated by phosphatidylinositol 3-kinase. Dev Biol. 2002;243:128–36. doi: 10.1006/dbio.2001.0557. [DOI] [PubMed] [Google Scholar]
- 34.Chi X, Hadjantonakis AK, Wu Z, Hyink D, Costantini F. A transgenic mouse that reveals cell shape and arrangement during ureteric bud branching. Genesis. 2009;47:61–6. doi: 10.1002/dvg.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishibe S, et al. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development. 2009;136:337–45. doi: 10.1242/dev.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanwar YS, et al. Role of membrane-type matrix metalloproteinase 1 (MT-1-MMP), MMP-2, and its inhibitor in nephrogenesis. Am J Physiol. 1999;277:F934–47. doi: 10.1152/ajprenal.1999.277.6.F934. [DOI] [PubMed] [Google Scholar]
- 37.Pohl M, Sakurai H, Bush KT, Nigam SK. Matrix metalloproteinases and their inhibitors regulate in vitro ureteric bud branching morphogenesis. Am J Physiol Renal Physiol. 2000;279:F891–900. doi: 10.1152/ajprenal.2000.279.5.F891. [DOI] [PubMed] [Google Scholar]
- 38.Koshikawa N, et al. Proteolytic processing of laminin-5 by MT1-MMP in tissues and its effects on epithelial cell morphology. Faseb J. 2004;18:364–6. doi: 10.1096/fj.03-0584fje. [DOI] [PubMed] [Google Scholar]
- 39.Helmbacher F, et al. Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron. 2003;39:767–77. doi: 10.1016/s0896-6273(03)00493-8. [DOI] [PubMed] [Google Scholar]
- 40.Izumiyama Y, et al. E1AF expression is closely correlated with malignant phenotype of tongue squamous cell carcinoma through activation of MT1-MMP gene promoters. Oncol Rep. 2005;13:715–20. [PubMed] [Google Scholar]
- 41.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–33. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 42.Oblander SA, et al. Distinctive functions of membrane type 1 matrix-metalloprotease (MT1-MMP or MMP-14) in lung and submandibular gland development are independent of its role in pro-MMP-2 activation. Dev Biol. 2005;277:255–69. doi: 10.1016/j.ydbio.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 43.Meyer TN, et al. Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: toward a model of branching through budding in the developing kidney. Dev Biol. 2004;275:44–67. doi: 10.1016/j.ydbio.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Toth M, et al. Complex pattern of membrane type 1 matrix metalloproteinase shedding. Regulation by autocatalytic cells surface inactivation of active enzyme. J Biol Chem. 2002;277:26340–50. doi: 10.1074/jbc.M200655200. [DOI] [PubMed] [Google Scholar]
- 45.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Qiao J, et al. Multiple fibroblast growth factors support growth of the ureteric bud but have different effects on branching morphogenesis. Mech Dev. 2001;109:123–35. doi: 10.1016/s0925-4773(01)00592-5. [DOI] [PubMed] [Google Scholar]
- 47.Schier AF. Chemokine signaling: rules of attraction. Curr Biol. 2003;13:R192–4. doi: 10.1016/s0960-9822(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 48.Maroni P, Bendinelli P, Matteucci E, Desiderio MA. HGF induces CXCR4 and CXCL12-mediated tumor invasion through Ets1 and NF-kappaB. Carcinogenesis. 2007;28:267–79. doi: 10.1093/carcin/bgl129. [DOI] [PubMed] [Google Scholar]
- 49.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Chi X, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takabatake Y, et al. The CXCL12 (SDF-1)/CXCR4 Axis Is Essential for the Development of Renal Vasculature. J Am Soc Nephrol. 2009 doi: 10.1681/ASN.2008060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–34. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 53.Michael L, Sweeney DE, Davies JA. The lectin Dolichos biflorus agglutinin is a sensitive indicator of branching morphogenetic activity in the developing mouse metanephric collecting duct system. J Anat. 2007;210:89–97. doi: 10.1111/j.1469-7580.2006.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang LG, Liu XM, Li ZR, Denstman S, Bloch A. Differential binding of nuclear c-ets-1 protein to an intron I fragment of the c-myb gene in growth versus differentiation. Cell Growth Differ. 1994;5:1243–51. [PubMed] [Google Scholar]
- 55.Sullivan J, Feeley B, Guerra J, Boxer LM. Identification of the major positive regulators of c-myb expression in hematopoietic cells of different lineages. J Biol Chem. 1997;272:1943–9. doi: 10.1074/jbc.272.3.1943. [DOI] [PubMed] [Google Scholar]
- 56.Gambarotta G, et al. Ets up-regulates MET transcription. Oncogene. 1996;13:1911–7. [PubMed] [Google Scholar]
- 57.Woolf AS, et al. Roles of hepatocyte growth factor/scatter factor and the met receptor in the early development of the metanephros. Journal of Cell Biology. 1995;128:171–84. doi: 10.1083/jcb.128.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos OF, et al. Involvement of hepatocyte growth factor in kidney development. Developmental Biology. 1994;163:525–9. doi: 10.1006/dbio.1994.1169. [DOI] [PubMed] [Google Scholar]
- 59.de Launoit Y, et al. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:research0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Min H, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–61. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–74. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 63.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–40. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 64.Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.