Abstract
Poly(ADP-ribose) polymerases (PARPs) convert NAD to polymers of ADP-ribose that are converted to free ADP-ribose by poly(ADP-ribose) glycohydrolase (PARG). The activation of the nuclear enzyme PARP-1 following genotoxic stress has been linked to release of apoptosis inducing factor from the mitochondria, but the mechanisms by which signals are transmitted between nuclear and mitochondrial compartments are not well understood. The study reported here has examined the relationship between PARG and mitochondria in HeLa cells. Endogenous PARG associated with the mitochondrial fraction migrated in the range of 60 kDa. Transient transfection of cells with PARG expression constructs with amino acids encoded by exon 4 at the N-terminus were targeted to the mitochondria as demonstrated by subcellular fractionation and immunofluorescence microscopy of whole cells. Deletion and missense mutants allowed identification of a canonical N-terminal mitochondrial targeting sequence consisting of the first 16 amino acids encoded by PARG exon 4. Sub-mitochondrial localization experiments indicate that this mitochondrial PARG isoform is targeted to the mitochondrial matrix. The identification of a PARG isoform as a component of the mitochondrial matrix raises several interesting possibilities concerning mechanisms of nuclear-mitochondrial cross talk involved in regulation of cell death pathways.
Keywords: Poly(ADP-ribose) glycohydrolase, PARG, mitochondrial targeting sequence, mitochondrial matrix, ADPR polymers
Introduction
Polymers of ADP-ribose (ADPR) are synthesized by a family of poly(ADP-ribose) polymerases (PARPs) encoded by a number of different genes [1, 2]. The best understood are the nuclear PARPs 1 and 2 that play a role in the maintenance of genomic integrity via promotion of DNA repair and cell recovery at low levels of genotoxic stress and promotion of cell death at higher levels of damage [3, 4]. The central role of ADPR polymer metabolism in modulating cell recovery or cell death has potentially important implications for the therapeutic targeting of this metabolism [5, 6].
Activation of PARP-1 has been specifically linked to the release of apoptosis inducing factor (AIF) from mitochondria, resulting in cell death [7, 8]. A number of possible mechanisms whereby PARP-1 is involved in nuclear-mitochondrial cross talk leading to AIF release have been proposed that include nuclear/cytoplasmic NAD depletion resulting in glycolysis blocks that deplete substrates for mitochondrial metabolism [9, 10], direct effects of ADPR polymers on mitochondria [11, 12], involvement of receptor-interacting protein-1, tumor necrosis factor receptor-associated factor and c-Jun N-terminal kinase [13] and involvement of calpains and Bax [14].
Poly(ADP-ribose) glycohydrolase (PARG) catalyzes the opposing arm of ADPR polymer cycles initiated by PARPs [15]. In contrast to multiple genes that encode proteins with PARP activity, only a single gene that encodes PARG activity clearly involved in ADPR polymer metabolism has been described [16]. A second enzyme with PARG activity has been described [17], but its functional significance is not yet clear. However, alternative splicing leads to multiple PARG gene transcripts, resulting in generation of a number of different PARG isoforms targeted to nuclear and extranuclear cell compartments [18]. A PARG isoform of approximately 111 kDa facilitates DNA repair via regulation of ADPR polymer levels following DNA damage [19, 20]. A number of studies suggest an association of PARG (and thus ADPR polymer metabolism) with mitochondria [16, 21-24]. The PARG gene shares a promoter with a gene encoding TIM23, a protein involved in import of proteins into mitochondria [16]. A hypomorphic mouse mutant derived from disruption of the PARG gene that contains only small PARG isoforms including an isoform with an N-terminus that begins with amino acids encoded by PARG exon 4 shows high levels of PARG associated with the mitochondria [21]. This same PARG isoform has been subsequently detected in wild type cells and shown to be associated with the mitochondrial fraction [22]. Activities capable of degrading ADPR polymers in vivo have been detected in the mitochondrial matrix [23]. PARG shows a strong association with the mitochondrial fraction in brain and other tissues from rodents [24]. In the present work, we have examined the relation between PARG and mitochondria in more detail in HeLa cells and we present here evidence that a specific PARG isoform is a valid and legitimate component of the mitochondrial matrix.
Methods and Materials
Cell culture and transfection methods
HeLa cells were cultured (37°C, 5% CO2) in Dulbecco's modified Eagle's Medium (DMEM, Sigma) supplemented with 10% bovine calf serum (BCS, Hyclone). For the overexpression of constructs encoding wild type and mutant PARG, cells were seeded in 150 mm diameter cell culture dishes or six-well plates (Sarstedt), and transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol. Alternatively, cells were transfected using a calcium phosphate transfection method [25].
Western blotting methods
Subcellular fractions and other protein samples were applied to 10% polyacrylamide gels, and separated by SDS-PAGE [26]. Samples were then transferred to PVDF membranes (Millipore) for analysis. Membranes were analyzed with anti-V5 (Invitrogen), anti-SMAC/Diablo (Abcam), anti-Hsp60 (Stressgen), anti-MnSOD (Stressgen), anti-Histones (Millipore), or anti-Lactate Dehydrogenase (Abcam) antibodies. Antibodies for the detection of endogenous PARG in total lysates and mitochondrial fractions were described previously [22]. Membranes were subsequently detected using horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories) and visualized with an enhanced chemiluminescent (ECL) reaction. Densitometric analysis of western blots was performed using Scion Image for Windows (Scion Corporation).
Deletion and site-directed mutagenesis
pΔE-C1hPARG59, a pEGFP-C1 (Clontech) plasmid containing the hPARG59 isoform [22] was created by deleting EGFP using the Nhe1 and Kpn1 restriction sites and primers shown in Table 1. Site-directed mutagenesis (Fig. 3) was performed using the Quickchange II-E mutagenesis kit (Stratagene), according to the manufacturer's protocol, using primers shown in Table 1. For generation of deletion mutants (Fig. 2), the entire plasmid was amplified by polymerase chain reaction using the Phusion high-fidelity DNA polymerase (Finnzymes) and deletion primers shown in Table 1, and then self-circularized with T4 DNA ligase (Fermentas).
Table 1.
Plasmids created for analysis and primers used
| “Abreviated name” - Full Mutant designation | |
|---|---|
| Primer sequence (5′→3′) Mutant sequence underlined |
|
| “2” - pΔE-C1hPARG59Δ2-4MTS | |
| CTAGTCGACCCTCGGTGTGGGATCC CTATGGTACCTTCGTAAGTGACATGCAATCG |
|
| “3” - pΔE-C1hPARG59Δ2-16MTS | |
| CTAGTCGACTCTGCCAATCACACAGTAAC CTATGGTACCTTCGTAAGTGACATGCAATCG |
|
| “4” - pΔE-C1hPARG59Δ2-24MTS | |
| CTAGTCGACCGGGTAGATCTTTTGCG CTATGGTACCTTCGTAAGTGACATGCAATCG |
|
| “5” - pΔE-C1hPARG59Δ15-26MTS | |
| GATCTTTTGCGAGCAGGAGAAGTTCC CAAGAGAGGCAGCCGGATCCCA |
|
| “6” - pΔE-C1hPARG59ΔR2A | |
| CAGATCCGCTAGCATGGCAAGAATGCCTCGGTGTGG CCACACCGAGGCATTCTTGCCATGCTAGCGGATCTG |
|
| “7” - pΔE-C1hPARG59ΔR3A | |
| GATCCGCTAGCATGAGAGCAATGCCTCGGTGTGGGATC GATCCCACACCGAGGCATTGCTCTCATGCTAGCGGATC |
|
| “8” - pΔE-C1hPARG59ΔR6A | |
| CATGAGAAGAATGCCTGCGTGTGGGATCCGGCTGC GCAGCCGGATCCCACACGCAGGCATTCTTCTCATG |
|
| “9” - pΔE-C1hPARG59ΔR10A | |
| GCCTCGGTGTGGGATCGCGCTGCCTCTCTTGAGAC GTCTCAAGAGAGGCAGCGCGATCCCACACCGAGGC |
|
| “10” - pΔE-C1hPARG59ΔR2A/R3A | |
| GATCCGCTAGCATGGCAGCAATGCCTCGGTGTGGGATC GATCCCACACCGAGGCATTGCTGCCATGCTAGCGGATC |
|
| “11” - pΔE-C1hPARG59ΔR2A/R3A/R6A/R10A | |
| GCCTGCGTGTGGGATCGCGCTGCCTCTCTTGAGAC GTCTCAAGAGAGGCAGCGCGATCCCACACGCAGGC |
|
| “12” - pΔE-C1hPARG59ΔL11D | |
| CGGTGTGGGATCCGGGACCCTCTCTTGAGACCAT ATGGTCTCAAGAGAGGGTCCCGGATCCCACACCG |
|
| “13” - pΔE-C1hPARG59ΔL11D/L13D | |
| TGGGATCCGGGACCCTGACTTGAGACCATCTGCC GGCAGATGGTCTCAAGTCAGGGTCCCGGATCCCA |
|
| “14” - pΔE-C1hPARG59ΔL11D/L13D/L14D | |
| GATCCGGGACCCTGACGACAGACCATCTGCCAATC GATTGGCAGATGGTCTGTCGTCAGGGTCCCGGATC |
|
| For deletion of EGFP from pEC1hPARG59 | |
| AGCTAGCATGAGAAGAATGCCTCGGTGTG CTATGGTACCTTCGTAAGTGACATGCAATCG |
|
| MTS-EGFP vector construction | |
| AGCAGAGCTGGTTTAGTGAACCGTCAGATC GCAGCTAGCTTCAAGTTTTGGGGTCGTGTAAAT |
|
Figure 3. Arginine and Leucine residues are important for PARG MTS function.
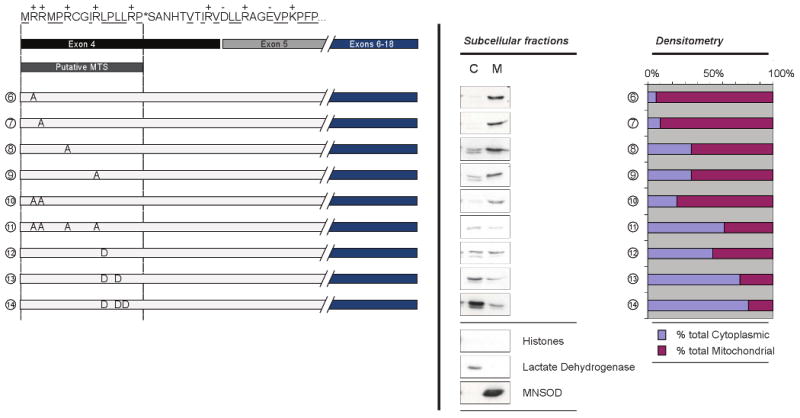
The putative PARG MTS sequence and exon structure is indicated at top left as in Fig. 2. Side-directed mutagenesis was utilized to identify amino acids essential for MTS function. Different arginine to alanine mutations are shown in mutants 6 to 11 and leucine to aspartate mutants are shown in mutants 12 to 14. Analysis of PARG using anti-V5 antibody and quantification were completed as described in Fig. 2.
Figure 2. PARG MTS is encoded by exon 4.
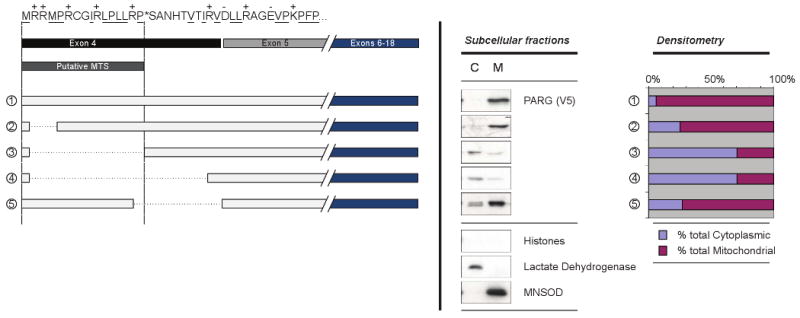
Deletion mutagenesis was used to characterize the predicted PARG MTS. The putative PARG MTS sequence and exon structure is indicated at top left. The location of positively charged residues is indicated (+) above and hydrophobic residues are underscored. Deleted sequences are indicated by the dotted lines. Representative western blots of cytosolic (C) and mitochondrial (M) fractions are shown center, with representative cellular markers indicating purity of the fractions (histones, lactate dehydrogenase, and MnSOD) shown center, bottom. For each sample, protein concentration was measured and 10 μg of each fraction was loaded onto an SDS-PAGE and PARG was detected using anti-V5 antibodies. At right, densitometry measurements are shown that compare the relative cytosolic and mitochondrial bands as a percent of the total signal intensity.
Fusion of putative MTS to EGFP
The pΔE-C1hPARG59 plasmid and the PARG mutant vectors were used as templates for the construction of vectors expressing PARG MTS-EGFP fusion proteins. Using the primers shown in Table 1, the MTS of hPARG59 and PARG mutants was amplified by polymerase chain reaction (PCR). The primers were designed to introduce NheI restriction sites into the PCR product. PCR products were subsequently subcloned into the pEGFP-C1 vector, giving rise to a vector expressing a fusion protein in which the first 94 amino acids of PARG were fused N-terminally to the EGFP. Cells were subsequently transfected for visualization by immunofluorescence microscopy.
Immunofluorescence microscopy
At 24 hours following transfection, cells seeded on coverslips were washed with phosphate buffered physiological saline (PBS) and 5% formaldehyde (Sigma). Cells were then fixed with 5% formaldehyde in PBS for 30 min at room temperature, protected from light with a foil covering. Fixed cells were washed three times in PBS, deactivated in 100 mM glycine for 1 min, washed three more times in PBS, and permeabilized for 4 min with 0.4% Triton X-100 in PBS. Following three more washes, coverslips were blocked in 3% bovine serum albumin (BSA, Sigma) in PBS for 30 min at room temperature. Coverslips were subsequently washed three more times with PBS, and incubated with anti-V5 (PARG) and anti-MnSOD (Stressgen) antibodies for 2 hrs at 37°C in a humid environment. Cells were then washed and incubated with FITC or TRITC-labeled secondary antibodies (Jackson ImmunoResearch). Alternatively, cells transfected with PARG-MTS-EGFP constructs were pretreated with Mitotracker (Invitrogen) prior to fixation, according to the manufacturer's protocol. Subsequently, cells were processed as described above. Following final washing, coverslips were mounted and DNA counterstained with Vectashield (Vector Laboratories) supplemented with 1μg/mL DAPI (Sigma). Images were captured by confocal laser microscopy (Zeiss LSM 510 META NLO system) and extracted with Zeiss LSM Image Browser software (Zeiss).
Subcellular and Submitochondrial fractionation
Mitochondria were isolated from cells 24 hours post-transfection using a mitochondrial isolation kit (Sigma) and a Potter-Elvehjem homogenizer (Fisher Scientific) for cell disruption. Briefly, following trypsinization, cells were washed in PBS and then in extraction buffer (50 mM HEPES, pH 7.5, containing 1 M mannitol, 350 mM sucrose, and 5 mM EGTA). Cells were incubated for 30 min on ice in extraction buffer supplemented with 2mg/mL BSA and a complete protease inhibitor cocktail (Roche). Cells were homogenized with 100 strokes in a Potter-Elvehjem homogenizer fitted to an overhead stirrer (IKA) set at 650 rpm. Lysates were subjected to differential centrifugation at 27g, 1,000g and 11,000g to obtain purified cellular fractions. For submitochondrial analysis, mitochondrial fractions were resuspended in a storage buffer (50 mM HEPES, pH 7.5, containing 1.25 M sucrose, 5 mM ATP, 0.4 mM ADP, 25 mM sodium succinate, 10 mM K2HPO4, and 5 mM DTT) and treated with 5μg/mL Proteinase K (Roche), and 0.1 - 0.4 mg/mL digitonin (Sigma) for 30 min at 37°C. Following heat-inactivation of Proteinase K (95°C for 10 min), samples were subsequently analyzed by SDS-PAGE and western blotting techniques.
Results
Mitochondrial fractions are enriched in smaller size PARG isoforms
The routine detection of PARG in HeLa cell extracts is limited by the low abundance of the protein. However, it was possible to detect PARG isoforms in total cell extracts and mitochondrial fractions using a polyclonal anti-peptide antibody directed against a C-terminal PARG peptide sequence [22]. This antibody has been used previously to detect endogenous isoforms of PARG in the range of approximately 100 to 110 kDa and 55 to 60 kDa [18], but their subcellular localization was not determined. Consistent with the previous study, detection of PARG in total HeLa cell extracts yielded bands in the same molecular weight ranges reported previously as well as immunoreactive material in the range of 30 kDa (Fig. 1A). Mitochondrial fractions showed PARG signals primarily in the range of approximately 55 to 60 kDa, indicating that endogenous mitochondrial PARG is comprised of the smaller PARG isoforms. This result is in agreement with previous studies involving transient transfection of cells with cDNAs expressing different PARG isoforms [22].
Figure 1. Association of endogenous PARG and overexpressed PARG with mitochondria.
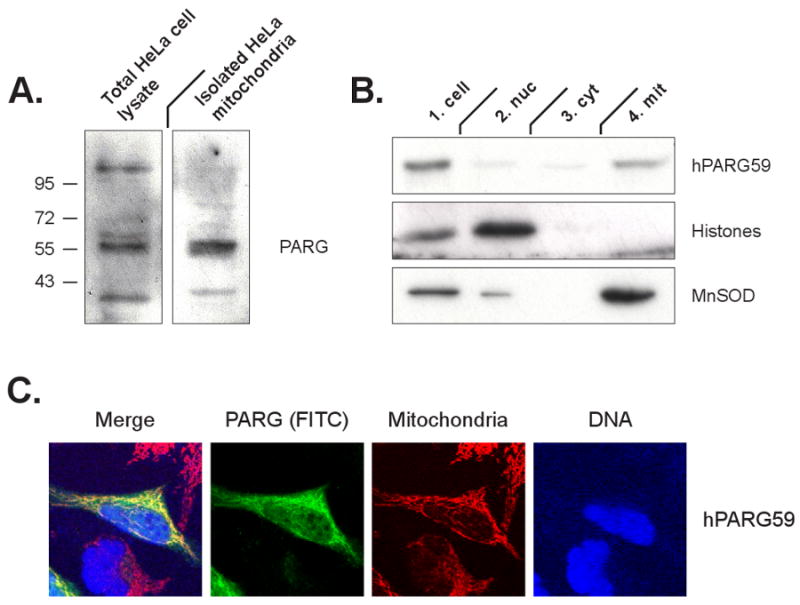
Panel A: Detection of endogenous PARG in total HeLa cell lysates and isolated mitochondria using an antibody recognizing the PARG C-terminus. Panel B: Immunodetection of PARG via the V5 tag following transfection of cells with the hPARG59 expression construct. Immunodetection of histones and MnSOD as indicators of purity of the mitochondrial fraction also is shown. Panel C: Immunodetection of PARG via the V5 tag (green) in whole cells following transfection with hPARG59 and using confocal microscopy. Mitochondrial colocalization was determined using the Mitotracker stain (red) and DNA was visualized with DAPI (blue).
PARG expressed from a plasmid containing a putative N-terminal mitochondrial targeting sequence is targeted to the mitochondria
To study the mechanisms by which PARG is targeted to mitochondria, a vector expressing hPARG59 [22] under the control of a CMV promoter was constructed. This vector expresses a PARG containing a putative N-terminal mitochondrial targeting sequence (MTS) that is encoded by exon 4 of the PARG gene. A V5 tag flanking the C-terminus has been added to the construct to facilitate PARG detection. HeLa cells were transfected with the hPARG59 vector and equal amounts of protein from the nuclear, cytosolic and mitochondrial fractions were analyzed, utilizing the V5 tag on PARG to assess the relative protein concentration of PARG in the different fractions. The results show that the expressed hPARG59 was targeted to the mitochondrial fraction (Fig. 1B). The purity of the mitochondrial fraction was confirmed by the absence of histones as an indicator of possible contamination with nuclei. Mitochondrial localization of hPARG59 was further confirmed microscopically in transfected cells by co-localization of the punctate cellular distribution the V5 tag with the Mitotracker dye (Fig. 1C).
Deletion mutagenesis indicates that PARG exon 4 encodes a mitochondrial targeting sequence
Deletion mutagenesis was used to examine the amino acid residues encoded by exon 4, to better understand their effect on mitochondrial targeting (Fig. 2). The amino acid sequence encoded by exon 4, the putative MTS, and the predicted site of cleavage are shown at the top of the left panel. Also shown are the deletion mutants constructed, the relative PARG concentration in cytosolic and mitochondrial fractions and the quantification of the relative content in cytosolic and mitochondrial fractions by densitometry. Results shown are a representative of multiple experiments performed. Immunoblotting of cytosolic and mitochondrial fractions for marker proteins revealed no detectable histones, indicating very low cross contamination with nuclei. Blots of the cytosolic marker lactate dehydrogenase and the mitochondrial marker MnSOD indicated very low levels of cross contamination of the cytosolic and mitochondrial fractions. Deletion of Arg2 and Arg3 (mutant 2) had little effect on mitochondrial targeting. However, deletion of the remaining portion of the predicted MTS up to the predicted cleavage point of the MTS (mutant 3) and further to the end of the region coded for by exon 4 (mutant 4) resulted in substantial loss of mitochondrial targeting. However, deletion of the predicted cleavage site up to the end of residues encoded by exon 4 (mutant 5) did not result in a substantial loss of mitochondrial targeting. The results indicate that the N-terminal 16 amino acids encoded by exon 4 play an important role in the mitochondrial PARG targeting.
Site directed mutagenesis indicates that both positively charged and hydrophobic amino acid residues are involved in the PARG mitochondrial targeting
A common feature of many MTS is an amphipathic alpha helix containing positively charged resides on one face of the helix and hydrophobic residues on the other face [22, 27]. In order to further evaluate the involvement of the PARG residues identified by deletion mutagenesis as pivotal in mitochondrial targeting, and to assess the contribution of the positively charged and hydrophobic residues to the mitochondrial localization of PARG, site-directed mutagenesis of Arg2, Arg3, Arg6, Arg10, as well as Leu11, Leu13, and Leu14 was completed. Figure 3 shows the mutants constructed, their relative concentration in cytosolic and mitochondrial fractions and quantification of the targeting. To examine the role of the positively charged residues, arginine to alanine mutants were made. In agreement with the deletion mutagenesis experiments, the R2A and R3A mutations alone (mutants 6 and 7) had little effect on the mitochondrial localization of the PARG protein. However, the R6A (mutant 8) and R10A (mutant 9) mutations each resulted in a larger decrease in mitochondrial targeting. The double R2A, R3A mutant (mutant 10) showed less mitochondrial targeting, suggesting additive contributions by Arg2 and Arg3 to mitochondrial targeting. Mutagenesis of Arg2, Arg3, Arg6, and Arg10 (mutant 11) showed a substantial loss of mitochondrial localization, similar that seen with the deletion of the sequence containing these amino acids (mutant 3, Fig. 2). In order to examine the hydrophobic contribution to targeting, a number of Leu to Asp mutations were generated to retain similar side-chain size but to convert the hydrophobic residue to a hydrophilic residue. Mutation of Leu 11 alone (mutant 12) substantially decreased mitochondrial localization and further mutagenesis of Leu 11 and Leu13 (mutant 13) and Leu 11, Leu 13, and Leu14 (mutant 14) almost completely abolished the mitochondrial targeting of the PARG protein. In some, but not all cases, the expressed PARG present in the cytosolic fractions appeared as a doublet, which may represent some proteolysis in that compartment.
Immunofluorescence microscopy of whole cells supports a role of exon 4 encoded amino acids in mitochondrial targeting
In order to further examine the role of the putative MTS in mitochondrial targeting, confocal microscopy was used. For these experiments, amino acids 1 to 94 of wild type hPARG59 and this segment containing the site directed mutants described in Fig. 3 were fused to an enhanced green fluorescent protein (EGFP). Confirming what we have reported previously [22], the hPARG59 N-terminal sequence localized to the mitochondria as evidenced by the punctate staining pattern and its co-localization with the mitotracker dye (Fig. 4, top panel). In support of the analyses with isolated mitochondria, substantial mitochondrial EGFP localization was observed in the R2A (mutant 6E) and R2A/R3A (mutant 10E) mutants. However, the R2A/R3A/R6A/R10A mutant (mutant 11E) showed diffuse cytoplasmic staining, similar to an EGFP control that did not contain the PARG sequence (result not shown). As was also seen in the Western blot analyses of isolated mitochondria, the single and multiple L to D mutations (mutants 12E, 13E, 14E) abrogated mitochondrial localization. The results of these experiments support the conclusion that amino acid residues encoded by exon 4 of the PARG gene are responsible for PARG mitochondrial targeting.
Figure 4. MTS identification is confirmed in whole cells.
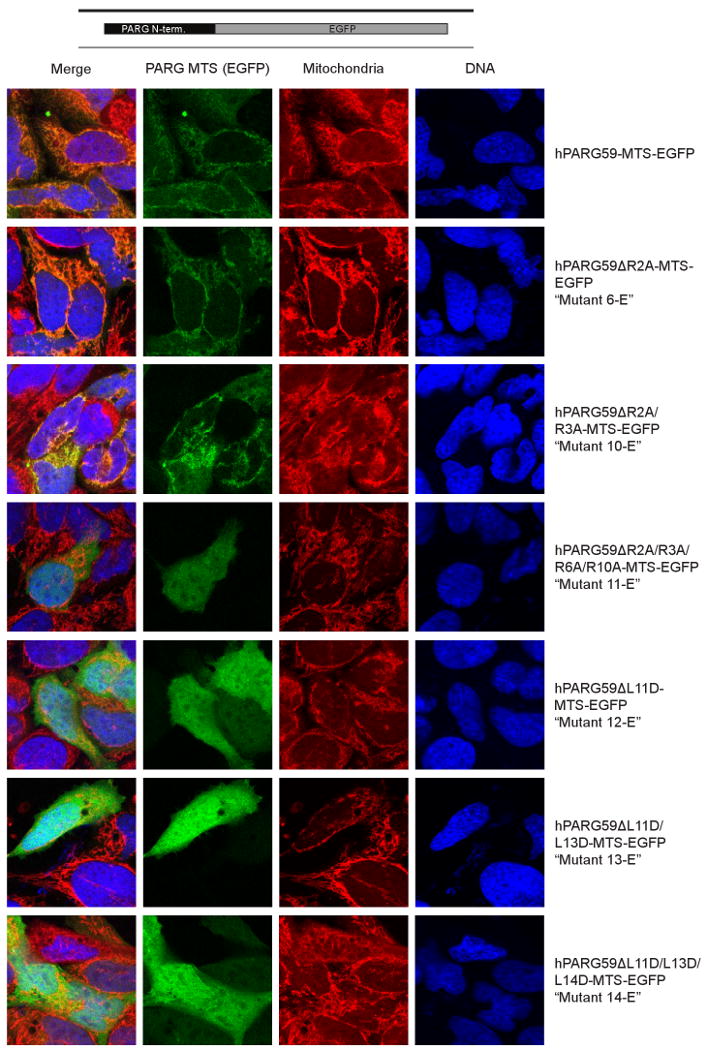
The region of hPARG59 encoding the first 94 amino acids of wild type hPARG59 and six mutant PARG isoforms were tagged to EGFP for analysis in whole cells. The mutant names (e.g.“Mutant 6-E”) correspond to the mutant numbers shown in Fig. 3. Localization of wild type and mutant sequences were detected by fluorescence of EGFP. Colocalization to mitochondria was determined through the staining of mitochondria with Mitotracker and DNA was counterstained with DAPI.
PARG is targeted to the mitochondrial matrix
In order to understand the potential role of PARG in mitochondria, experiments were completed to assess the location of PARG within the mitochondria. Using varying concentrations of digitonin in combination with protease treatment, the localization of PARG was compared with proteins of known mitochondrial location (Fig. 5). Smac/Diablo was used as an inter membrane space (IMS) marker protein and Hsp 60 was used as a matrix protein marker. Treatment with 0.1 mg/ml digitonin achieved removal of the outer mitochondrial membrane allowing protease access to Smac/Diablo but not Hsp 60. Treatment with 0.3 to 0.4 mg/ml digitonin achieved removal of the inner membrane for protease access to Hsp 60. For these experiments, both hPARG59 that contains amino acids encoded by exon 5 and hPARG55 that does not contain the residues encoded by exon 5 [22] were examined. In both cases, the results show that the PARG signal mirrored that of the matrix protein marker, although slight differences between the experiments shown in digitonin concentrations needed for complete protease sensitivity were observed. These results indicate that mitochondrial PARG localizes to the mitochondrial matrix.
Figure 5. PARG is localized to the mitochondrial matrix.
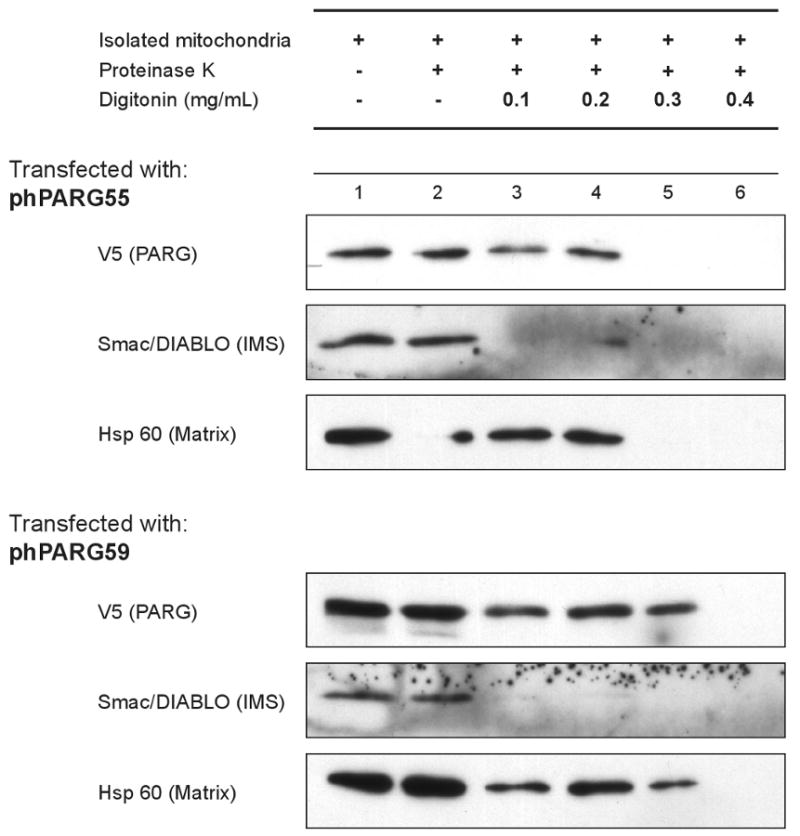
Mitochondria isolated from HeLa cells transfected with either hPARG55 or hPARG59 were subjected to proteinase K and digitonin treatment at the concentrations shown. Equal volumes of isolated mitochondria were exposed and then loaded onto an SDS-PAGE for analysis and detection of PARG by anti-V5, Smac/DIABLO via Anti-Smac/DIABLO, and Hsp60 via Anti-Hsp60 antibodies.
Discussion
The ability of cells exposed to genotoxic stress to recover or engage in programmed cell death depending upon the degree of damage is fundamentally important to the maintenance of genomic integrity of multi-cellular organisms. Mitochondrial metabolism is central to cellular responses to genotoxic stress as the release of mitochondrial proteins play important roles as effectors of programmed cell death [28]. The existence of cross talk between PARP-1 and mitochondrial metabolism in this regard was first shown by the studies of Yu et al. that established that activation of PARP-1 following high levels of damage is required for mitochondrial AIF release [11]. The involvement of ADPR polymer metabolism in modulation of cell recovery or cell death has potentially important therapeutic consequences as PARP inhibitors show promise for conditions such as cancer that evade programmed cell death [5, 6] and for conditions such as ischemia-reperfusion injury where excessive cell death leads to severe impairment or death [8].
Previous studies have provided evidence that PARG activity is an integral component of PARP-1 dependent cell death that can either enhance or protect against cell death. Partial silencing of PARG does not affect PARP-1 dependent cell death induced by MNNG [29] but protects against H2O2 induced cell death [30]. PARG inhibitors have been shown to provide partial protection against MNNG induced cell death [31]. PARG gene disruption that results in the loss of the normal nuclear isoform of PARG confers protection against renal ischemia/reperfusion injury [32] but increases sensitivity to alkylating agents, ionizing radiation, and endotoxic stress [21].
The nuclear-mitochondrial cross talk involving ADPR metabolism has raised the possibility that ADPR polymer metabolism in the mitochondrial compartment may be a component of PARP-1 dependent cell death. There have been numerous reports of association of PARP activity, and thus presumably ADPR polymer metabolism, with mitochondria (reviewed in [33]), but this question is still unresolved as it has been difficult to rule out cross contamination of mitochondrial fractions with nuclei. Previous studies have reported the association of PARG activity with mitochondrial fractions [21, 24]. A PARG isoform of approximately 60 kDa was first identified in PARG gene disrupted animals [21], but this led to the discovery that this isoform also is also present in wild type cells [22]. The deletion and site-directed studies presented here (Figs. 3-5) provide compelling evidence that this small PARG isoform, containing amino acids encoded by exon 4 at the N-terminus of the protein, is a legitimate mitochondrial protein targeted to the mitochondria by the presence of a N-terminal MTS with properties similar to MTS sequences of other mitochondrial proteins [27].
The studies reported here demonstrate that PARG is primarily a component of the mitochondrial matrix (Fig. 5), although our studies could not rule out the possibility that a minor fraction of PARG is associated with the outer mitochondrial membrane or the mitochondrial intermembrane space. A previous study has described PARG activity in the mitochondrial matrix [23] and our studies provide a mechanism by which PARG is targeted to this compartment. The predominant mitochondrial matrix location of PARG differs from AIF and other cell death proteins that are located in or facing the mitochondrial intermembrane space [28].
While most human and mouse PARG isoforms share sequence homology, there is a species difference in the short mitochondrial PARG isoforms as the amino acids encoded by exon 5 of the PARG gene are present in mouse cells but absent in human cells [18]. Indeed, many of the experiments shown in Figs. 2 and 3 were completed before the discovery that mitochondrial isoform of PARG does not contain amino acids encoded by exon 5. However, multiple pieces of evidence indicate that the presence or absence of amino acids encoded by exon 5 does not affect the function of the MTS that results in targeting this PARG isoform to the mitochondrial matrix compartment. In a previous study [22], we have shown that both hPARG59 constructs containing exon 5 encoded amino acids and hPARG55 constructs that do not contain these amino acids are both targeted to the mitochondria. The results with mutant 5 in Fig. 2 show that a sizable sequence of amino acids on the carboxy terminal side of the MTS can be deleted without affecting MTS function. Finally, our results in Fig. 5 show that the presence or absence of exon 5 encoded amino acids does not affect mitochondrial targeting or location within the mitochondrion. While this species difference in mitochondrial PARG does not affect mitochondrial targeting, further study will be needed to determine if this difference has other functional significance.
The transcript for the mitochondrial PARG isoform previously detected [22] contains two alternative sites of protein translation initiation, one that would contain a number of amino acids N-terminal to the MTS and a second that would place the MTS at the N-terminus of the protein. The constructs used for the studies shown in Figs. 2 to 5 have used a translation initiation site that places the MTS at the N-terminus of the protein. The MTS is present at the N-terminus of almost all proteins targeted to mitochondria [27]. In a previous study, both constructs were expressed and the construct with both potential initiation sites yielded two protein bands while the construct with the initiation site placing the MTS at the N-terminus was also efficiently expressed as a single protein band [22]. These data indicate that the initiation site resulting in the MTS of PARG at the N-terminus can be used in cells. Whether the expression of the isoform that places the MTS internally is targeted to the mitochondria will require further study.
A prior study that has reported association of PARG with the mitochondrial fraction in rodent tissues [24] shows several differences from those reported here. First, PARG associated with the mitochondrial fraction in brain tissue was in the range of 100 kDa, which contrasts with the smaller isoform described here. Typically MTS sequences are N-terminal [27]. A PARG isoform in the molecular weight range of 100 kDa would not likely contain the MTS encoded by exon 4 at the N-terminus. Second, most of the PARG associated with the mitochondrial fraction was extracted with salt under conditions where mitochondrial marker proteins were resistant to extraction, although some PARG was resistant to extraction. These differences suggest the possibility of multiple associations of PARG with mitochondria as large isoforms may be associated with the outer mitochondrial membrane in some tissues while smaller isoforms containing an N-terminal MTS are targeted to the mitochondrial matrix.
The presence of PARG in the mitochondrial matrix raises interesting questions concerning its function(s) in mitochondrial metabolism that will require further study to answer. Polymers of ADPR are the physiological substrate for PARG and it is possible that the function of mitochondrial PARG is hydrolysis of ADPR polymers generated by mitochondrial PARPs. Nuclear PARPs 1 and 2 function in DNA repair [2] and it has been previously reported that PARP inhibitors also inhibit repair of mitochondrial DNA [34], which supports the possibility that mitochondria contain functional cycles of ADPR polymer synthesis catalyzed by mitochondrial PARPs and PARG. There have been previous reports of PARP activity associated with mitochondrial fractions [33] and modification of mitochondrial proteins by ADPR polymers has been described [35]. The presence of a mitochondrial PARG isoform dictates additional searches for mitochondrial PARPs and studies that can rule out the possibility of nuclear contamination accounting for the PARP activity.
A second possibility for the function of a mitochondrial matrix PARG is that it catalyzes hydrolysis of ADPR polymers generated by nuclear PARPs that are exported from the nucleus to the mitochondria following high levels of genotoxic stress. Evidence has been presented indicating that ADPR polymers can exit the nucleus and cause AIF release [12]. In this setting, it is possible that mitochondrial PARG may play a protective role in preventing inappropriate release of AIF or could promote AIF release by generating free ADPR that has been shown to activate membrane calcium channels [36, 37] and thus alter mitochondrial calcium homeostasis. Finally, it cannot be ruled out at present that mitochondrial matrix PARG may play a role in mitochondrial metabolism that does not involve ADPR polymer hydrolysis. Nevertheless, the definitive identification of PARG as a legitimate component of mitochondria presented here dictates a closer examination of the possibility that ADPR polymer cycles plays a role in mitochondrial metabolism.
Acknowledgments
This research was supported in part by NIH Grants CA043894 (to MKJ), HD048837 (to RGM), CA106677, CA27502, and ES06694, and by a pre-doctoral fellowship from the American Foundation for Pharmaceutical Education (to CJW). DNA sequence analyses were performed by the University of Arizona Genetic Analysis and Technology Core Service Facility. Confocal microscopy was completed in the Microscopy Facility of the University of Arizona Southwest Environmental Health Sciences Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 3.Oei SL, Keil C, Ziegler M. Poly(ADP-ribosylation) and genomic stability. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2005;83:263–269. doi: 10.1139/o05-039. [DOI] [PubMed] [Google Scholar]
- 4.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 5.Curtin NJ. PARP inhibitors for cancer therapy. Expert Rev Mol Med. 2005;7:1–20. doi: 10.1017/S146239940500904X. [DOI] [PubMed] [Google Scholar]
- 6.Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anti-cancer agents in medicinal chemistry. 2007;7:515–523. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- 7.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 8.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Annals of the New York Academy of Sciences. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying W, Garnier P, Swanson RA. NAD+ repletion prevents PARP-1-induced glycolytic blockade and cell death in cultured mouse astrocytes. Biochem Biophys Res Commun. 2003;308:809–813. doi: 10.1016/s0006-291x(03)01483-9. [DOI] [PubMed] [Google Scholar]
- 10.Ying W, Alano CC, Garnier P, Swanson RA. NAD+ as a metabolic link between DNA damage and cell death. J Neurosci Res. 2005;79:216–223. doi: 10.1002/jnr.20289. [DOI] [PubMed] [Google Scholar]
- 11.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Huang S, Liu ZG, Han J. Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. J Biol Chem. 2006;281:8788–8795. doi: 10.1074/jbc.M508135200. [DOI] [PubMed] [Google Scholar]
- 14.Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J, Susin SA. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol. 2007;27:4844–4862. doi: 10.1128/MCB.02141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Enzymes in Poly(ADP-Ribose) Metabolism. In: Bürkle A, editor. In Poly(ADP-Ribosyl)ation. Landes Bioscience/Eurekah.com; Georgetown, TX: 2004. pp. 1–12. [Google Scholar]
- 16.Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase (PARG) gene and the common promoter sequence it shares with inner mitochondrial membrane translocase 23 (TIM23) Gene. 2003;314:181–190. doi: 10.1016/s0378-1119(03)00738-8. [DOI] [PubMed] [Google Scholar]
- 17.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 18.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 19.Gao H, Coyle DL, Meyer-Ficca ML, Meyer RG, Jacobson EL, Wang ZQ, Jacobson MK. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp Cell Res. 2007;313:984–996. doi: 10.1016/j.yexcr.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 20.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poitras MF, Koh DW, Yu SW, Andrabi SA, Mandir AS, Poirier GG, Dawson VL, Dawson TM. Spatial and functional relationship between poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in the brain. Neuroscience. 2007;148:198–211. doi: 10.1016/j.neuroscience.2007.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook JaR, David W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- 26.Gallagher SR, editor. One-Dimensional SDS Gel Electrophoresis of Proteins. John Wiley & Sons, Inc.; Hoboken: 2002. [Google Scholar]
- 27.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annual review of biochemistry. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 28.Smith DJ, Ng H, Kluck RM, Nagley P. The mitochondrial gateway to cell death. IUBMB life. 2008;60:383–389. doi: 10.1002/iub.44. [DOI] [PubMed] [Google Scholar]
- 29.Cohausz O, Blenn C, Malanga M, Althaus FR. The roles of poly(ADP-ribose)-metabolizing enzymes in alkylation-induced cell death. Cell Mol Life Sci. 2008;65:644–655. doi: 10.1007/s00018-008-7516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blenn C, Althaus FR, Malanga M. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. Biochem J. 2006;396:419–429. doi: 10.1042/BJ20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Formentini L, Arapistas P, Pittelli M, Jacomelli M, Pitozzi V, Menichetti S, Romani A, Giovannelli L, Moroni F, Chiarugi A. Mono-galloyl glucose derivatives are potent poly(ADP-ribose) glycohydrolase (PARG) inhibitors and partially reduce PARP-1-dependent cell death. British journal of pharmacology. 2008;155:1235–1249. doi: 10.1038/bjp.2008.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ, Thiemermann C. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolase are protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 2005;16:712–719. doi: 10.1681/ASN.2004080677. [DOI] [PubMed] [Google Scholar]
- 33.Scovassi AI. The poly(ADP-ribosylation) story: a long route from Cinderella to Princess. Rivista di biologia. 2007;100:351–360. [PubMed] [Google Scholar]
- 34.Druzhyna N, Smulson ME, LeDoux SP, Wilson GL. Poly(ADP-ribose) polymerase facilitates the repair of N-methylpurines in mitochondrial DNA. Diabetes. 2000;49:1849–1855. doi: 10.2337/diabetes.49.11.1849. [DOI] [PubMed] [Google Scholar]
- 35.Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, Jenkins LW, Szabo C, Clark RS. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. Journal of neurochemistry. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- 36.Buelow B, Song Y, Scharenberg AM. The Poly(ADP-ribose) polymerase PARP-1 is required for oxidative stress-induced TRPM2 activation in lymphocytes. J Biol Chem. 2008;283:24571–24583. doi: 10.1074/jbc.M802673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perraud AL, Takanishi CL, Shen B, Kang S, Smith MK, Schmitz C, Knowles HM, Ferraris D, Li W, Zhang J, Stoddard BL, Scharenberg AM. Accumulation of free ADP-ribose from mitochondria mediates oxidative stress-induced gating of TRPM2 cation channels. J Biol Chem. 2005;280:6138–6148. doi: 10.1074/jbc.M411446200. [DOI] [PubMed] [Google Scholar]


