Abstract
The structure of an uncharacterized member of the enolase superfamily from Oceanobacillus iheyensis (GI: 23100298; IMG locus tag Ob2843; PDB Code 2OQY) was determined by the New York SGX Research Center for Structural Genomics (NYSGXRC). The structure contained two Mg2+ ions located 10.4 Å from one another, with one located in the canonical position in the (β/α)7β-barrel domain (although the ligand at the end of the fifth β-strand is His, unprecedented in structurally characterized members of the superfamily); the second is located in a novel site within the capping domain. In silico docking of a library of mono- and diacid sugars to the active site predicted a diacid sugar as a likely substrate. Activity screening of a physical library of acid sugars identified galactarate as the substrate (kcat = 6.8 s−1, KM = 620 μM; kcat/KM = 1.1 × 104 M−1 s−1), allowing functional assignment of Ob2843 as galactarate dehydratase (GalrD-II) The structure of a complex of the catalytically impaired Y90F mutant with Mg2+ and galactarate allowed identification of a Tyr 164-Arg 162 dyad as the base that initiates the reaction by abstraction of the α-proton and Tyr 90 as the acid that facilitates departure of the β-OH leaving group. The enzyme product is 2-keto-3-deoxy-D-threo-4,5-dihydroxyadipate, the enantiomer of the product obtained in the GalrD reaction catalyzed by a previously characterized bifunctional L-talarate/galactarate dehydratase (TalrD/GalrD). On the basis of the different active site structures and different regiochemistries, we recognize that these functions represent an example of apparent, not actual, convergent evolution of function. The structure of GalrD-II and its active site architecture allow identification of the seventh functionally and structurally characterized subgroup in the enolase superfamily. This study provides an additional example that an integrated sequence/structure-based strategy employing computational approaches is a viable approach for directing functional assignment of unknown enzymes discovered in genome projects.
Members of the enolase superfamily catalyze mechanistically diverse reactions, each initiated by abstraction of the α-proton of a carboxylate substrate by an active site base to form an enolate intermediate (1, 2). The active sites are located at the interface between an N-terminal α+β capping domain and a C-terminal (β/α)7β-barrel domain (modified (β/α)8- or TIM-barrel2). Residues responsible for binding the essential Mg2+ and the acid/base catalysts are located at the C-terminal ends of the β-strands of the barrel domain or in the loops that connect the β-strands with the following α-helices; loops in the capping domain contain residues that provide shape and polarity determinants for the active site, thereby determining the identity of the substrate. A conserved strategy is used to stabilize the enolate intermediate: one or both carboxylate oxygens of the substrate is coordinated to a Mg2+ so that the increased negative charge in the enolate can be stabilized. The superfamily is divided into subgroups on the basis of the identities of the ligands for the Mg2+ (located at the ends of the third, fourth, and fifth β-strands of the barrel domain) and the acid/base catalysts (located at the ends of the second, third, sixth, and/or seventh β-strands of the barrel domain). Six subgroups currently are recognized (designated by the name of the paradigm enzyme/structure): enolase, mandelate racemase (MR), cis,cis-muconate lactonizing enzyme (MLE), D-glucarate dehydratase (GlucD), D-mannonate dehydratase (ManD), and β-methylaspartate ammonia lyase (3).
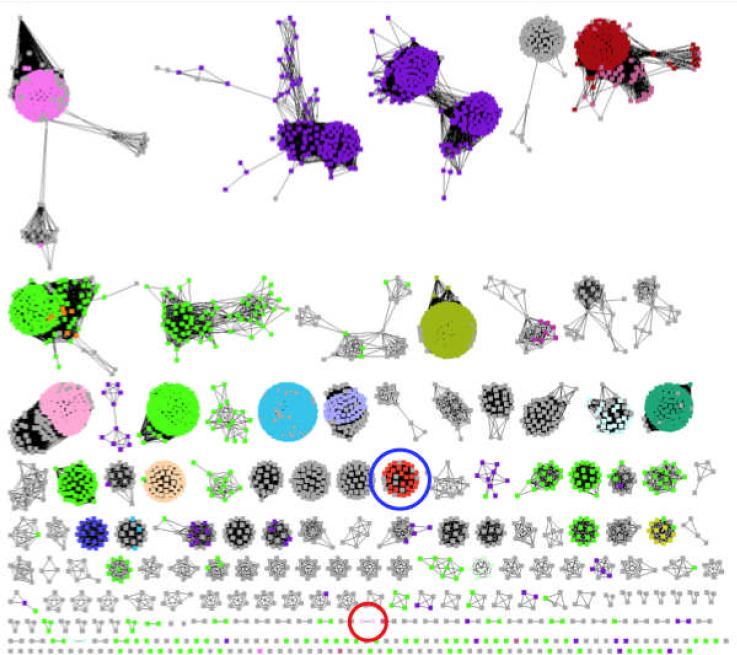
To date, nineteen distinct chemical reactions are catalyzed by members of the enolase superfamily, with these involving elimination of water or ammonia, intramolecular β-elimination (cycloisomerization), and 1,1-proton transfer (epimerization or racemization). As illustrated in the sequence similarity network (4) shown in Figure 1, the enolase superfamily can be partitioned into sequence “clusters” that aid identification of isofunctional families. In Figure 1 the members of each of the separate clusters share a BLASTP e-value of < 10−80, corresponding to > 35% sequence identity). The sequences in this network are color-coded by function, with grey sequences having unknown or uncertain function. The results of these and other bioinformatic analyses indicate that at least 50% of the currently known members of the enolase superfamily have unknown, uncertain, or incorrect functional assigments. If the structural bases for divergent evolution of function in the superfamily are to be understood and, perhaps, utilized for rational (re)design of enzymatic activities, the functions of all members of the superfamily must be assigned.
Figure 1.
Sequence similarity network for the enolase superfamily. Sequences are shown schematically as nodes (dots); BLASTP connections with E-values ≤ 10−80 are shown as edges (lines). At this E-value (sequence identities > ~35%), most of the resulting clusters of sequences are expected to be isofunctional families. Sequences that have unknown functions are colored grey. The cluster that contains Ob2843 is circled in red; the cluster that contains TalrD/GalrD is circled in blue.
Our goal is to use the sequences and/or structures of functionally uncharacterized members to enable computation-directed prediction of their substrate specificities, thereby focusing experimental efforts to accomplish their in vitro functional assignments. In the enolase superfamily, we have used comparative protein structure modeling and docking to guide discovery of a novel N-succinylamino acid racemase (5) and dipeptide epimerases with novel substrate specificities (6) in the absence of X-ray structures. In the mechanistically diverse amidohydrolase superfamily, Shoichet, Raushel, and coworkers have used in silico docking of “high-energy intermediates” to X-ray structures of an uncharacterized member to direct discovery of the 5-methylthioadenosine/S-adenosyl-homocysteine deaminase function for an uncharacterized protein from Thermotoga maritima (7). These examples establish the utility of an integrated sequence/structure-based computational strategy to direct experimental assignment of function.
Herein we describe computation-facilitated assignment of the galactarate dehydratase function to a divergent member of the enolase superfamily from Oceanobacillus iheyensis (GI:23100298; IMG locus tag Ob2843). In the sequence similarity network in Figure 1, Ob2843 is “clustered” (highlighted with red circle) with one additional presumed orthologue from Bacillus clausii KSM-K16 (GI:56964777). The structure of Ob2843 (PDB Code 2OQY) was determined in the presence of Mg2+, but the absence of an organic ligand, by the New York SGX Research Center for Structural Genomics (NYSGXRC; PSI-2) as one of their community-nominated targets. In silico screening of a virtual library of acid sugars to the active site of Ob2843 predicted that a diacid sugar was a plausible substrate; activity screening with a physical library of acid sugars identified galactarate as the substrate.
Dehydration of galactarate (as well as of L-talarate) was previously assigned as the enzymatic function of a divergent family by physical library screening (GalrD/TalrD; highlighted with blue circle in Figure 1) (8). However, the “old” (GalrD/TalrD) and “new” (GalrD-II) galactarate dehydratases differ in regioselectivity so the products obtained from (meso-)galactarate are enantiomers. The structure of the catalytically impaired Y90F mutant of GalrD-II was determined in the presence of galactarate and Mg2+, thereby allowing identification of novel active site residues: 1) an Arg-x-Tyr dyad at the end of the second β-strand of the barrel domain is the base that initiates the reaction, 2) a Tyr in the capping domain is the acid that facilitates departure of the 3-OH leaving group, and 3) a His (not Asp, Glu, or Asn as in other characterized members of the superfamily) located at the end of the fifth β-strand as a ligand for the essential Mg2+. This active site motif defines the seventh subgroup in the enolase superfamily (GalrD-II). The assignment of the GalrD-II function to Ob2843 provides additional evidence that an integrated sequence/structure-based strategy to direct functional assignment (using computational prediction to focus experimental activity measurements) is a viable approach for enabling nontrivial functional assignments.
MATERIALS AND METHODS
All 1H NMR spectra were recorded on a Varian Unity INOVA 500NB MHz spectrometer. All compounds used were the highest grade commercially available.
Cloning, Expression and Purification of GI:23100298 from Oceanobacillus iheyensis
The gene encoding the protein (IMG locus tag Ob2843) was synthesized (Codon Devices Inc., Cambridge, MA), cloned into a TOPO vector (pSGX3) and transformed into E. coli TOP10 competent cells. The protein was purified by growing the transformed cells in High Yield (Medicillon, Chicago) medium at 37 °C until the OD600 reached ~1. The temperature was then reduced to 22 °C; the culture was induced with IPTG, supplemented with selenomethione buffer and allowed to grow for 21 hr. The cells were harvested by centrifugation (11 min at 6500 × g) and resuspended in 50 mM Tris-HCl, pH 7.5, containing 500 mM NaCl, 20 mM imidazole and 0.1% Tween 20. The cells were lysed by sonication, and the lysate was clarified by centrifugation (30 min at 38,900 × g). The protein was purified by 1) adding Ni-NTA beads to the lysate, 2) incubating the mixture at 4 °C for 30 min, and 3) transferring the lysate to a drip column and eluting with 50 mM Tris-HCl, pH 7.8, containing 500 mM NaCl, 10 mM methionine, 10% glycerol and 500 mM imidazole. The sample was then equilibrated into gel filtration buffer (10 mM Hepes, pH. 7.5, containing 150 mM NaCl, 10% glycerol and 5 mM DTT) and passed over an S200 gel gel filtration column. The purified protein (as determined by SDS-PAGE) was dialyzed into gel filtration buffer.
For additional experiments, the gene in the pSGX3 vector was transformed into E. coli strains XL1-Blue for transformation and BL21(DE3) for expression. Purified protein was obtained by growing transformed BL21(DE3) cells at 37 °C for 24 hours in Luria-Burtani (LB) broth supplemented with 50 μg/mL kanamycin and induced with 500 μM IPTG when the OD600 reached ~0.5. The cells were harvested by centrifugation (10 min at 4,500 × g), resuspended in binding buffer (20 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 500 mM NaCl, 5 mM imidazole), and lysed by sonication. The lysate was clarified by centrifugation (60 min at 15,000 × g) and loaded onto a chelating Sepharose Fast Flow column (American Biosystems) that was charged with Ni2+. The C-terminal His-tagged protein was purified using a linear gradient of 0 to 1 M imidazole.
Site-Directed Mutagenesis
Mutants were generated using a variation of the overlap extension method. PCR reactions (50 μL) to generate megaprimers contained 1 ng of the gene encoding wild type Ob2843 in the pSGX3 plasmid, 5 μL 10x PCR buffer, 4 mM MgCl2, 2 mM dNTPs, 40 pmol of each primer, 1 unit of Taq DNA polymerase (Invitrogen) and 0.5 units of Pfu polymerase (Stratagene). The 5’-megaprimer was constructed using the T7pro primer and an antisense primer encoding the desired mutation. The 3’-megaprimer was constructed using the T7term primer and a sense primer encoding the desired mutation. The PCR cycle was as follows: 95 °C for 4 min followed by 26 cycles of 95 °C for 45 sec, 55 °C for 45 sec and 72 °C for 2 min and 15 sec followed by 7 min at 72 °C. Primers were purified by 1% agarose gel electrophoresis followed by gel extraction (Qiagen). The second reaction (50 μL) contained 5 μL of 10X PCR buffer, 4 mM MgCl2, 2 mM dNTPs, 40 pmol each of T7pro and T7term, 200 pmol of each megaprimer, 1 unit of Taq DNA polymerase and 0.5 units of Pfu polymerase. The same PCR cycle as above was utilized for this reaction. The R162N and Y164F mutants were digested with XhoI (Stratagene) and EcoRI (Stratagene) and subcloned into the pET15b vector. The H45Q mutant was double digested with XhoI and NdeI (Stratagene) and subcloned back into pSGX3. Overlap extension failed for the Y90F mutant so an alternate PCR reaction was utilized: 5 μL 10X PCR buffer, 0.5 mM MgCl2, 2 mM dNTPs, 200 pmol of each megaprimer, 1 ng of the pSGX3 plasmid encoding wild-type enzyme and 2.5 units of Pfu Turbo polymerase (Stratagene). The PCR cycle was: 95 °C for 30 sec followed by 18 cycles of 95 °C for 30 sec, 55 °C for 1 min and 68 °C for 6 min and 32 sec followed by 68 °C for 7 min. Proteins were expressed and purified as described for the wild type protein. Due to low expression and solubility, the R162N and Y164F mutants required purification on a Dowex DEAE column (equilibrated with 20 mM Tris-HCl, pH 7.9, 5 mM MgCl2, and 100 mM NaCl and purified on a 0 to 1 M linear gradient of NaCl) and dialysis into binding buffer prior to Ni-affinity purification.
Screen for Dehydratase Activity
Ob2843 was screened for dehydratase activity with a library of mono- and diacid sugars. Briefly, reactions (50 μL) were performed in acrylic, UV-transparent 96-well plates (Corning) in 20 mM Tris-HCl, pH 7.9, containing 5 mM MgCl2 and 10 mM substrate. Ob2843 (1 μM) was added to each well, and the reactions were allowed to proceed at 30 °C overnight. The reactions were quenched with a five-fold volume excess of 1% sodium acetate/1% semicarbazide-HCl for a minimum of one hour before the absorbance at 250 nm (ε = 10,200 M−1cm−1) was quantitated with a SpectraMax Plus384 multiplate reader (Molecular Devices).
Kinetic Assay for Galactarate Dehydratase Activity
The rate of galactarate dehydration was quantitated by performing an end-point semicarbazide quenching assay. For wild-type enzyme, 40 nM enzyme was incubated with 0.1-10 mM galactarate in 50 mM Hepes, pH 8.0, containing 5 mM MgCl2. Aliquots were taken at one, three, and five minutes and quenched in a five-fold volume excess of 1% sodium acetate/1% semicarbazide-HCl for a minimum of one hour before the absorbance at 250 nm was quantitated with a Perkin-Elmer Lambda14 UV/Vis spectrophotometer. The mutant proteins were assayed by incubating either 1 or 5 μM enzyme with 0.5-40 mM galactarate using the same buffer conditions as described for wild type.
Polarimetric Assay to Determine Galactarate Regiospecificitiy
The regiochemical preference of galactarate dehydration was determined by reacting 10 mM galactarate with 3 μM enzyme in 50 mM Hepes, pH 8.0, containing 5 mM MgCl2 at room temperature. The change in optical rotation was quantitated at 589 nm in a Jasco P-1010 polarimeter using a 10 cm path length cuvette and a 10 sec integration time.
Stereochemical Assignment of Solvent Hydrogen Incorporation
Ob2843 was exchanged into D2O by dilution of 1 mL of 200 μM Ob2843 with 10 mL of D2O and concentration to 1 mL in a 10 mL Amicon filter with a Millipore 10,000 MW polyethersulfone ultrafiltration membrane; this procedure was repeated twice. A dehydration reaction (800 μL, 30°C) was then performed with 10 mM galactarate in 50 mM d11-Tris-DCl, pD 7.9, containing 5 mM MgCl2 and 1 μM Ob2843 overnight. The reaction was adjusted to pD 2.0 with DCl before 1H NMR spectra were taken.
Kinetic Analysis of Aldol Cleavage Activity
The enantiomeric pair of products from galactarate dehydration were generated by incubating 50 mM galactarate in the presence of 1 μM of either Ob2843 or TalrD/GalrD (8), respectively, in 50 mM Tris-HCl, pH 7.9, containing 5 mM MgCl2. Completion of the reaction was confirmed by 1H NMR, and the enzymes were removed by centrifugation (3000 × g, 2 hr) through a Centricon centrifuge filtration device. Aldolase activity was then measured by performing reactions (1 mL) in the presence of 50 mM Hepes, pH 8.0, 5 mM MgCl2, 0.16 mM NADH, 30 units of lactate dehydrogenase and 1 μM of either the O. iheyensis or B. clausii enzyme. Retroaldol cleavage activity was monitored by following the decrease in absorbance at 340 nm (ε = 6,220 M−1cm−1).
Crystallization and Data Collection
Five different crystal forms (Table 1) were grown via hanging drop vapor diffusion at room temperature: 1) SeMet-substituted wild type Ob2843 complexed with Mg2+ (SeMet-Ob2843•Mg2), 2) wild type Ob2843 (containing Met) complexed with Mg2+ (Ob2843•Mg2+), 3) a tetragonal form of wild type complexed with Mg2+ and L-malate (Ob2843•Mg2+•L-malate), 4) a triclinic form of Ob2843•Mg2+•L-malate, and 5) the Y90F mutant complexed with Met with Mg2+ and galactarate (Y90F•Mg2+•galactarate).
Table 1.
Data collection and refinement statistics.
| SeMet-Ob2843•Mg2+ | Ob2843•Mg2+ | Ob2843•Mg2+•L-malate | Ob2843•Mg2+•L-malate | Y90F•Mg2+•Galactarate | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P1 | I4 | I4 | P1 | I4 |
| No. of molecules/aymmetric unit | 8 | 2 | 2 | 8 | 2 |
| Cell dimensions | |||||
| a, b, c (Å) | 104.6,104.6,104.5 | 121.2,121.2,120.9 | 121.2,121.2,122.1 | 105.6,105.7,105.7 | 120.0,120.0,122.2 |
| α, β,γ (°) | 109.5,109.5,109.5 | 109.3,109.5,109.7 | |||
| Resolution limit (Å) | 2.0 | 1.8 | 1.9 | 2.2 | 1.8 |
| No. of unique reflections | 220117 | 79723 | 69160 | 170773 | 79510 |
| Rmerge | 0.067 | 0.065 | 0.084 | 0.079 | 0.087 |
| I / σI | 18.9 | 17.1 | 21.5 | 15.1 | 26.9 |
| Completeness (%) | 95.5 | 98.9 | 99.8 | 95.7 | 99.8 |
| Refinement | |||||
| Resolution (Å) | 25.0-2.0 | 25.0-1.8 | 25.0-1.9 | 25.0-2.2 | 25.0-1.8 |
| Rcryst | 0.215 | 0.208 | 0.227 | 0.224 | 0.194 |
| Rfree | 0.229 | 0.234 | 0.259 | 0.241 | 0.223 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 | 0.007 | 0.005 |
| Bond angles (°) | 1.3 | 1.3 | 1.3 | 1.4 | 1.3 |
| No. atoms | |||||
| Protein | 23944 | 6002 | 6194 | 24776 | 6194 |
| Waters | 626 | 578 | 246 | 506 | 488 |
| Inhibitor | 18 | 72 | 28 | ||
| Mg2+ ions | 16 | 4 | 2 | 8 | 4 |
| PDB Code | 2OQY | 3FYY | 3ES7 | 3ES8 | 3HPF |
For SeMet-Ob2843•Mg2+, the protein solution contained SeMet-Ob2843 (10 mg/mL) in 10 mM Hepes, pH 7.5, containing 150 mM NaCl, 10 mM methionine, 10% glycerol, and 10 mM MgCl2; the precipitant contained 25% PEG 3350, 0.1 M Hepes, pH 7.5, and 0.2 M NaCl. Crystals appeared in 1-2 days and exhibited diffraction consistent with the space group P1, with eight molecules per asymmetric unit.
For Ob2843•Mg2+, the protein solution contained Ob2843 (12 mg/mL) in 10 mM Hepes, pH 7.5, containing 150 mM NaCl, 5 mM DTT, 10% glycerol, and 10 mM MgCl2; the precipitant contained 1.0 M K/Na tartrate, 0.1 M Tris, pH 7.0, and 0.2 M Li2SO4. Crystals appeared in 3 days and exhibited a diffraction pattern consistent with space group I4, with two molecules per asymmetric unit.
For the tetragonal form of Ob2843•Mg2+•L-malate, the protein solution contained Ob2843 (12 mg/mL) in 10 mM Hepes, pH 7.5, containing 150 mM NaCl, 5 mM DTT, 10% glycerol, and 10 mM MgCl2; the precipitant contained 20% PEG 3350, and 0.15 M D/L-malate, pH 7.0. Crystals appeared in 4-5 days and exhibited a diffraction pattern consistent with space group I4, with two molecules per asymmetric unit.
For the triclinic form of Ob2843•Mg2+•L-malate, the protein solution and the precipitant content were identical to those used for tetragonal crystal form of the same complex. The crystals of the triclinic form appeared in 6 days and exhibited diffraction consistent with the space group P1, with 8 molecules per asymmetric unit.
For Y90F•Mg2+•galactarate, the protein solution contained the Y90F mutant of Ob2843 (7.2 mg/mL) in 20 mM Tris, pH 7.9, containing 100 mM NaCl, 5 mM methionine, 10% glycerol, and 50 mM galactarate; the precipitant contained 25% M PEG 3350, 0.1 M Tris, pH 8.5, and 0.2 M NaCl. Crystals appeared in 4-5 days and exhibited diffraction consistent with the space group I4, with 2 molecules per asymmetric unit.
Prior to data collection, all five crystal forms were transferred to cryoprotectant solutions composed of their mother liquors supplemented with 20% glycerol and flash-cooled in a nitrogen stream. The X-ray diffraction data sets (Table 1) were recorded at the NSLS X4A beamline (Brookhaven National Laboratory) using an ADSC CCD detector. Diffraction intensities were integrated and scaled with programs DENZO and SCALEPACK (9). Data collection statistics are provided in Table 1.
Structure Determination and Model Refinement
The structure of SeMet-substituted Ob2843 (column 1 in Table 1) was determined via single wavelength anomalous dispersion with SOLVE (10); 49 of 56 possible selenium sites were located and used to calculate initial phases which were improved by density modificaiton and NCS averaging with RESOLVE (11), yielding an interpretable map for eight monomers in the asymmetric unit for space group P1. Subsequent iterative cycles of refinement were performed using manual rebuilding with TOM (12), refinement with CNS (13), automatic model rebuilding with APR (14), and solvent building with the CCP4 suite (15). None of the C-terminal segments 375-391 were visible in the electron density maps and are not included in the final models. The structure had two clearly visible Mg2+ ions bound to each polypeptide chain in the asymmetric unit. The first Mg2+ ion (metal site A) is coordinated by the side chains of Asp 193, Glu 221, His 246 and two water molecules; this site is positionally conserved with those in other members of the enolase superfamily, although His has not been observed previously as a ligand. The second Mg2+ ion (metal site B) is coordinated by the side chains of Asp 42, His 45, Thr 297, two water molecules, and the main chain carbonyl oxygen of Thr 297; metal site B had not been observed previously. The distance between the Mg2+ ions in the two metal sites A and B is 10.4 Å.
The remaining structures (Table 1, columns 2-5) were determined via molecular replacement with fully automated molecular replacement pipeline BALBES (16), using only input diffraction and sequence data. The structure of wild type Ob2843•Mg2+ was used by BALBES as the search model for all additional structure determinations. Partially refined structures of these crystal forms (Table 1, columns 2-5) were produced by BALBES without any manual intervention. Subsequently, several iterative cycles of refinement were performed for each crystal form including: manual model rebuilding with TOM, refinement with CNS, automatic model rebuilding with ARP, and solvent building with the CCP4 suite.
The C-terminal segment 376-391 is disordered in the wild type Ob2843•Mg2+ structure and was not included in the final refinement model. Each of the polypeptides in the asymmetric unit contains two Mg2+ ions bound in metal sites designated A and B (above).
The tetragonal and triclinic crystal forms of wild type Ob2843•Mg2+•L-malate both have well ordered electron density for bound L-malate molecules associated with each polypeptide chain. The structures also have well-defined electron density through residue 387 in the C-terminal chain segment in each polypeptide (residues 388-391 were not visible in the electron density maps). Arg 385 in the C-terminal segment contacts the L-malate ligand in each protomer. However, for both wild type OB2843•Mg2+•L-malate structures, Mg 2+ is located only in metal site A in each polypeptide.
The structure of the Y90F•Mg2+•galactarate complex has a well-defined C-terminal segment through residue 387, well-ordered electron density for galactarate, and Mg2+ ions in both metal sites for both protomers comprising the asymmetric unit.
In all five structures, the polypeptides are arranged as dimers, with the polypeptides in each dimer related by noncrystallographic two-fold axis common to all five structures.
Final crystallographic refinement statistics are provided in Table 1.
In silico Ligand Docking
The computational procedures were described in detail in an earlier publication (17). Briefly, we used Glide v4.5 (Schrödinger LLC) to dock two in silico metabolite libraries against the active site of Ob2843 structures. The first ligand library consisted of 19,132 (18) metabolites obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (19). The second, more focused library contained all known substrates of enolase superfamily enzymes as well as chemically similar ligands which have not yet been shown to be substrates. Specifically, we included all possible mono- and di-carboxylate sugars and their dehydration products, including phosphorylated acid sugars, and all possible L-amino acid-containing dipeptides. The total size of this focused library was 546 ligands; many of these are not included in the KEGG library. This focused library was created using the Build module of Maestro v8.0 (Schrödinger LLC). Both libraries were converted into a dockable form using LigPrep v2.1 (Schrödinger LLC).
Docking consisted of three steps: (1) protein preparation, (2) grid generation, and (3) ligand docking. During the protein preparation step, waters were removed, hydrogens were added, protonation states of titratable residues were optimized using the protein preparation wizard module available within the Maestro v8.0 graphics user interface, and the protein was energy minimized such that the heavy atoms of the protein were not allowed to move beyond 0.3 Å. We used default settings during the grid generation and ligand docking steps. In addition to using the Glide SP scoring function, we also subjected the docking poses to rescoring using a molecular mechanics/Generalized Born implicit solvent (MM-GBSA) energy function (17), specifically using the OPLS-AA force field (20). Briefly, each ligand was energy minimized in the enzyme active site, and relative binding affinities were estimated by subtracting the energies of the free ligand and protein from that of the protein-ligand complex. Energy minimization and evaluation are carried out using Protein Local Optimization Program (PLOP; commercially distributed by Schrödinger LLC as Prime) (17).
RESULTS AND DISCUSSION
The sequence of Ob2843 shares 64% sequence identity (83% sequence similarity) with a presumed orthologue from Bacillus clausii KSM-16 but < 30% sequence identity with all other members of the enolase superfamily. As described herein, we used bioinformatics and the structure of Ob2843 to direct its functional assignment as a galactarate dehydratase. This function in the context of the active site structure provides novel insights into structure/function relationships in the superfamily, as expected by the divergence of the sequence of Ob2843 from all but one other sequence in the enolase superfamily.
Bioinformatics: Operon/Genome Context of Ob2843 and its Orthologue
The genome context of Ob2843 is indicative of acid sugar metabolism (Figure 2; using img.jgi.doe.gov). The gene encoding Ob2843 is separated by one gene from that encoding a member of the dihydrodipicolonate synthase superfamily member (Ob2845, GI:23100300); enzymes in this superfamily utilize Schiff base chemistry in reactions involving 2-ketoacids, i.e., products of acid sugar dehydration, as do aldolases, dehydratases, and/or decarboxylases that have been identified in pathways involving acid sugar dehydratases in the enolase superfamily. In addition, the gene cluster also encodes a putative “aldehyde dehydrogenase” as well as putative transporters and transcriptional control genes of the GntR family (Figure 2). Homologous genes are located near the gene encoding the presumed orthologue of Ob2843 in B. clausii. Therefore, we expected that Ob2843 likely would be an acid sugar dehydratase. As described below, this prediction was corroborated by in silico metabolite screening against an experimentally determined structure.
Figure 2.
Genome context of Ob2843 from O. iheyensis and its orthologue from B. clausii.
An alignment of the sequence of Ob2843 with those of the nine functionally assigned acid sugar dehydratases in the enolase superfamily [D-glucarate dehydrtatase (GlucD; GlucD subgroup (21)), D-mannonate dehydratase (ManD; ManD subgroup (3)), L-fuconate dehydratase (FucD; MR subgroup (22)), D-tartrate dehydratase (TarD; MR subgroup (23)), L-talarate/galactarate dehydratase (TalrD/GalrD; MR subgroup (8)), D-gluconate dehydratase (GluD; MR subgroup (24)), D-arabinonate (AraD; MR subgroup (25)), D-galactonate dehydratase (GalD; MR subgroup (26)), and L-rhamnonate dehydratase (RhamD; MR subgroup (27))] strongly suggested that the active site architecture and catalytic strategy of Ob2843 would be unique:
The active site of Ob2843 was not expected to possess homologues of either residue in the His-Asp dyad located at the ends of the seventh and sixth β-strands of the (β/α)7β-barrel domain as observed in the dehydratases in the GlucD and MR subgroups.
The active site was not expected to contain an electrophilic Lys at the end of the second β-strand that stabilizes the enolate anion intermediate and often occurs as a Lys-x-Lys motif in the dehydratases in the GlucD and MR subgroups, where the first Lys is the electrophile and the second is an acid/base catalyst. Instead, the alignment suggested that an Arg is located at the end of the second β-strand, with a Tyr located two residues toward the C-terminus from the Arg to form an Arg-x-Tyr motif (vide infra); ManD contains an Arg at this position but forms an Arg-Tyr dyad with a distal Tyr in the loop that follows the second β-strand (3).
The active site was expected to contain homologues of the ligands for the essential Mg2+ found in the ManD and MR subgroups (Asp, Glu, and Glu located at the ends of the third, fourth, and fifth β-strands, respectively). However, the structure of Ob2843 revealed that this expectation was incorrect (vide infra).
Structure of Ob2843
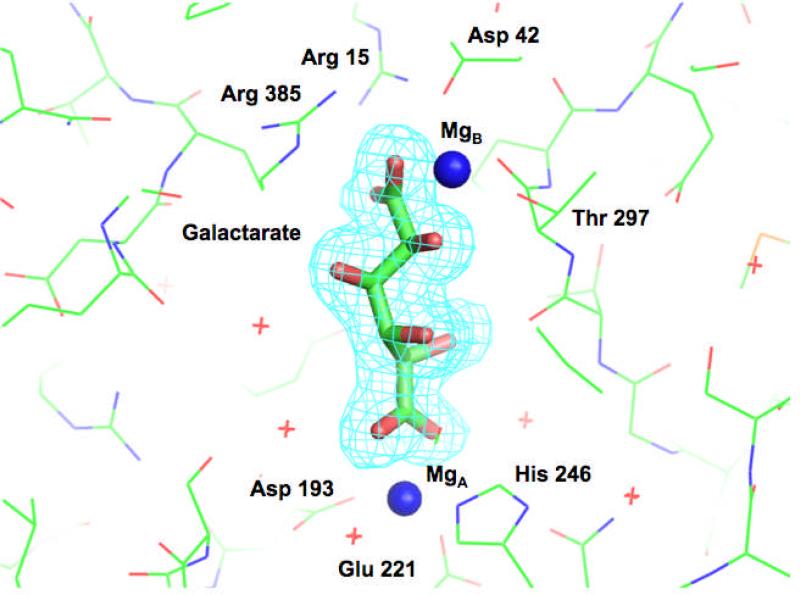
The structure of Ob2843 from Oceanobacillus iheyensis (GI:23100298; PDB Code 2OQY) was determined by the NYSGXRC (a Large Scale PSI-2 Center) in the presence of Mg2+ but in the absence of an organic ligand. Four additional structures were subsequently determined: wild type containing Met in the presence of Mg2+ (PDB Code 3FYY), two crystal forms of wild type in the presence of Mg2+ and L-malate (PDB Codes 3ES7 and 3ES8), and the Y90F mutant in the presence of Mg2+ and galactarate (PDB Code 3HPF). Representative electron density for the galactarate ligand in the last structure is shown in Figure 3.
Figure 3.
Representative electron density map for the galactarate ligand in the complex of the Y90F mutant with Mg2+ and galactarate (PDB Code 3HPF).
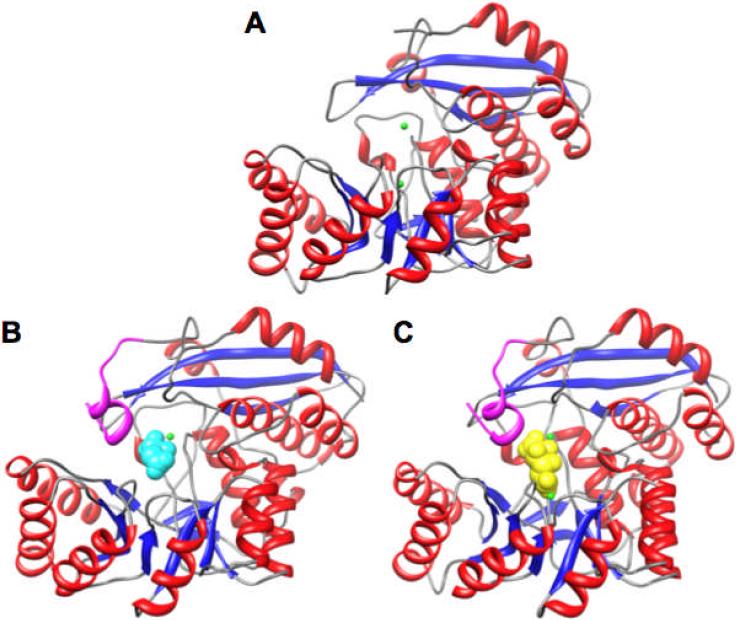
The two structures obtained with Mg2+ but in the absence of an organic ligand, 2OQY (space group P1), with eight polypeptides in the asymmetric unit, and 3FYY (space group I4) with two polypeptides in the asymmetric unit, superimpose well (r.m.s.d.= 0.19 Å for 374 Cα pairs). Trp 374 is the “last” residue visible in 2OQY; Asp 375 is the “last” residue visible in 3FYY. The polypeptides adopt the two-domain architecture observed in other members of the enolase superfamily (Figure 4). However, the 20s and 50s loops are shorter than those observed in members of the MR and MLE subgroups; these loops provide many of the substrate specificity determinants in those subgroups. The C-terminal ordered residues (Trp 374 in 2OQY and Asp 375 in 3FYY) are proximal to the active site, with the remainder of the C-terminus of the 391-residue polypeptide disordered (Figure 4, panel A).
Figure 4.
Structures of GalrD-II from O. ihenyensis. Panel A, SeMet-substituted in the presence of Mg2+ (PDB Code 2OQY). Panel B, wild type in the presence of Mg2+ and L-malate (PDB Code 3ES7). Panel C, Y90F mutant of GalrD-II in the presence of Mg2+ and galactarate (PDB Code 3HPF). The Mg2+ ions are highlighted in green, L-malate is highlighted in cyan (panel B), and galactarate is highlighted in yellow (panel C). The segment of the polypeptide shown in the structure that is ordered in the structures in panels B and C but disordered in Panel A but is highlighted in magenta; this segment contains Arg 385.
The polypeptides in both 2OQY and 3FYY contain two Mg2+ ions (Figure 5, panel A). Three polypeptide-derived ligands are present in the Mg2+ site (site A) that is positionally conserved with those in all other members of the enolase superfamily: the carboxylate group of Asp 193 located at the end of the third β-strand and the carboxylate group of Glu 221 at the C-terminus end of the fourth β-strand of the barrel domain. However, the third ligand is provided by Nε of His 246 at the end of the fifth β-strand, not a Glu as in the ManD and MR subgroups or the carboxamide oxygen of an Asn as in the GlucD group. The Glu that we expected to provide the third ligand is located immediately N-terminal to the His in a Glu-His motif. The second Mg2+ site (site B) is located in the capping domain, with four ligands provided by the polypeptide: the carboxylate group of Asp 42, the side chain of His 45, and both the side chain oxygen and backbone carbonyl oxygen of Thr 297. The distance between the Mg2+ ions in sites A and B is 10.4 Å.
Figure 5.
The active site of GalrD-II. Panel A, Se-Met GalrD-II in the presence of Mg2+ (PDB Code 2OQY). Panel B, Wild type (S-Met) GalrD-II in the presence of Mg2+ and L-malate (PDB Code 3ES7). Panel C, Y90F mutant of GalrD-II in the presence of Mg2+ and galactarate (PDB Code 3HPF). The Mg2+ ions are highlighted in green, L-malate is highlighted in cyan (panel B), and galactarate is highlighted in yellow (panel C). The active site residues are labelled only in panel B for clarity.
The only other member of the enolase superfamily known to coordinate two Mg2+ ions in the active site is enolase (28), with the second nonconserved ion in a different location. The polypeptide chain contributes two ligands for the second Mg2+ ion: the backbone carbonyl and side chain oxygen of Thr 39 (PDB Code 1ONE). Both carboxylate oxygens of the 2-phosphoglycerate/phosphoenolpyruvate substrate are coordinated to the Mg2+ ion in the conserved binding site; one carboxylate oxygen (μ-oxo bridge) as well as two nonesterified phosphoryl oxygens of the phosphate group are also ligands of the second Mg2+ ion. The distance between the Mg2+ ions is 4.2 Å. In enolase, the Mg2+ ions participate in stabilization of the enolate intermediate; in Ob2843, the Mg2+ ions are too distant (10.4 Å) to both stabilize an enolate intermediate (vide infra).
The structures determined in the presence of L-malate and Mg2+ [tetragonal (PDB Code 3ES7) and trinclinic (PDB Code 3ES8) space groups; r.m.s.d.= 0.18 Å for 387 Cα pairs] each contain one Mg2+ ion located in the B site as well as a single molecule of L-malate coordinated to the Mg2+ via the 2-OH group and one carboxylate oxygen; no Mg2+ is located in the A site (Figure 5, panel B). The oxygens of the carboxylate group coordinated to the Mg2+ also are hydrogen-bonded to Arg 15; the “remote” carboxylate group is hydrogen bonded to water molecules within the active site. In these structures, the C-termini of the polypeptide chains are ordered through Ala 387 (Figure 4, panel B), with these residues further defining the active site cavity. In particular, Nε of Arg 385 is hydrogen-bonded (3.0 Å) to the carboxylate group of L-malate that is coordinated to the Mg2+ in site B (Figure 5, panel B).
All of these structures suggest the identities of the acid/base catalysts for the dehydration reaction. As expected from the sequence alignment, Tyr 164 and Arg 162 are located at the end of the second β-strand and hydrogen-bonded to one another in the L-malate liganded structures to form a Tyr-Arg dyad that is reminiscent of the acid/base Tyr-Arg dyad in ManD; however, in ManD Arg 147 is located at the end of the second β-strand (homologous to Arg 162 in Ob2843) but Tyr 159 is located sequence-distal on a loop that follows the second β-strand. Two additional Tyr residues (Tyr 89 and Tyr 90) are located on the opposite face of the active site of Ob2843, suggesting the involvement of one or both of these as acid/base catalysts.
In silico Ligand Docking to the Unliganded Active Site
We have used in silico metabolite docking to identify putative substrates for uncharacterized enzymes in the enolase superfamily in both retrospective (17) and prospective (5, 6) tests. These successes were achieved with substrate-liganded structures (in retrospective tests) and homology-modeled structures using substrate-liganded structures as templates (in prospective predictions). In contrast, our results with unliganded X-ray structures generally have been poor ((17); unpublished results), primarily because the 20s loop in the capping domain, which contains critical specificity determinants, is almost always disordered or “open” in unliganded structures. Nonetheless, we attempted metabolite docking using the active site in 2OQY, because the active site architecture of this highly divergent member is dramatically different from all other structurally characterized members of the enolase superfamily. In particular, no loop equivalent to the 20s loop is present, although we suspected (and later confirmed, vide infra) that other conformational changes accompany ligand binding. We also hypothesized that the additional Mg2+ in the active site (site B) would both orient potential substrates and provide essential specificity discrimination.
We first docked 19,132 metabolites from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (18, 19) into the unliganded active site using the docking software Glide v4.5 (Schrödinger LLC) (29, 30). We did not apply the MM-GBSA scoring as we have in other work, because the results are much more sensitive to small changes in binding site structure than the “softer” Glide scoring function (less sensitive to minor steric clashes). We then visually inspected the predicted poses for the top 200 ligands (~1% of the hit list). These top hits included many compounds that could not be substrates because they either did not contain the required carboxylate group with an α-proton or because the carboxylate group was not positioned near the Mg2+ ion in site A. However, 33% of the top 200 hits, as well as 16 of the top 25 hits, were phosphorylated acid sugars or similar metabolites containing either two carboxylate groups or one carboxylate group and one phosphate group. Not surprisingly, the acidic/anionic groups usually interacted with and bridged the two Mg2+ ions, with 4 or 5 single bonds (from the carbon of the carboxylate group(s) or the phosphorus of the phosphate group) separating the acidic/anionic groups.
We then performed a more focused study using a library containing all known substrates for members of the enolase superfamily plus chemically similar ligands that are not known to be substrates. This library contained monoacid and diacid sugars, uronic acids, and their dehydration products, phosphorylated monoacid sugars and their dehydration products, and LL-dipeptides (substrates for dipeptide epimerases in the MLE subgroup); 546 ligands were included in this library, many of which are represented in the KEGG library. By design, the top hits were all plausible substrates; however, some did not have plausible docking poses, because the acidic groups did not coordinate the Mg2+ ion in site A in a manner consistent with catalysis. After filtering based on visual inspection to eliminate the implausible docking poses, the top 15 compounds were assembled in the hit list shown in Table 2. Almost all are 6-carbon diacid sugars or products obtained by dehydration of diacid sugars. The predicted binding modes suggested a novel mechanism of molecular recognition, bridging the Mg2+ ions in sites A and B, with the distance between the two Mg2+ ions largely responsible for substrate discrimination.
Table 2.
Top ranking metabolites from in silico ligand docking.
| Rank after Filtering | Rank before Filtering | Metabolite |
|---|---|---|
| 1 | 6 | 3-Keto-D-glucarate |
| 2 | 7 | 2-Keto-D-glucarate |
| 3 | 9 | 5-Keto-4-deoxy-D-mannarate |
| 4 | 11 | 5-Keto-4-deoxy-D-glucarate / 5-Keto-4-deoxy-galactarate |
| 5 | 12 | 2-Keto-3-deoxy-L-idarate |
| 6 | 17 | L-Gularate |
| 7 | 18 | 3-Phosphoglycerate |
| 8 | 22 | D-Alluronate |
| 9 | 24 | D-Mannarate |
| 10 | 25 | 3-Phosphoglycerate |
| 11 | 27 | Xylarate |
| 12 | 28 | 2,3-Dideoxy-D-glucarate |
| 13 | 31 | L-Glucarate |
| 14 | 34 | D-Talarate |
| 15 | 37 | L-Arabarate |
Ob2843 is a Galactarate Dehydratase
Ob2843 was screened for dehydration activity using a library of 66 mono- and diacid sugars (22); a single member of that library, the diacid sugar galactarate, was dehydrated.
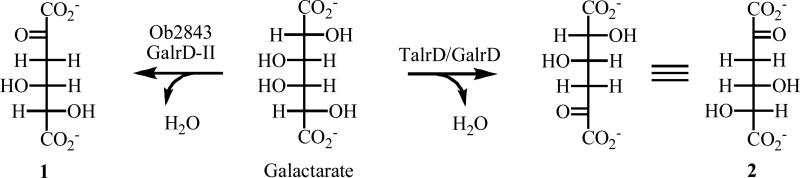
The kinetic constants were determined in an end-point assay: kcat = 6.8 ± 1.2 s−1, KM = 620 ± 380 μM; kcat/KM = 1.1 × 104 M−1s−1. These values are similar to those for other acid sugar dehydratases in the enolase superfamily, so we assign the galactarate dehydratase function (GalrD) to Ob2843 (Scheme I).
Scheme I.
Regiochemistry of Galactarate Dehydration
We previously characterized a promiscuous acid sugar dehydratase in the MR subgroup that catalyzes both L-talarate dehydratase (TalrD) and GalrD reactions via a common enolate intermediate (8); this family is circled in blue in the sequence similarity network in Figure 1. The dehydration of L-talarate is initiated by the abstraction of the 2-proton by the characteristic His-Asp dyad at the ends of the seventh and sixth β-strands, respectively, of the barrel domain, with the conjugate acid of the His also facilitating departure of the 3-OH group (syn-elimination). The dehydration of galactarate is initiated by a Lys (in a Lys-x-Lys motif at the end of the second β-strand) in an anti-elimination reaction. These enzymes also catalyze epimerization of galactarate and L-talarate via protonation of the enolate anion intermediate on the opposite face from which the 2-proton is abstracted.
Ob2843 shares 23% sequence identity TalrD/GalrD, with the X-ray structures described in an earlier section suggesting a different catalytic strategy. Thus, the GalrD function assigned to Ob2843 appears to be an additional example of convergent evolution of function with the enolase superfamily.
However, galactarate is a meso-compound, with two regiochemically distinct α-protons that can be abstracted and β-OH groups that can be eliminated. Dehydration from opposite “ends” of galactarate would produce enantiomeric products, 2-keto-D-threo-4,5-dihydroxyadipate and 2-keto-L-threo-4,5-dihydroxyadipate, raising the possibility that the reactions catalyzed by the previously described TalrD/GalrD and Ob2843 might be regiochemically distinct, catalyzing formation of enantiomeric products.
We measured the formation of the products of galactarate dehydration obtained with TalrD/GalrD and Ob2843 using polarimetry. Galactarate does not rotate plane-polarized light (meso); the enantiomeric dehydration products rotate light in opposite directions. As shown in Figure 6, Ob2843 produces a product with a negative rotation (2-keto-D-threo-4,5-dihydroxyadipate; 1 in Scheme I) and TalrD/GalrD produces a product with a positive rotation (2-keto-L-threo-4,5-dihydroxyadipate; 2 in Scheme I), establishing that the products are enantiomers and that the enzymes differ in their regiochemistry of dehydration. Thus, unlike the two families of cis,cis-muconate lactonizing enzymes that produce the same 4S-muconolactone product in reactions that are accomplished by opposite (syn and anti) addition of the remote carboxylate to the proximal double bond, TalrD/GalrD and Ob2843 utilize the same substrate but do not produce the same product (Scheme I). Thus, these functions represent apparent, not actual, convergent evolution of the same function. Therefore, we designate the “new” reaction catalyzed by Ob2843 as GalrD-II.
Figure 6.
Time-course for dehydration of galactarate measured by polarimetry for TalrD/GalrD from S. typhimurium (red) and GalrD-II from O. iheyensis (blue).
Stereochemical Course of the GalrD-II-Catalyzed Reaction
Acid sugar dehydration reactions incorporate a solvent-derived hydrogen at the β-carbon, the position of the leaving OH group. In our previous studies of acid sugar dehydratases, we demonstrated that some replace the OH group with retention of configuration [GlucD (31), GalD (26), ManD (3), RhamD (27), TalrD/GalrD (8)], one with inversion of configuration [FucD (22)], and one with racemization [TarD; the enolate anion is released to solvent where it is protonated nonenzymatically (23)].
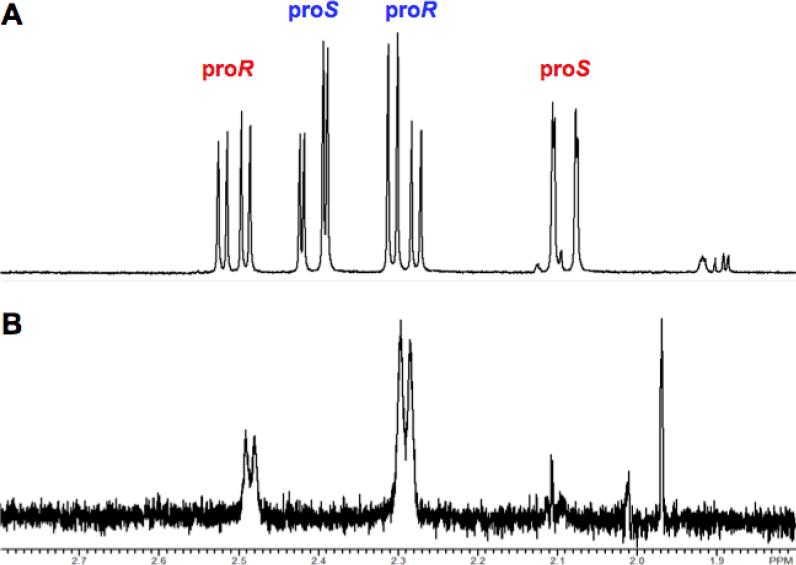
We reported that the dehydration of galactarate by TalrD/GalrD proceeds with retention of configuration at the β-carbon (8). Those studies required assignment of the 1H NMR chemical shifts of the prochiral hydrogens in the β-methylene group. Because the products of the TalrD/GalrD and GalrD-II reactions are enantiomers, those assignments can be used to determine the stereochemical course of the GalrD-II reaction: the assignments of the resonances of the proR and proS protons for the TalrD/GalrD-derived product are reversed in the GalrD-II-derived product (Figure 7, panel A). When the GalrD-II-catalyzed reaction is performed in D2O, the proS hydrogen is replaced with deuterium (Figure 7, panel B), establishing that the β-OH group is replaced by a solvent-derived hydrogen with retention of configuration, i.e., the acid that facilitates the departure of the OH group is likely the acid that catalyzes the formation of the α-keto-β-methylene product.
Figure 7.
Stereochemical course of the replacement of the leaving OH group with solvent-derived hydrogen. Panel A, 1H NMR resonances of the prochiral methylene protons of the hemiketals of the 2-keto-3-deoxy-D-threo-4,5-dihydroxyadipate product obtained by reaction in H2O. Panel B, 1H NMR resonances of the prochiral methylene protons of the hemiketals of the 2-keto-3-deoxy-D-threo-4,5-dihydroxyadipate product obtained by reaction in H2O.
Site-Directed Mutants
On the basis of the previously described structures, we constructed the H45Q mutant (ligand for the Mg2+ in site B), Y90F mutant (possible acid catalyst for departure of the β-OH group, and both the R162N and Y164F mutants (participants in the possible Tyr-Arg dyad base catalyst); their kinetic constants are reported in Table 3.
Table 3.
Kinetic constants for mutants.
| Enzyme | kcat (s−1) | KM (mM) | kcat/KM (M−1s−1) |
|---|---|---|---|
| Wild type | 6.8 ± 1.2 | 0.62 ± 0.38 | 1.1 × 104 |
| H45Q | 0.0013 ± 0.003 | 4.4 ± 2.7 | 0.29 |
| Y90F | 0.0061 ± 0.006 | 2.0 ± 0.48 | 3.1 |
| R162N | -a | -a | 0.65 |
| Y164F | ndb | nd | nd |
Velocity independent of substrate concentration.
Not detectable.
Each mutant significantly impacted the kinetic constants, as expected if their proposed roles in the mechanism were correct. The H45Q mutant had no detectable activity, consistent with the importance of the Mg2+ in site B. The Y164F mutant had no detectable activity, with the R162N mutant retaining a small amount of activity; these phenotypes are consistent with the importance of the Tyr-Arg dyad. The activity of the Y90F mutant was detectable, but the values of kcat and kcat/KM were both decreased, consistent with importance of Tyr 90.
An Aldolase Specific for the D-Enantiomer of 2-Keto-3-Deoxy-threo-4,5-Dihydroxyadipate
As noted previously, the genes encoding GalrD-II in both O. iheyensis and B. clausii are proximal to a gene encoding a homologue of dihydropicolinate synthase. These proteins were prepared by the NYSGXRC and assayed for (retro)aldolase activity by quantitating pyruvate production using 2-keto-L-threo-4,5-dihydroxyadipate (prepared with TalrD/GalrD) and 2-keto-D-threo-4,5-dihydroxyadipate (prepared with GalrD-II) as substrates. No activity was observed with the product prepared with TalrD/GalrD, although for both aldolases the kinetic constants for the enantiomeric product prepared by GalrD-II were less than might have been expected for an evolved catalytic function (e.g., kcat = 0.13 s−1, KM = 0.16 mM; and kcat/KM = 800 M−1 s−1 for the aldolase from O. iheyensis).
We considered that a possible explanation for the low retroaldolase activity is that galactarate is not the “true” substrate of GalrD-II and that another molecule containing a different anionic group at the distal end is the “true” substrate, e.g., D-galactonate 6-phosphate. Because our acid sugar library does not contain phosphorylated acid sugars (their syntheses are not straightforward), we examined whether the aldolases would catalyze the condensation of pyruvate and glycolaldehyde phosphate or DL-glyceraldehyde 3-phosphate. Each aldolase was incubated with pyruvate and either glycolaldehyde phosphate or DL-glyceraldehyde 3-phosphate; 1H NMR spectra were recorded after overnight incubation. No reactions were observed. Because the equilibrium constants for aldolase-catalyzed reactions favor condensation, the absence of a reaction provides evidence that galactarate, not a phosphorylated five- or six-carbon diacid, is the “true” substrate for GalrD-II.
Structure of the Y90F•Mg2+•Galactarate Complex
The structure of the catalytically impaired Y90F mutant complexed with Mg2+ and galactarate was determined at 1.8 Å resolution (PDB Code 3HPF; Figure 4, panel C). As in the structures determined in the presence of one Mg2+ ion (in site B) and L-malate (PDB Codes 3ES7 and 3ES8), Ala 387 is the last ordered residue in the galactarate complex with the Y90F mutant (the final four residues are disordered). This contrasts with the structures determined in the presence of Mg2+ (PDB Codes 2OQY and 3FYY) in which Asp 375 is the last ordered residue. Both oxygens of one carboxylate group are ligands for the Mg2+ ion in site A; one oxygen of the second carboxylate group as well as its proximal OH group are ligands of the Mg2+ ion in site B (Figure 5, panel C). The polypeptide-derived ligands for the Mg2+ ions in sites A and B are those observed in the unliganded 2OQY and 3FYY structures.
The phenolic OH group of Tyr 164 is hydrogen-bonded (3.2 Å) to the guanidinium group of Arg 162 which, in turn, is also hydrogen-bonded to one oxygen of the carboxylate group that is coordinated to the Mg2+ ion in site A. The phenolic OH group is also located 3.2 Å from the α-carbon of the substrate, appropriately positioned to abstract the α-proton to generate the Mg2+-stabilized enolate intermediate (mechanism is shown in Figure 8). Thus, we conclude that the Tyr 164-Arg 162 dyad is the base that initiates the dehydration reaction.
Figure 8.
Mechanism of the reaction catalyzed by GalrD-II.
That the β-OH group is located 4.2 Å from the ζ-carbon of Phe 90 suggests that Tyr 90 is the acid catalyst that facilitates the vinylogous β-elimination reaction, consistent with the significantly decreased activity observed for the Y90F mutant. Because the replacement of the β-OH group by solvent hydrogen occurs with retention of configuration, Tyr 90 likely is also the acid catalyst for ketonization of the enol intermediate obtained by the vinylogous β-elimination. The Tyr 164-Arg 162 dyad and Phe 90 are located on opposite faces of the active site; therefore, the dehydration is an anti-elimination reaction.
The arrangement of galactarate vis-a-vis the base/acid catalysts is consistent with the expected regiochemistry for the dehydration reaction from the meso-substrate and allows the correct prediction of the enantiomeric configuration of the dehydration product, e.g., 2-keto-D-threo-4,5-dihydroxyadipate (1), confirming the mechanistic relevance of the structure of this mutant enzyme-substrate complex.
The structure of this complex also discloses the identities of active site residues that determine the configuration of the diacid sugar substrate. Numbering from the carboxylate group that is coordinated to the Mg2+ in site A, the 2-OH group (that becomes the 2-keto group of the product) is hydrogen-bonded to the backbone carbonyl oxygen of Thr 296, the 3-OH group (leaving group) is predicted to be hydrogen-bonded to the phenolic OH group of Tyr 90 (the general acid catalyst), the 4-OH group is hydrogen-bonded to a water that participates in an active site hydrogen-bonding network, and the 5-OH group is a ligand of the Mg2+ in site B as well as hydrogen-bonded to the OH group of Thr 297. These interactions explain the strict specificity of GalrD-II for its galactarate substrate.
Retrospective Docking Using the Y90F•Mg2+•Galactarate Structure
In the prospective docking, we correctly predicted a 6-carbon diacid sugar is a substrate. However, galactarate ranked only within the top 17.8% of the hit list obtained by docking the KEGG ligand database against the unliganded structure. Although many of the top hits were chemically similar to galactarate, galactarate ranked too low in the hit list to merit visual inspection of the predicted pose. Similarly, galactarate itself ranked only 60 of 546 ligands in the focused library docking (unfiltered results), i.e. the top-ranked 6-carbon diacid sugars and their dehydration products had different stereochemistries (Table 2).
Although predicting the exact stereochemistry of the substrate is challenging, we wanted to understand whether the larger limitation was the 1) use of an unliganded structure (including unknown conformational changes associated with ligand binding in this novel binding site) or 2) limitations of the docking and scoring functions. The first key observation was that, in the predicted docking pose, galactarate was rotated 180° (Figure 9, panel B) from the pose observed in the galactarate-liganded Y90F structure (Figure 9, panel A), i.e., the predicted pose is the regiochemical reverse of the experimentally determined structure, with 2-keto-L-threo-4,5-dihydroxyadipate the predicted product. In addition, the carboxylate group coordinated to the Mg2+ in site A is a monodentate, rather than bidentate, ligand. However, the distal carboxylate group and its α-OH group are correctly predicted to be ligands for the Mg2+ in site B. Although the rotated pose of galactarate could result from poor sampling and/or scoring, it also could arise from incomplete hydrogen bonding interactions in the unliganded structure. In the galactarate-liganded Y90F structure, Arg 385 forms a hydrogen bonding interaction with the distal carboxylate group of the galactarate; however, residues 375–391 were disordered in the unliganded structure that was used in the docking.
Figure 9.
Panel A, the active site of the Y90F mutant of GalrD-II in the presence of Mg2+ and galactarate (PDB Code 3HPF). Panel B, the docking pose of galactarate obtained using the apo structure (PDB Code 2OQY). Panel C, the docking pose of galactarate obtained using the “wild type” holo structure (with Tyr 90) constructed from the coordinates of the Y90F•Mg2+•galactarate complex (PDB Code 3HPF). The Mg2+ ions are highlighted in green; galactarate is highlighted in yellow. Arg 385, missing in the apo structure (panel B), provides a hydrogen-bonding interaction to the substrate (panel C).
Using the galactarate-liganded Y90F structure (Figure 9, panel A), we retrospectively examined whether the docking protocol could recover the correct pose for galactarate and improve its rank. We prepared a “wild-type” galactarate-liganded structure by mutating Phe 90 in the mutant structure to Tyr 90 using the software Maestro v9.0.111 and then docked the acid sugar library using Glide (29, 30).
When docked against the “wild type” holo structure, the conformation of galactarate superimposes almost perfectly on the conformation observed in the crystal structure (Figure 9, panel C). However, although the docking geometry was correctly identified by Glide, it did not rank the substrate at the top of the docking hit list using the docking scoring function. Specifically, galactarate ranked 87 out of the 546 metabolites in the “focused” library. However, when we applied MM-GBSA rescoring (17), galactarate, the substrate, ranked at the top of the hit list (Table 4). By contrast, retrospectively applying MM-GBSA rescoring to the prospective docking results using the apo structure would have made galactarate rank somewhat lower than it did using the Glide scoring function (results not shown), as we expected. That is, as we have seen elsewhere, the MM-GBSA rescoring approach is powerful for predicting substrate specificity but is sensitive to the details of the active site structure. We conclude that a major limitation of the prospective docking can be attributed to one key residue, Arg 385, being disordered in the unliganded structure, rather than problems intrinsic to the docking and scoring methods. Because the “missing” portion of the binding site in the unliganded structure was part of a large (16 residue) disordered region, it would have been very difficult to predict its precise interaction with an unknown ligand. Although in the long-term we seek to predict such large induced-fit (or “induced structure”) effects, the lesson learned from these studies is that the results with an unliganded structure can still be useful in guiding screening by focusing attention on a small subset of potential substrates, in this case six-carbon diacid sugars. One key to success was filtering the rersults to remove compounds that do not contain the correct reactive functional groups (carboxylates with α-protons in the enolase superfamily) and compounds whose reactive groups are predicted to bind in grossly incorrect orientations for catalysis.
Table 4.
Retrospective docking results against the “wild type” holo structure, using the 546 compound library of plausible substrates for members of the enolase superfmaily.
| Rank using MM-GBSA scoring | Rank using Glide SP scoring | Metabolite |
|---|---|---|
| 1 | 87 | Galactarate |
| 2 | 8 | L-Altrarate |
| 3 | 55 | D-Gularate/L-Glucarate |
| 4 | 35 | 5-Keto-4-deoxy-D-mannarate |
| 5 | 81 | 2,3-Dideoxy-D-glucarate |
| 6 | 4 | 2-Deoxy-D-glucarate |
| 7 | 48 | 2-Keto-3-deoxy-D-glucarate |
| 8 | 51 | 5-Keto-4-deoxy-D-glucarate |
| 9 | 6 | 5-Keto-4-Deoxy-L-mannarate |
| 10 | 9 | L-Talarate/L-Altrarate |
| 11 | 13 | L-Gularate |
| 12 | 40 | L-Allarate |
| 14 | 5 | L-Mannarate |
| 15 | 61 | D-Allarate |
| 16 | 54 | 3-Deoxy-D-glucarate |
The Seventh Subgroup in the Enolase Superfamily
On the basis of 1) the absence of a 20s loop with specificity determinants, 2) the identities and positions of the acid/base catalysts, 3) the identification of a His at end of the fifth β-strand in the barrel domain as a ligand for the Mg2+ ion that stabilizes the enolate intermediate (site A), and 4) the discovery of a second Mg2+ ion that provides specificity determinants for the distal “end” of the galactarate substrate, we recognize the GalrD-II subgroup as the seventh functionally and structurally defined subgroup in the enolase superfamily. Dehydration of acid sugars is now catalyzed by members of four subgroups: GlucD in the GlucD subgroup; FucD, TarD, TalrD/GalrD, GalD, and RhamD in the MR subgroup, ManD in the ManD subgroup, and GalrD-II in the GalrD-II subgroup. We will not be surprised if acid sugar dehydration is catalyzed by other yet-to-be-recognized subgroups with divergent active site architectures.
Summary
We used bioinformatics and an experimentally determined structure of the unliganded form of Ob2843 (PDB Code 2OQY) to guide its functional assignment as a novel galactarate dehydratase (GalrD-II). Although the C-terminal end of the polypeptide was disordered, including Arg 385 that provides a specificity determinant, the results of in silico ligand docking correctly predicted a diacid sugar substrate, which subsequently was identified as galactarate by physical library screening. Because the kinetic constants are similar to those of other characterized acid sugar dehydratases in the enolase superfamily, the galactarate dehydratase (GalrD-II) function was assigned. GalrD-II possesses a novel active site architecture that allows identification of the seventh sugbroup in the enolase superfamily. Although dehydration of galactarate is also catalyzed by a member of the MR subgroup (TalrD/GalrD), the products of the GalrD-II and TalrD/GalrD reactions are enantiomers so these enzymes represent apparent, not actual, convergent evolution of function in the enolase superfamily. This work provides further compelling evidence that an integrated sequence/structure-based strategy employing computational approaches is a viable approach for directing functional assignment of unknown enzymes discovered in genome projects.
ACKNOWLEDGEMENTS
We thank the members of the NYSGXRC protein production team for expression and purification of proteins used in this study.
Footnotes
This research was supported by NIH P01 GM071790 (to J.A.G., M.P.J., and S.C.A.) and NIH U54 GM074945 (to S.K.B.). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). M.P.J. is a consultant to Schrödinger LLC. The X-ray coordinates and structure factors for wild type GalrD-II from Oceanobacillus iheyensis (SeMet-labeled) liganded with Mg2+, wild type GalrD-II (SMet) liganded with Mg2+, wild type GalrD-II (SMet) liganded with Mg2+ and L-malate in two space groups, and the Y90F mutant of GalrD-II (SMet) liganded with Mg2+ and galactarate have been deposited in the Protein Data Bank (PDB Codes 2OQY, 3FYY, 3ES7, 3ES8, and 3HPF, respectively).
Abbreviations: AraD, D-arabinonate dehydratase; FucD, L-fuconate dehydratase; GalD, D-galactonate dehydratse; GalrD-II, galactarate dehydratase; GlucD, D-glucarate dehydratase; GluD, D-gluconate dehydratase; LB, Luria-Bertani broth; ManD, D-mannonate dehydtrase; MLE, muconate lactonizing enzyme; MR, mandelate racemase; NYSGXRC, New York SGX Research Center for Structural Genomics; TalrD/GalrD, L-talarate/galactarate dehydratase; TarD, D-tartrate dehydratase; TIM, triose phosphate isomerase.
REFERENCES
- 1.Babbitt PC, Hasson MS, Wedekind JE, Palmer DR, Barrett WC, Reed GH, Rayment I, Ringe D, Kenyon GL, Gerlt JA. The enolase superfamily: a general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry. 1996;35:16489–16501. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 2.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 3.Rakus JF, Fedorov AA, Fedorov EV, Glasner ME, Vick JE, Babbitt PC, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans. Biochemistry. 2007;46:12896–12908. doi: 10.1021/bi701703w. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, Kalyanaraman C, Fedorov AA, Fedorov EV, Glasner ME, Brown S, Imker HJ, Babbitt PC, Almo SC, Jacobson MP, Gerlt JA. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat Chem Biol. 2007;3:486–491. doi: 10.1038/nchembio.2007.11. [DOI] [PubMed] [Google Scholar]
- 6.Kalyanaraman C, Imker HJ, Fedorov AA, Fedorov EV, Glasner ME, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP. Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening. Structure. 2008;16:1668–1677. doi: 10.1016/j.str.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yew WS, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. Evolution of Enzymatic Activities in the Enolase Superfamily: l-Talarate/Galactarate Dehydratase from Salmonella typhimurium LT2(,). Biochemistry. 2007;46:9564–9577. doi: 10.1021/bi7008882. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, Abelson JN, Simon MI, editors. Methods in Enzymology. Academic Pres; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 10.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones AT. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 13.Brunger AT, Adams PD, Clore GM, DeLAno WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Lamzin VS, Wilson KS. Automated refinement for protein crystallography. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 15.CCP4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalyanaraman C, Bernacki K, Jacobson MP. Virtual screening against highly charged active sites: identifying substrates of alpha-beta barrel enzymes. Biochemistry. 2005;44:2059–2071. doi: 10.1021/bi0481186. [DOI] [PubMed] [Google Scholar]
- 18.Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30:402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen WL, Tirado-Rives J. The OPLS Force Field for Proteins. Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 21.Hubbard BK, Koch M, Palmer DR, Babbitt PC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: characterization of the (D)-glucarate/galactarate catabolic pathway in Escherichia coli. Biochemistry. 1998;37:14369–14375. doi: 10.1021/bi981124f. [DOI] [PubMed] [Google Scholar]
- 22.Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry. 2006;45:14582–14597. doi: 10.1021/bi061687o. [DOI] [PubMed] [Google Scholar]
- 23.Yew WS, Fedorov AA, Fedorov EV, Wood BM, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: D-tartrate dehydratase from Bradyrhizobium japonicum. Biochemistry. 2006;45:14598–145608. doi: 10.1021/bi061688g. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed H, Ettema TJ, Tjaden B, Geerling AC, van der Oost J, Siebers B. The semi-phosphorylative Entner-Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem J. 2005;390:529–540. doi: 10.1042/BJ20041711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouns SJ, Walther J, Snijders AP, van de Werken HJ, Willemen HL, Worm P, de Vos MG, Andersson A, Lundgren M, Mazon HF, van den Heuvel RH, Nilsson P, Salmon L, de Vos WM, Wright PC, Bernander R, van der Oost J. Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J Biol Chem. 2006;281:27378–27388. doi: 10.1074/jbc.M605549200. [DOI] [PubMed] [Google Scholar]
- 26.Wieczorek SW, Kalivoda KA, Clifton JG, Ringe D, Petsko GA, Gerlt JA. Evolution of Enzymatic Activities in the Enolase Superfamily: Identification of a “New” General Acid Catalyst in the Active Site of D-Galactonate Dehydratase from Escherichia coli. J. Am. Chem. Soc. 1999;121:4540–4541. [Google Scholar]
- 27.Rakus JF, Fedorov AA, Fedorov EV, Glasner ME, Hubbard BK, Delli JD, Babbitt PC, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase. Biochemistry. 2008;47:9944–9954. doi: 10.1021/bi800914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen TM, Wedekind JE, Rayment I, Reed GH. A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution. Biochemistry. 1996;35:4349–4358. doi: 10.1021/bi952859c. [DOI] [PubMed] [Google Scholar]
- 29.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 30.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 31.Gulick AM, Hubbard BK, Gerlt JA, Rayment I. Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D- glucarate dehydratase from Escherichia coli. Biochemistry. 2001;40:10054–10062. doi: 10.1021/bi010733b. [DOI] [PubMed] [Google Scholar]