Abstract
Switches between different phenotypes and their underlying states of gene transcription occur as cells respond to intrinsic developmental cues or adapt to changing environmental conditions. Post-translational modification of the master regulatory transcription factors that define the initial phenotype is a common strategy to direct such transitions. Emerging evidence indicates that the modification of key transcription factors by the small polypeptide ubiquitin plays a central role in many of these transitions1, 2. However, the molecular mechanisms by which ubiquitination regulates the switching of promoters between active and inactive states are largely unknown. Ubiquitination of the yeast transcriptional repressor α2 is necessary to evoke the transition between mating-types3, and here, we dissected the impact of this modification on α2 dynamics at its target promoters. The ubiquitination of α2 does not alter DNA occupancy by depleting the existing pool of the transcription factor, despite its well-characterized function in directing repressor turnover. Rather, α2 ubiquitination plays a direct role in the rapid removal of the repressor from its DNA targets. This disassembly of α2 from DNA depends on the ubiquitin-selective AAA-ATPase Cdc48. Our findings expand the functional targets of Cdc48 to active transcriptional regulatory complexes in the nucleus, a far broader role than previously anticipated. These data reveal an ubiquitin-dependent extraction pathway for dismantling transcription factor-DNA complexes and provide an archetype for the regulation of transcriptional switching events by ubiquitination.
Cell identity in Saccharomyces cerevisiae is determined by a set of master regulatory transcription factors that dictate cellular phenotype by regulating mating-type-specific gene expression programs 4. These regulators, and the gene expression states they control, must be labile since transitions between the two haploid mating-types occur readily and are evident phenotypically within the span of a single cell cycle5. For example, the α2 regulator represses the transcription of one cell type-specific gene set to help define the α cell phenotype. Upon the genetic switch of mating-type information, α2 expression is lost; the existing pool of the α2 repressor is rapidly turned over (α2 half-life is ~5 min6–8) and its regulatory targets are de-repressed, allowing the transition to a new cellular phenotype3. This rapid destruction of α2 is governed by the ubiquitin (Ub)-proteasome pathway7, 9, 10, and if this process is impaired, phenotypic switching is delayed3.
Since α2 promoter occupancy is critical for its functions, it is the elimination of this promoter-bound pool of α2 that is of paramount importance to the mating-type transition. To explore the role of ubiquitination in directing mating-type switching, we first determined how long the DNA operator-bound fraction of α2 occupies its binding sites by chromatin immunoprecipitation (ChIP) following the repression of new α2 synthesis with cycloheximide. After cycloheximide treatment, α2 was lost in a complex manner: initially, the level of occupancy remained relatively constant and then decayed rapidly only after a >5 min lag (Fig. 1a). This apparent biphasic decay contrasts with the behavior of the bulk population of the α2 protein, which is degraded with first-order kinetics (Fig. 1a). The discrepancy in the behavior of these two pools of α2 was surprising, since we expected that the DNA-bound and the free populations would be highly dynamic—as is the case with other transcription factors11–14 —and in rapid equilibrium, such that the rate-determining step for the removal of α2 from its DNA sites would be the rate of α2 turnover.
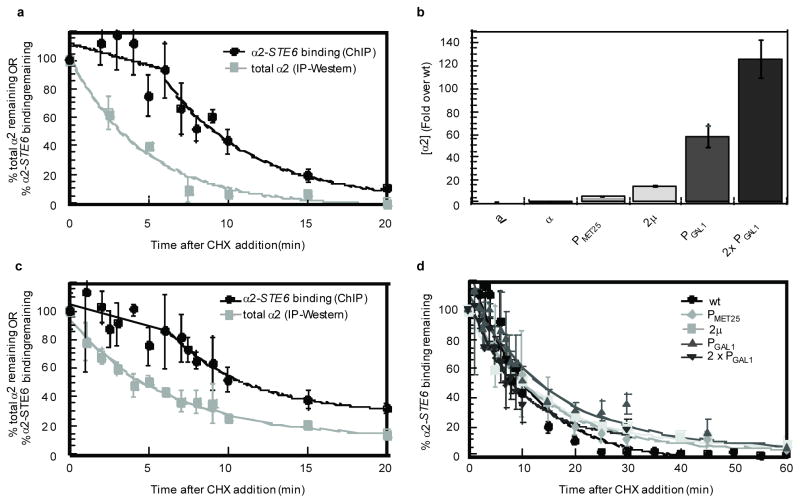
Figure 1. Increased expression of α2 does not affect its dissociation from DNA.
a, Dissociation of α2 from its DNA-binding site in the STE6 promoter compared to α2 degradation kinetics in wild-type cells, after the addition of cycloheximide (CHX). b, Relative levels of steady-state α2 expression in cells where α2 is expressed from a multi-copy vector (2μ) or from several different promoters. Cells containing two copies of a PGAL1-α2 construct are denoted 2 × PGAL1. c, The dissociation of α2 from its DNA-binding site in the STE6 promoter compared to the degradation kinetics of the α2 protein after the addition of CHX to cells over-expressing α2 from the GAL1 promoter. Note the similarity to a, despite the over-expression of α2. d, Dissociation of α2 from its STE6 DNA-binding site after addition of CHX to cells expressing varying amounts of α2. The time-course was expanded and the data from all of the different over-expression conditions is plotted together to highlight the observation that no increase in the DNA-binding lag time was apparent in cells expressing increased amounts of α2, which is in contrast to the specific prediction of a model in which α2 is depleted below its dissociation constant. The data for wild-type and PGAL1-α2 cells from a and c respectively are included for comparison. Note that the DNA-binding lag time is not apparent in d because a number of these experiments did not sample multiple times during the first five minutes of the time-course. For this reason and for simplicity, all of the dissociation curves in d were fit to an exponential decay. For easier visualization of the individual trends, the data in d is split into two parts and plotted again in Supplemental Fig. S1. Error bars represent s.e.m., n =2–8; min= minutes.
Since the loss of α2 from DNA did not mirror the decay of the entire pool of the protein, we considered two alternative interpretations. In the first scenario, α2 does indeed associate with its DNA target sites in a highly dynamic fashion but there is an excess of α2 protein in cells. The observed lag in the loss of α2 from DNA would then reflect the time required to degrade this excess pool of α2 down to a concentration below that necessary for effective association with its DNA-binding sites (its dissociation constant). In the second scenario, the fraction of α2 bound to its DNA operators and co-factors is a substrate for ubiquitination, but here the modification facilitates the removal of the protein from DNA. In this case, the lag observed in the cycloheximide-chase – ChIP experiments would reflect the time α2 remains in a DNA-bound complex before remodeling occurs.
To distinguish between these models, we determined the effects of α2 concentration on promoter occupancy. If the lag in the loss of α2 occupancy represents the time required to degrade α2 below its dissociation constant, then increasing the amount of α2 in cells should increase the length of the lag in a manner directly related to the level of α2 expression and the in vivo half-life of the protein. For example, if the level of α2 is increased 4-fold in cells, then the lag time observed in these cells should increase by two half-lives (or 10 min). To vary its steady-state levels, α2 was expressed in cells from a multi-copy vector or from promoters of different strengths. Using this system, we obtained a broad range of α2 levels (from ~4-fold to ~125-fold greater than the endogenous level; Fig. 1b), without altering the rate of α2 turnover (Fig. 1c and data not shown)6. Remarkably, the large increases in the quantity of α2 expressed in cells did not alter the fraction of α2 that remained bound to a target gene promoter after the repression of new α2 synthesis (Figs. 1c and 1d). For example, in cells over-producing α2 from the GAL1 promoter, which express ~60-fold more protein (Fig. 1b), the dissociation of α2 appears nearly identical to cells expressing endogenous levels of the protein (Fig. 1c). Since dramatically increasing the amount of α2 does not enhance the DNA-binding lag time, these results suggest that the concentration of α2 in wild-type cells may already be well below that necessary to saturate its DNA-binding sites in vivo. Indeed, measurements of the amount of endogenous α2 in cells indicate that the concentration of α2 is ~3-fold less than its dissociation constant, as determined by in vitro DNA-binding assays15–20.
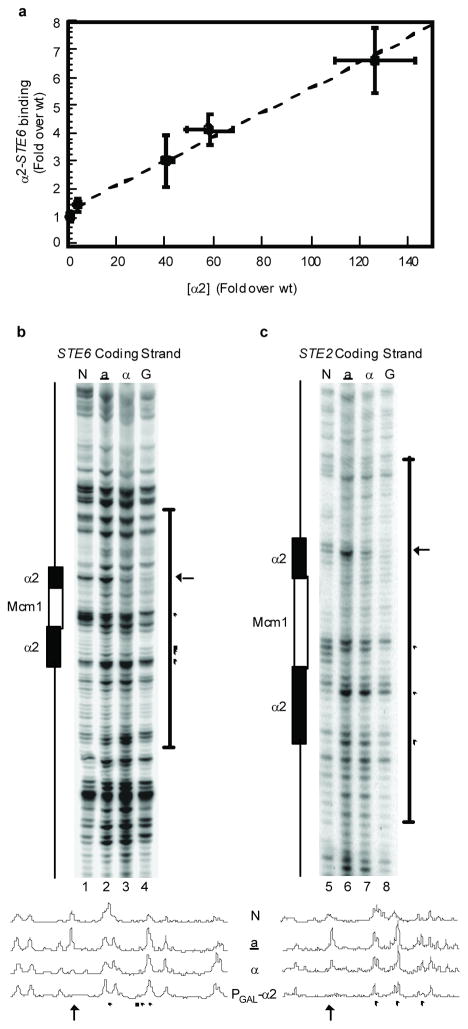
To examine this inference directly, we determined the relationship between α2 protein levels and binding to its sites in target gene promoters in vivo using ChIP assays. In cells expressing increased amounts of α2, the level of association with the STE6 promoter increased in a dose-dependent manner (Fig. 2a). Similar increases in DNA-binding upon α2 over-expression were observed at five other known genomic targets of the protein (supplemental Fig. S2), indicating that the concentration of endogenous α2 is well below its in vivo dissociation constant. To confirm this observation with another assay that is independent of potential antibody avidity effects, we performed genomic footprinting analyses that similarly probe DNA-protein interactions in vivo but do not utilize immunoprecipitation. Maps of α2-target promoters were obtained at nucleotide-resolution using dimethyl sulfate (DMS) modification followed by primer extension. When compared with a cells that do not express α2, α cells show protection of a guanine residue in the α2 binding site upstream of STE6 and STE2 (arrows in Figs. 2b and 2c), in agreement with previous reports15, 21. However, this protection from DMS attack was not absolute on either α2 target, and was strongly increased in cells expressing higher levels of α2 (Figs. 2b and 2c). In addition, a number of other residues within the α2 binding sites in both promoters gain protection from methylation or show enhanced DMS modification in cells whereα2 expression is augmented (Figs. 2b and 2c). The results of these genomic footprinting assays are consistent with those of the ChIP experiments (Fig. 2a) and demonstrate that α2 does not fully occupy its DNA-binding sites in vivo. Thus, the concentration of α2 in MATα cells must be below its in vivo dissociation constant. Taken together with the observations described above (Fig. 1c and 1d), these findings are strongly inconsistent with a role for the Ub system in the depletion of α2 below its dissociation constant.
Figure 2. Over-expression of α2 leads to increased DNA-binding.
a , Increased expression of α2 correlates with increased binding to the STE6 promoter. α2 was expressed in cells from its endogenous promoter, the Ptet07, PGPD, or PGAL1 promoters, or in cells carrying two copies of a PGAL1-α2 construct. Error bars indicate s.e.m.; n =6–12. b, Genomic DMS footprinting of the STE6 promoter coding strand. A schematic showing the position of the α2-Mcm1 operator is diagrammed on the left. N= naked DNA, a= a cells, α= α cells, G=α cells with α2 expressed from the GAL1 promoter. The arrow indicates increased protection of a G residue in the α2-binding site from methylation by DMS. ● indicates additional α2-related increases in protection from methylation due to α2 expression levels. ■ indicates α2-related enhancement of methylation due to α2 expression levels. c, similar to b, but of the STE2 promoter coding strand. Line graphs shown below are vertical line quantitations of the band intensities from the bracketed regions.
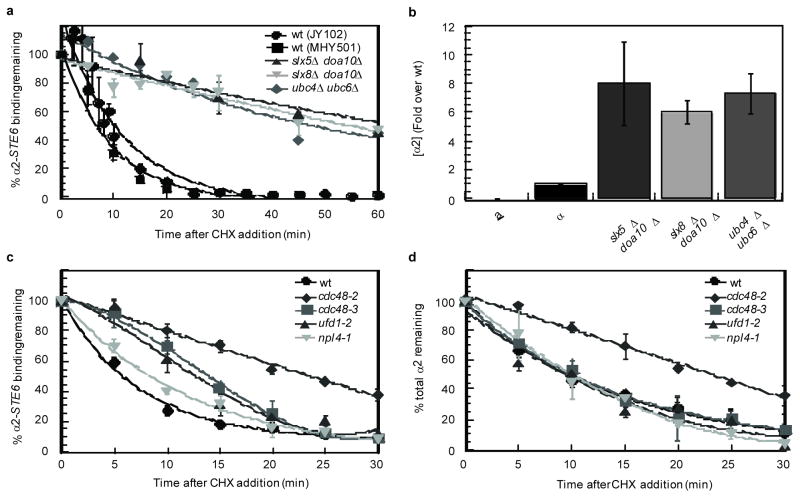
Since the dynamics of α2-target promoter interactions were not influenced by α2 concentration, we next determined if the ubiquitination of α2 plays a role in the removal of the protein from its DNA targets. At least two different ubiquitination pathways target α2 for rapid turnover7, 8 (Y. Xie and M. Hochstrasser, personal communication). When both of these pathways were disrupted, the binding of α2 to its DNA target sites strongly persisted after α2 expression was inhibited (Fig. 3a). For example, in cells lacking the E2 ubiquitin-conjugating enzymes Ubc4 and Ubc6, ~50% of the ChIP signal from α2 binding to the STE6 promoter remained 60 min after α2 expression was blocked. Similar results were observed in doubly mutant cells deleted for the genes encoding the E3 Ub ligases Doa10 and Slx5–Slx8 (Fig. 3a). These observations contrast greatly with the behavior of wild-type cells and with that of singly mutant cells that show wild-type levels of ubiquitinated α27: these strains exhibited background levels of α2 binding to STE6 promoter DNA only 20 min after the loss of α2 expression (Fig. 3a and data not shown). The α2 protein was stabilized in all of these double mutant backgrounds, resulting in a ~7-fold increase in the steady-state level of the protein (Fig. 3b) and a ~2-fold increase in target gene binding (data not shown). However, the strong persistence of α2 occupancy that is observed cannot be explained simply by such a modest increase in the level of the protein or target gene binding, since much higher levels of α2 expressed from strong promoters did not lead to any appreciable change in the dynamics of the α2-DNA interaction (Fig. 1b–1d). Therefore, these results argue that α2 ubiquitination plays a direct role in the remodeling of this transcription factor complex.
Figure 3. Cdc48 and the ubiquitination machinery of α2 are actively involved in its dissociation from DNA.
a, Dissociation of α2 from its DNA-binding site in the STE6 promoter in cells deficient in α2 ubiquitination after the addition of CHX. The data for wild-type cells in the JY102 background is the same as that presented in Fig. 1. For simplicity, the dissociation curves for wild-type cells were fit to an exponential decay. Note that these ubiquitination-deficient strains increase the amount of α2-DNA binding ~2-fold relative to wild-type. b, Steady-state α2 expression levels in cells that are deficient in ubiquitination of α2. Note that these ubiquitination-deficient strains increase the steady-state level of α2 ~7-fold, while cells containing two copies of a PGAL1-α2 construct have ~125-fold more α2 than wild-type and do not show any effect on the dissociation kinetics. c, Dissociation of α2 from its DNA-binding site in the STE6 promoter after the addition of CHX at the non-permissive temperature (37°C). In the time-courses shown in a and c, the DNA-binding lag time is not apparent because most of these experiments did not sample multiple times during the first five minutes of the time-course. To control for the specificity of these dissociation defects in cdc48 mutants, we determined the dynamics of α2 binding in cdc48-3 strains grown at the permissive temperature (30°C) and observed that α2 dissociated from its DNA targets with wild-type kinetics (data not shown). d, Degradation of α2 after the addition of CHX at the non-permissive temperature (37°C). Error bars represent s.e.m., n =2–4; time-courses are fit to an exponential decay, except for the DNA dissociation time-course of cdc48-3 and ufd1-2, which are best fit by a polynomial (3); min= minutes.
One mechanism utilized by cells for the remodeling of transcriptional regulators on DNA involves molecular chaperones. For example, protein-DNA complexes containing nuclear hormone receptors in mammalian cells are actively disassembled by the molecular chaperone p23 (Sba1 in yeast)22. Therefore, we examined the dynamics of α2-target promoter interactions in wild-type and sba1Δ cells but observed no differences (A. DeSimone and J.D.L., data not shown), suggesting that other chaperones may function to disassemble α2-containing complexes. Given that the removal of α2 from its cognate promoters depends on the ubiquitination machinery that targets α2 (Fig. 3a), we determined if Cdc48, a Ub-selective AAA ATPase, plays a role in remodeling these complexes. Perhaps the best-characterized function for Cdc48 is in dislocating substrates into the cytosol during endoplasmic reticulum-associated degradation, but it also has been implicated in a growing number of additional cellular pathways where it appears to use the energy of ATP hydrolysis to generate mechanical force that can disassemble protein complexes or extract proteins from intracellular structures23–30. If the Cdc48 complex similarly promotes the dissociation of α2 from its DNA targets, then loss-of-function mutations in CDC48 should lead to persistent α2-target gene binding after α2 synthesis is blocked. To test this notion, α2 occupancy of the STE6 promoter was determined by cycloheximide-chase – ChIP experiments in congenic wild-type and temperature-sensitive cdc48 mutant cells. The association of α2 with this target promoter was quickly diminished in wild-type cells, but persisted in the absence of functional Cdc48. The dynamics of α2 binding were strongly affected in cdc48-2 cells and were altered in a more complex manner in cdc48-3 and cdc48-14 mutants: soon after α2 synthesis was blocked, STE6 occupancy remained relatively high and was similar to that observed in cdc48-2 mutants, but then decayed more rapidly (Fig. 3c and Supplemental Fig. S3). The activity of Cdc48 is linked to specific cellular pathways by a set of ubiquitin-binding adaptors and one of the best characterized is the Ufd1-Npl4 complex30. In ufd1-2 mutants, α2 binding dynamics were similar to that seen in cdc48-3 strains, while a much more modest defect was observed in two different npl4 strains (Fig. 3c and data not shown). Interestingly, the stability of the entire steady-state population of α2 was increased in cdc48-2 mutants but was not altered by the cdc48-3, cdc48-14, or any of the adaptor mutations (Fig. 3d and Supplemental Fig. S4). The finding that α2 exhibited persistent occupancy of its target promoter in cdc48-3, cdc48-14, or ufd1-2 mutants in the absence of any increase in the stability of the protein provides particularly compelling evidence that α2 is not lost from its DNA sites by simple biochemical dissociation; instead, these results indicate that the rapid disassembly of theα2-DNA complex requires the activity of the Ub-selective AAA-ATPase Cdc48. Furthermore, the differential effects of these cdc48 mutants on α2 suggest that Cdc48 mediates two different aspects of α2 metabolism: its dissociation from target promoters and its destruction by the Ub-proteasome pathway.
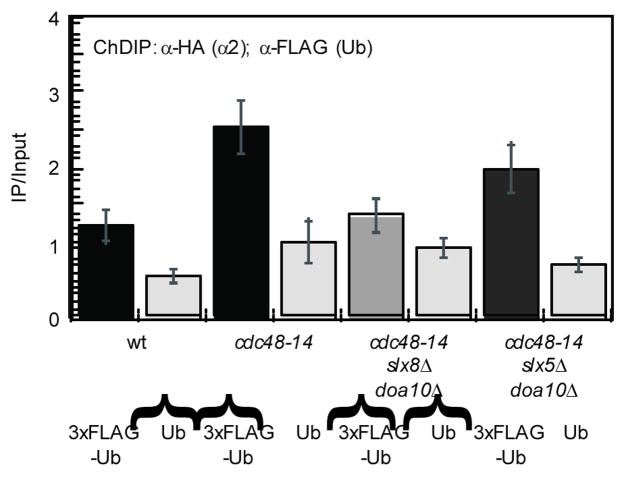
The observation that the Cdc48 complex is necessary for the dissociation of α2 from its DNA targets suggests that the promoter-bound fraction of α2 is ubiquitinated, since most, if not all, functions of Cdc48 are dependent on the ability of the AAA-ATPase to bind to ubiquitinated proteins30. To directly test this hypothesis, we measured the levels of modified α2 by a chromatin double immunoprecipitation (ChDIP) assay and observed that ubiquitinated α2 was modestly enriched at the STE6 promoter relative to the untagged Ub background (Fig. 4). The involvement of Cdc48 in the dissociation of α2 from its DNA target sites suggests that the amount of ubiquitinated α2 on promoter DNA may be enhanced when the function of Cdc48 is abrogated. In agreement with this idea, the level of STE6 promoter DNA associated with ubiquitinated α2 was increased when ChDIP was performed with chromatin from cdc48-14 cells (Fig. 4). Since other chromatin-associated proteins are ubiquitinated (for example, histone H2B31) and potentially could be co-precipitated with α2, it was important to determine that the enrichment observed at STE6 was due to ubiquitinated α2 and not another factor. If the ChDIP signal truly represents ubiquitinated α2, it should be diminished when cells are deleted for the genes encoding Doa10 and Slx5–Slx8, the Ub ligase enzymes that target α28 (Y. Xie and M. Hochstrasser, personal communication). Indeed, when ChDIP was performed with chromatin from cdc48-14 slx8Δ doa10Δ cells, the signal was reduced to near-background levels (Fig. 4). Interestingly, the ChDIP signal was only modestly reduced when cdc48-14 slx5Δ doa10Δ cells were analyzed, implying that Slx8 has at least residual Ub ligase activity in vivo even in the absence of its heterodimeric partner Slx5, similar to its activity in vitro32. In addition, we determined that the enrichment of STE6 promoter DNA containing ubiquitinated α2 in cdc48-14 cells was similar to that found in strains containing cdc48-14 and any of the single Ub ligase deletions (Supplemental Fig. S5). Because α2 ubiquitination is reduced only when both of its ubiquitin-conjugation pathways are disrupted7, these results are further evidence that the ChDIP signals represent ubiquitinated α2. Taken together, these observations indicate that the functionally engaged pool of α2 is ubiquitinated in vivo, consistent with its CDC48-dependent removal from DNA.
Figure 4. α2 bound to its specific target sites is ubiquitinated.
Quantitation of DNA co-immunoprecipitated with α2HA, eluted, and then co-precipitated with either 3xFLAG-tagged ubiquitin or untagged ubiquitin. ChDIP was performed with wild-type, cdc48-14, cdc48-14 slx8Δ doa10Δ, and cdc48-14 slx5Δ doa10Δ cells grown at the non-permissive temperature (37°C) and the ChDIP values are normalized to the untagged ubiquitin negative control in cdc48-14 cells. All of the ChDIP values are statistically distinct from cdc48-14 + 3xFLAG-Ub at a significance level of p<0.015 (Student’s t-test), except for those from cdc48-14 slx5Δ doa10Δ + 3xFLAG-Ub cells. Error bars represent s.e.m., n=3–6.
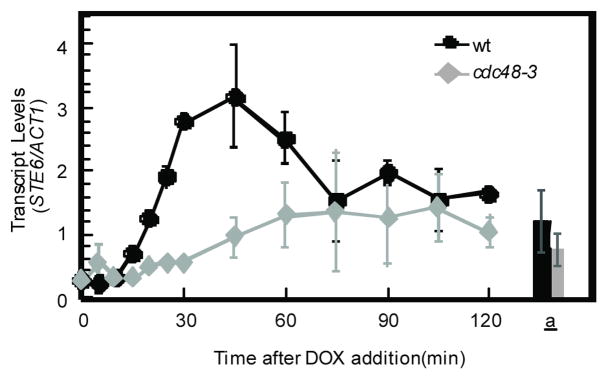
The data presented thus far suggest a model in which the pool of α2 functionally engaged on its target promoters is ubiquitinated and that this modification targets the AAA-ATPase Cdc48 to these protein-DNA complexes for their rapid disassembly. A strong prediction of this model is that the activity of Cdc48 would be required for the timely de-repression of α2-target genes, since the persistent binding of α2 in the absence of Cdc48-dependent dissociation should lead to the continued repression of these promoters. To examine this hypothesis, we analyzed the de-repression of the STE6 mRNA in wild-type and cdc48-3 strains in which the sole copy of α2 was expressed from a doxycycline (DOX)-repressible promoter. The cdc48-3 mutant was chosen specifically because the rapid turnover of α2 is not altered in this strain (Fig. 3d). In wild-type cells, the expression of STE6 was rapidly de-repressed, showing a burst of transcripts that peak 45 min after the addition of DOX before decaying to the steady-state levels observed in MATa cells. In contrast, the de-repression of STE6 transcription was severely blunted in cdc48-3 cells: no burst of STE6 transcripts was apparent and the accumulation of mRNA only slowly approached the level of STE6 in MATa cells (Fig. 5). Thus, the rapid disassembly of α2-promoter DNA complexes by Cdc48 is required for the robust and timely de-repression of an α2-target gene.
Figure 5. The rapid dissociation of α2 from promoter DNA is required for the timely de-repression of an α2-target gene.
The de-repression of STE6 transcription after the doxycycline-induced loss of α2 was determined by quantitative reverse transcription-PCR. Doxycycline was added to cells containing a PtetO2-α2 construct to inhibit α2 synthesis and the amount of STE6 mRNA was normalized to that of ACT1 mRNA. Wild-type and cdc48-3 cells containing PtetO2-α2 were grown at the non-permissive temperature (37°C). The amount of STE6 mRNA in wild-type MATa and MATa cdc48-3 strains are shown for comparison (bars). Error bars represent s.e.m., n =2–3.
Like many other transitions in cellular identity, the switching of yeast cells from one mating-type to another is dependent upon two processes: a block to the continued expression of the master regulatory proteins of the initial state and the rapid inactivation of the existing pool of the same regulators. Perhaps the most efficient means of inactivating a protein is to induce its destruction, and not surprisingly, many cell- and organism-based phenotypic transitions are dependent upon the Ub-proteasome pathway33, 34. Our studies on the transcriptional repressor α2, however, indicate that ubiquitination contributes to cellular dynamics through an additional pathway that is independent of protein stability. Given the growing number of transcription factors that have been shown or inferred to be targets of ubiquitination and the increasing links between components of the ubiquitination machinery and transcriptional activity2, the previously unappreciated Ub-mediated remodeling of promoters is likely to profoundly impact the regulation of gene expression in eukaryotes.
Supplementary Material
Acknowledgments
We are grateful to Yang Xie and Mark Hochstrasser for sharing data and reagents in advance of publication, for providing strains and plasmids, and for many helpful discussions. We thank Randy Hampton and Stefan Jentsch for providing strains, Rachel Whitaker and Rob Reenan for the sequencing of cdc48 alleles, Janet Mead and Drew Vershon for the His-tagged α2 construct, Steve Gregory and Al Dahlberg for their help with the footprinting experiments, and Tricia Serio for numerous helpful discussions. Our ideas were also shaped by discussions with Keith Wilkinson and Ray Deshaies. The manuscript was improved by comments from Tricia Serio, Judith Bender, Yang Xie, Mark Hochstrasser, Rob Reenan, Alec DeSimone, and an anonymous reviewer. This work was supported by a grant from the National Institutes of Health (GM71764 to JDL) and by a Basil O’Connor Starter Scholar Research Award from the March of Dimes.
Footnotes
AUTHOR CONTRIBUTIONS
A.J.W. performed all of the experiments. A.J.W. and J.D.L. conceived, designed, and analyzed the experiments and prepared the manuscript.
COMPETING INTERESTS
The authors declare that they have no competing financial interest.
References
- 1.Dennis AP, O’Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Laney JD, Hochstrasser M. Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev. 2003;17:2259–2270. doi: 10.1101/gad.1115703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 5.Nasmyth K, Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987;237:1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- 6.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 8.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter-Ruoff B, Wolf DH, Hochstrasser M. Degradation of the yeast MATα2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M, Ellison MJ, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci U S A. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 12.Stenoien DL, et al. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol Cell Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 14.Karpova TS, et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 15.Keleher CA, Goutte C, Johnson AD. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988;53:927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- 16.Smith DL, Johnson AD. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992;68:133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- 17.Vershon AK, Johnson AD. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell. 1993;72:105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- 18.Vershon AK, Jin Y, Johnson AD. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes Dev. 1995;9:182–192. doi: 10.1101/gad.9.2.182. [DOI] [PubMed] [Google Scholar]
- 19.Zhong H, Vershon AK. The yeast homeodomain protein MATalpha2 shows extended DNA binding specificity in complex with Mcm1. J Biol Chem. 1997;272:8402–8409. doi: 10.1074/jbc.272.13.8402. [DOI] [PubMed] [Google Scholar]
- 20.Zhong H, McCord R, Vershon AK. Identification of target sites of the alpha2-Mcm1 repressor complex in the yeast genome. Genome Res. 1999;9:1040–1047. doi: 10.1101/gr.9.11.1040. [DOI] [PubMed] [Google Scholar]
- 21.Ganter B, Tan S, Richmond TJ. Genomic footprinting of the promoter regions of STE2 and STE3 genes in the yeast Saccharomyces cerevisiae. J Mol Biol. 1993;234:975–987. doi: 10.1006/jmbi.1993.1652. [DOI] [PubMed] [Google Scholar]
- 22.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 23.Acharya U, et al. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 24.Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 25.Hetzer M, et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 26.Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 27.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 28.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 29.Ramadan K, et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 30.Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 33.Deshaies RJ. The self-destructive personality of a cell cycle in transition. Curr Opin Cell Biol. 1995;7:781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 34.DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.