Abstract
The structural basis for nucleotide incorporation fidelity remains an open question for all nucleic acid polymerases. Addressing this question for the viral RNA-dependent RNA polymerase (RdRp) is of particular, practical significance because it is a determinant of sensitivity to antiviral nucleosides and may be a determinant of viral virulence. All polymerases are thought to employ the same catalytic mechanism, but the rate of nucleotide incorporation can vary substantially. Here we review some of the recent work with the RdRp that leads us to suggest that structure provides only a partial understanding of RdRp function and dynamics may be the missing link.
Introduction
RNA viruses represent the largest class of existing and emerging human pathogens. Few vaccines have been licensed to prevent RNA virus infections, and only one broad-spectrum antiviral (ribavirin) has been licensed to treat RNA virus infections in humans. Unfortunately, this single antiviral agent is not effective against all RNA viruses [1]. Replication of the RNA virus genome is catalyzed by the virus-encoded RNA-dependent RNA polymerase (RdRp) [2]. Like other viral polymerases, the RdRp is an important target for the development of antiviral therapeutics. Rational design of RdRp-specific, nucleotide-based inhibitors and/or substrates would benefit from the knowledge of how catalytic efficiency and nucleotide specificity are engineered into the active site of the enzyme. Addressing these very fundamental issues regarding polymerase function is among the most active areas of polymerase enzymology. Herein we review some of the most recent studies of RdRp structure and function. We conclude that existing gaps in our ability to connect RdRp structure to RdRp function may require investigations into RdRp dynamics.
Structures of the RdRp are unable to explain the reduced catalytic efficiency of antiviral nucleoside triphosphates
RdRps have a semi-classical structural organization that is often compared to a right hand (Figure 1a). The enzyme not only has the fingers, palm, and thumb subdomains found in other classes of nucleic acid polymerases but also has a fingertips subdomain that leads to the formation of a completely encircled active site. Seven conserved structural elements, termed motifs A–G, exist. Motifs A–E are located in the palm subdomain and contribute to aspects of nucleotide and nucleic acid binding, as well as catalysis. Motif C is the most well known motif as it contains the GDD sequence that is a hallmark of viral RdRps. The function of motif D is only now becoming clear and will be discussed below. Motifs F and G are located in the fingertips and fingers subdomains, respectively. Motif F contains basic amino acid residues that appear to be responsible for ground-state binding of the triphosphate moiety of the incoming nucleotide [2]. The function of motif G is not absolutely clear, although substitutions of residues in this motif inhibit polymerase activity [3].
Figure 1.
Limited flexibility observed of the RdRp during the catalytic cycle. RdRp from foot-and-mouth disease virus (FMDV) is employed here but is a representative for most viral RdRps [8••]. (a) FMDV RdRp in the absence of ligands. The conserved structural motifs are highlighted by different colors (motif A, magenta; motif B, yellow; motif C, orange; motif D, blue; motif E, cyan; motif F, green; and motif G, gold). (b) Kinetic parameters: Kd,app and kpol for correct (ATP), incorrect (GTP), and ambiguous (RTP) nucleotide incorporation catalyzed by FMDV RdRp. Parameters were obtained from Ref. [31]. (c) Close-up view of the RdRp active sites in the presence of ligands: RNA and UTP (left, pdb 2E9Z); RNA and RTP (right, pdb 2E9R). The 3′-terminal nucleotide of RNA primer is shown as red sticks; the nucleotide and the nucleotide analog are shown as black sticks. (d) Active site residues within 5 Å of the RNA terminus and incoming nucleotides are shown as balls and sticks.
RdRps have often been characterized as error-prone polymerases. However, it is now clear that the low fidelity is primarily due to the absence of proofreading exonuclease activity [4]. RNA viruses with genomes longer than 10 000 nt, coronaviruses, for example, may encode a proofreading activity [5••]. Transition mutations do occur with a frequency of 1 in 10 000, primarily due to GMP misincorporation opposite uridine (Figure 1b). RdRps do not distinguish correct nucleotides from incorrect nucleotides or nucleotide analogs, such as ribavirin triphosphate (RTP), at the binding step as the apparent dissociation constants (K d,app) for all nucleotides are quite similar (Figure 1b), consistent with motif F interactions with the triphosphate governing nucleotide binding [4]. The primary fidelity checkpoint for the RdRp occurs after binding and represents a conformational change that has been attributed to the reorientation of the triphosphate into a position for efficient catalysis [4, 6]. Changes in the equilibrium constant for this conformational-change step manifest as a change in the maximal rate constant for nucleotide addition (k pol) (Figure 1b).
One of the most exciting advances in the field of RdRp enzymology recently was the solution of structures for the RdRp from foot-and-mouth disease virus (FMDV) in complex with primed template and various correct nucleotides and RTP [7, 8••]. Because the fingertips of the RdRp constrain movement of the fingers subdomain relative to the thumb subdomain, reorganization of these subdomains did not occur as observed in polymerases that lack a fingertips subdomain [8••]. This observation suggests that subdomain movements are not responsible for rate-limiting, conformational-change steps observed by kinetic analysis of the single-nucleotide-addition cycle. The most substantial difference between a complex with a correct nucleotide (UTP in Figure 1c) relative to a complex with an inefficiently incorporated nucleotide analog (RTP in Figure 1c) was the position of residues, such as Lys-369, on motif D. The complexes are essentially superimposable when atoms involved in catalysis are interrogated (Figure 1d). As discussed below, Lys-369 (Lys-359 in poliovirus (PV) RdRp) of motif D is a dynamic element that influences the rate of catalysis. Structural studies may fail to identify the basis of fidelity because they are unable to capture the trajectory of motif D during a catalytic cycle.
Remote sites control active site, fidelity checkpoints
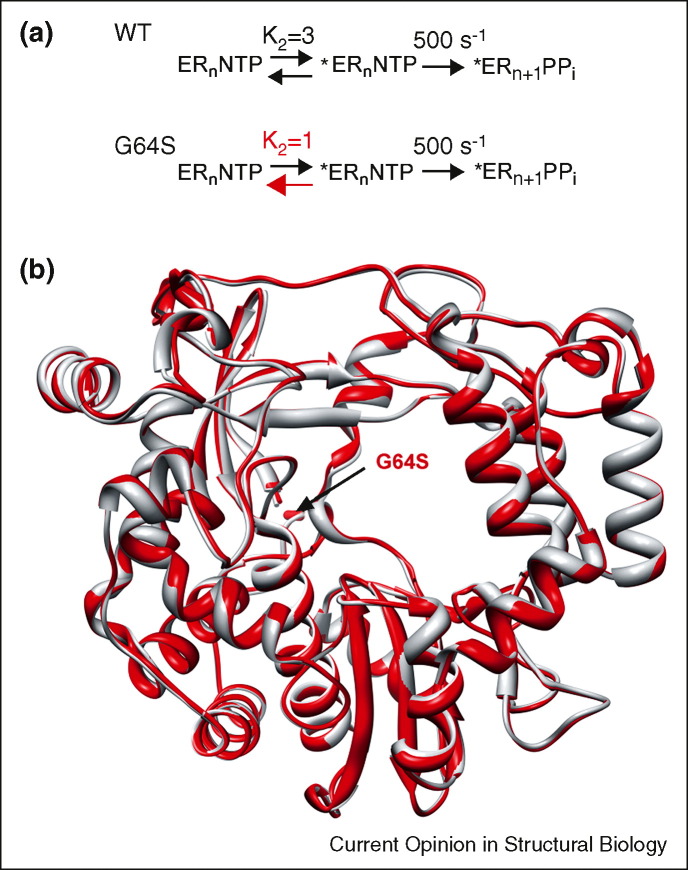
Several surprises regarding the mechanistic basis for RdRp fidelity came from studies of PV that were aimed at identifying a high-fidelity polymerase by selecting for PV mutants that exhibit reduced sensitivity to ribavirin [9, 10]. The first surprise was that a site remote from the catalytic center controlled fidelity. Gly-64 of the PV RdRp, a residue in the fingers subdomain, was changed to Ser. Gly-64 has an indirect connection to motif A in the active site; this motif contributes to interactions in the ribose-binding site and coordination of Mg2+ ions required for catalysis [9]. A second surprise was that this same mutant was isolated independently by two different laboratories, suggesting that few solutions may exist to development of resistance to this class of antiviral nucleosides, a distinct contrast to other classes of nucleoside-based RdRp inhibitors [11].
Mechanistic studies of the PV G64S polymerase revealed that this substitution decreased the equilibrium constant for the conformational-change step (K 2) preceding chemistry (Figure 2a) [9]. The more difficult it is for the enzyme to achieve the catalytically competent conformation, the more faithful the nucleotide-addition cycle will be. Structural studies of the G64S polymerase in complex with substrates have not been completed, but the structure of the unliganded enzyme provided no insight into changes in the organization of motif A as might have been predicted (Figure 2b). Because conformational changes appear to be altered by the G64S substitution, analysis of the dynamics during the nucleotide-addition cycle may be most insightful.
Figure 2.
Structure is unable to reveal the basis for RdRp fidelity. (a) The conformational-change step (K2) is used to alter fidelity of G64S PV RdRp [9]. (b) Crystal structure of the high-fidelity mutant G64S in PV-3Dpol is essentially identical to the WT (rmsd of 0.33 Å) [26•].
Biological studies of PV encoding the G64S substitution have shown that the fidelity of the PV RdRp appears to be important for pathogenesis. The ability for G64S PV to cause disease was attenuated in a very permissive mouse model [12•, 13•]. The attenuated phenotype was shown to be a reflection of the reduced population diversity produced by a high-fidelity polymerase by using chemical mutagenesis to restore virulence to the G64S PV population [13•]. Importantly, G64S PV and related mutants confer long-term protection of mice from a lethal challenge with wild-type PV [14••]. Therefore, an understanding of how fidelity is engineered into the RdRp structure has practical implication for the development of vaccines.
A determinant of nucleotide incorporation efficiency resides on a highly mobile structural element in all nucleic acid polymerases
Nucleic acid polymerases employ a two-metal-ion mechanism for catalysis [15]. One metal (B in Figure 3a) enters the active site with the nucleotide; the other (A in Figure 3a) enters after the nucleotide. Metal B likely influences organization of the triphosphate in the active site. However, metal A lowers the pK a value for the 3′-OH of the primer and contributes to charge neutralization that forms during the transition state. The acceptor for the proton from the primer 3′-OH is not known. Recent studies of the nucleotidyl transfer reaction catalyzed by the RdRp and other polymerases have made the unexpected observation that two protons are transferred during the reaction, not just one proton [16]. Moreover, it is now clear that the second proton derives from a basic amino acid (general acid) of the polymerase and is transferred to the pyrophosphate leaving group (Figure 3a) [17••]. Protonation of the pyrophosphate leaving group is not absolutely essential but contributes 50-fold to 1000-fold to the rate constant for nucleotide addition. The general acid is located on motif D of RdRps and reverse transcriptases (RTs), on helix O of A-family polymerases (e.g. T7 RNA polymerase), helix P of B-family polymerases (e.g. RB69 DNA polymerase) and the trigger loop of multisubunit RNA polymerases (Figure 3a) [17••].
Figure 3.
Substantial flexibility of a functionally important element observed in all classes of nucleic acid polymerases. (a) Extending the two-metal-ion mechanism of nucleotidyl transfer to include general acid catalysis. The positions of general acids of the model polymerases are shown. (b) Conserved structural motif D is a dynamic element that approaches the triphosphate moiety of the incoming nucleotide. To the left are shown ribbon representations of the superimposed RdRps from poliovirus (PV), foot-and-mouth disease virus (FMDV), Norwalk virus (NV), and human rhinovirus (HRV) types 14/16/1B, as well as the RT from the human immunodeficiency virus type 1 (HIV1) (amino acid residues 1–315 of the catalytic p66 subunit). The displayed structures were taken from the following PDB files (PV: 1RA6, 1RA7; FMDV: 1U09, 1WNE, 2E9Z; NV: 1SH0, 1SH1, 3BSO; HRV14: 1XR5; HRV16: 1XR7; HRV1B: 1XR6; HIV1-RT: 1RT1, 1RTD). Motif D in the various polymerases is color coded as indicated in the lower right panel. The GTP substrate (from PV binary complex) is shown as sticks at the catalytic cleft. (c) Dynamics of structural elements containing the general acid in DNA-dependent polymerases. Comparison of enzyme–primer/template complexes to enzyme–primer/template–nucleotide complexes reveals movement of the general acid in response to nucleotide binding. Movement of helix P (left) in RB69 DdDp created from PDB entries: 1IG9, 2DTU. Movement of helix O in T7 DdRp (center) created from PDB entries: 1S76, 1MSW. Movement of the trigger loop in yeast RNA polymerase II created from PDB entries: 2E2H, 2YU9, 1Y1V. The conformations corresponding to the ternary complexes are gray. The figure is adapted from Ref. [17••].
The general acid is Lys-359 in PV RdRp, Lys-369 in FMDV RdRp, and Lys-220 in the RT from human immunodeficiency virus (HIV). This residue is absolutely conserved in all RdRps and RTs. Motif D is the most dynamic structural element of RdRps and RTs (Figure 3b). Surprisingly, none of the structures available for RdRps or RTs, presumably poised for catalysis, reveal a conformation that is consistent with the ability of this residue to function as a general acid. The general acid (Lys-374) of the RdRp from a Norovirus (NV) comes the closest (Figure 3b).
It is not at all clear why the FMDV RdRp and HIV RT structural studies have not succeeded in capturing the general acid and motif D in its active conformation. Studies of the other polymerases have successfully captured the general acid in the active conformation (Figure 3c). Interestingly, all of the structural elements that contain the general acid undergo substantial changes in conformation after nucleotide binding (Figure 3c). This conformational change may be related to the rate-limiting step (K 2) that is used for the fidelity of nucleotide addition. Consistent with this possibility, amino acid substitutions in helix O of A-family polymerases and the trigger loop of multisubunit RNA polymerases contribute to fidelity [18, 19, 20, 21, 22, 23•]. If this is also the case for motif D of the RdRp, then this motif may represent an important target to alter fidelity of the viral RdRp for viral attenuation and vaccine development as done for the G64S PV RdRp. Motif D may represent a universal target for the control of viral fidelity and viral attenuation in all RNA viruses. Also worth noting is the observation that one of the attenuating mutations in the Sabin vaccine strain for PV type 1 converts Thr-362 of the RdRp to Ile [24]. Given its proximity to Lys-359, the Thr-to-Ile substitution at position 362 may impact Lys-359 function. In conclusion, dynamics of motif D will clearly impact function. We suggest that motif D dynamics may hold the key to our understanding of RdRp fidelity and our ability to tune RdRp fidelity for applications such as vaccine development.
Structure of an RdRp precursor cannot explain the absence of polymerase activity
Picornaviruses produce a single polyprotein that is post-translationally processed by viral proteases to produce a variety of incompletely and terminally processed products. For example, in addition to the 3C protease and 3D RdRp, a 3C–3D fusion protein (3CD) is also made. Like 3C, 3CD has protease activity, but unlike 3D, 3CD lacks RdRp activity. The molecular basis for this difference has been of great interest to picornavirologists. A recent suggestion was that the fingers subdomain of 3D was derived from a ‘chameleon’ sequence [25]. As long as the amino terminus of 3D is free to bury itself into the fingers subdomain, then the fingers subdomain would fold into conformation suitable for RdRp function [25]. When the amino terminus of 3D is fused to 3C, however, a new conformation that is inconsistent with RdRp function would be realized. The first structure of a picornaviral 3CD protein was solved recently [26•]. As shown in Figure 4 , 3CD is a composite of 3C and 3D. The only hypothesis that could be put forward to explain the absence of RdRp activity in 3CD was that the dynamics of the 3D domain of 3CD may be negatively impacted [26•].
Figure 4.
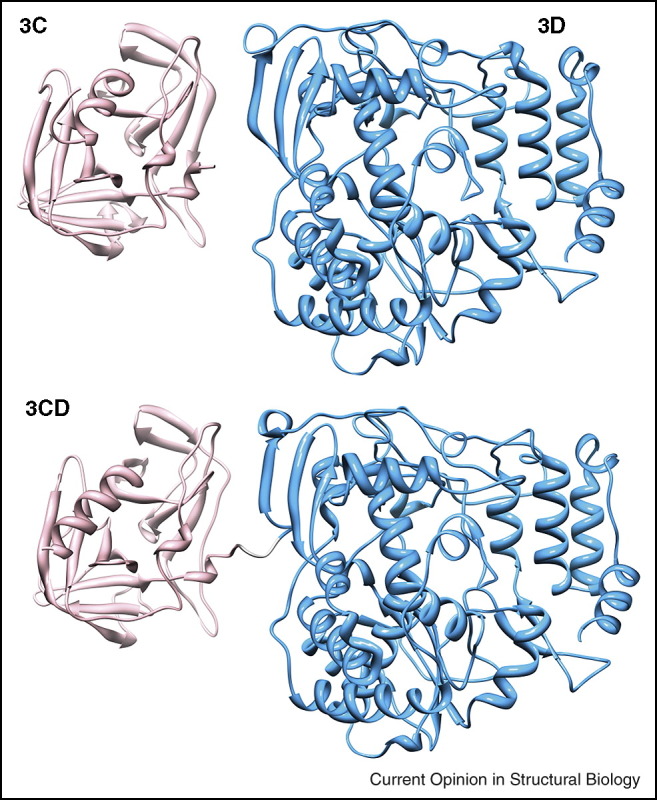
Structure is unable to reveal the basis for the absence of activity for polymerase precursors. The polymerase precursor 3CD (pdb 2IJD) is a composite of 3C (pdb 1L1N, pink) and 3Dpol (pdb 1RA6, light blue) [26•].
Processing intermediates have the ability to expand the viral proteome, potentially creating new functions or, as described here, inactivating some functions. Moreover, a single amino acid change of a residue that influences protein dynamics can be sufficient to alter the global dynamics of a protein and potentially its function [27]. If this is the case, then the combination of a viral quasispecies and variable extents of polyprotein processing creates unlimited possibilities for the acquisition and evolution of function by a virus with limited coding capacity.
Future directions: exploiting RdRp structures for the discovery of dynamics–function relationships
In spite of all of the structural information now available for the RdRp (see Supplementary Table 1), many gaps exist in our understanding of RdRp function. We would like to suggest that dynamics may represent the missing link to our complete understanding of RdRp function (Figure 5 ). Given the numerous high-resolution structures available for the RdRp in the same and different families, it should be possible to use molecular dynamics (MD) simulations to determine the extent to which dynamics coevolves with structure and function. In addition, the impact of mutations or fusions on dynamics should be tractable by using MD. Indeed, MD has become extraordinarily powerful [28]. A recent study on HIV RT reported the use of a 360 ns simulation to explain the impact of a non-nucleoside RT inhibitor on HIV RT dynamics [29•]. It should also be possible to use hydrogen-deuterium exchange mass spectrometry (HDXMS) to evaluate RdRp dynamics. HDXMS should permit analysis of RdRp dynamics in complex with substrates, as well as when fused to other proteins. Again, HDXMS studies of HIV RT have recently been used to identify regions of RT that change conformation in the absence of substrates [30•]. Finally, the molecular weight of most viral RdRps is in the 50–65 kDa range. Enzymes in this size range can now be studied by using nuclear magnetic resonance spectroscopy, which offers an empirical approach to the evaluation of dynamics on the widest range of timescales.
Figure 5.
Dynamics: the missing link to a complete understanding of RdRp function.
Conclusion
RdRp fidelity may be a tractable target for viral attenuation and vaccine development. Our current understanding of the basis of RdRp fidelity is incomplete even with the availability of multiple crystal structures. Computational and empirical studies of RdRp dynamics may complete our understanding of RdRp function.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
Our work on RdRp mechanism has been supported by grant AI045818 from NIAID/NIH.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.sbi.2009.10.012.
Contributor Information
Craig E Cameron, Email: cec9@psu.edu.
Ibrahim M Moustafa, Email: I.moustafa@psu.edu.
Jamie J Arnold, Email: jja5@psu.edu.
Appendix A. Supplementary data
References
- 1.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng K.K., Arnold J.J., Cameron C.E. Structure–function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikonov A., Juronen E., Ustav M. Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2008;283:24089–24102. doi: 10.1074/jbc.M803422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first demonstration that an exonuclease contributes to replication fidelity of RNA virus.
- 6.Arnold J.J., Gohara D.W., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrer-Orta C., Arias A., Perez-Luque R., Escarmis C., Domingo E., Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template–primer RNA. J Biol Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 8••.Ferrer-Orta C., Arias A., Perez-Luque R., Escarmis C., Domingo E., Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci U S A. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of structures of RdRp elongation complexes.
- 9.Arnold J.J., Vignuzzi M., Stone J.K., Andino R., Cameron C.E. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeiffer J.K., Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci U S A. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaulieu P.L. Recent advances in the development of NS5B polymerase inhibitors for the treatment of hepatitis C virus infection. Expert Opin Ther Pat. 2009;19:145–164. doi: 10.1517/13543770802672598. [DOI] [PubMed] [Google Scholar]
- 12•.Pfeiffer J.K., Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes RdRp fidelity as a determinant of viral virulence.
- 13•.Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes RdRp fidelity and the corresponding quasispecies as a determinant of viral virulence and pathogenesis.
- 14••.Vignuzzi M., Wendt E., Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]; This report describes RdRp fidelity as a target for viral attenuation and vaccine development.
- 15.Brautigam C.A., Steitz T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 16.Castro C., Smidansky E., Maksimchuk K.R., Arnold J.J., Korneeva V.S., Gotte M., Konigsberg W., Cameron C.E. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Gotte M., Konigsberg W., Cameron C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the discovery of a general acid for nucleotidyl transfer by polymerases.
- 18.Carroll S.S., Cowart M., Benkovic S.J. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 19.Bell J.B., Eckert K.A., Joyce C.M., Kunkel T.A. Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J Biol Chem. 1997;272:7345–7351. doi: 10.1074/jbc.272.11.7345. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki M., Avicola A.K., Hood L., Loeb L.A. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J Biol Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 21.Padilla R., Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002;30:e138. doi: 10.1093/nar/gnf138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Kaplan C.D., Larsson K.M., Kornberg R.D. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the use of the trigger loop for the fidelity of nucleotide addition by multisubunit RNA polymerases.
- 24.Tardy-Panit M., Blondel B., Martin A., Tekaia F., Horaud F., Delpeyroux F. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J Virol. 1993;67:4630–4638. doi: 10.1128/jvi.67.8.4630-4638.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson A.A., Peersen O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Marcotte L.L., Wass A.B., Gohara D.W., Pathak H.B., Arnold J.J., Filman D.J., Cameron C.E., Hogle J.M. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first structure for a biologically relevant fusion of the RdRp.
- 27.Cameron C.E., Benkovic S.J. Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant. Biochemistry. 1997;36:15792–15800. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- 28.Klepeis J.L., Lindorff-Larsen K., Dror R.O., Shaw D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 29•.Ivetac A., McCammon J.A. Elucidating the inhibition mechanism of HIV-1 non-nucleoside reverse transcriptase inhibitors through multicopy molecular dynamics simulations. J Mol Biol. 2009;388:644–658. doi: 10.1016/j.jmb.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the use of molecular dynamics simulations to study the basis for the inhibition of HIV RT by a non-nucleoside inhibitor.
- 30•.Seckler J.M., Howard K.J., Barkley M.D., Wintrode P.L. Solution structural dynamics of HIV-1 reverse transcriptase heterodimer. Biochemistry. 2009 doi: 10.1021/bi900790x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the use of hydrogen-deuterium exchange to study HIV RT dynamics.
- 31.Arias A., Arnold J.J., Sierra M., Smidansky E.D., Domingo E., Cameron C.E. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J Virol. 2008;82:12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.