Abstract
Human immunodeficiency virus type 1 (HIV-1) infections and the resulting acquired immunodeficiency syndrome (AIDS) pandemic remain a global challenge in the absence of a protective vaccine and because of rapid selection of drug-resistant viral variants in response to all currently available antiviral therapies. Development of new and highly active antiviral agents would greatly facilitate effective clinical management of HIV-1 infections and delay the onset of AIDS. Recent advances in our understanding of intracellular immunity conferred by host cytidine deaminases APOBEC3G (A3G) and APOBEC3F (A3F), and the mechanism by which the virally encoded Virion Infectivity Factor (Vif) protein induces their proteasomal degradation, provide fresh opportunities for the development of novel antiviral treatments. Interestingly, the interactions between Vif-A3G and Vif-A3F that overcome this host defense mechanism are structurally distinct, and provide two potential targets for antiviral drug development. This review provides an overview of the current knowledge of APOBEC3/Vif interactions and recent efforts to target these interactions for antiviral drug development.
Introduction
The spread of human immunodeficiency virus type 1 (HIV-1) infections has resulted in the acquired immunodeficiency syndrome (AIDS) pandemic. Approximately 33 million people are currently infected with HIV-1, and each year about 2.1 million people die of AIDS while 2.5 million are newly infected (www.unaids.org). In the absence of an effective vaccine or antiviral treatments, AIDS is likely to expand and continue to claim the lives of millions for decades. Despite heroic efforts over the last 25 years, a protective vaccine is not currently on hand, and the recent suspension of the Merck vaccine trial suggests that an effective vaccine is not likely to be available in the near future [1, 2]. Since the approval of AZT in 1987 [3], approximately 30 anti-HIV drugs or drug combinations have been approved for clinical use. Combination antiviral drug therapy has been effective in controlling HIV-1 replication and the onset of AIDS in the developed nations. However, successful management of HIV-1 infection has been hampered by the selection of drug resistant viral variants in response to all approved antiviral agents. Furthermore, successful application of antiviral drug therapy has been hindered by its high cost, toxicity, and lack of patient adherence. Thus, new and more effective antiviral drugs are needed for the clinical management of HIV-1 infection and AIDS.
HIV-1 life cycle
A thorough understanding of HIV-1 replication has greatly facilitated the development of antiviral therapeutics. HIV-1 primarily infects CD4+ T cells, macrophages, and dendritic cells and compromises the human immune system. Viral infection is initiated by binding of the envelope glycoprotein to the primary (CD4) and secondary (CXCR4 and CCR5) cell surface receptors and fusion of the viral envelope with the cell membrane (Figure 1). During uncoating of the viral nucleoprotein complex, the viral single-stranded genomic RNA is converted to double-stranded DNA by viral reverse transcriptase (RT). The viral DNA and associated proteins form a preintegration complex that is transported to the nucleus, and the viral DNA integrates into the host chromosome to form a provirus. The viral RNA transcribed from the proviral DNA is either packaged into virions as genomic RNA or translated to produce viral structural polyproteins Gag, Gag-Pol, and Env, regulatory proteins Tat and Rev, and accessory proteins virion infectivity factor (Vif), Vpr, Vpu, and Nef. The viral proteins and viral genomic RNAs are transported to the plasma membrane, where progeny virions are assembled and released. The viral polyproteins are processed into their mature components by the virally encoded protease (PR) at the time of virus budding or soon thereafter to form mature infectious virions.
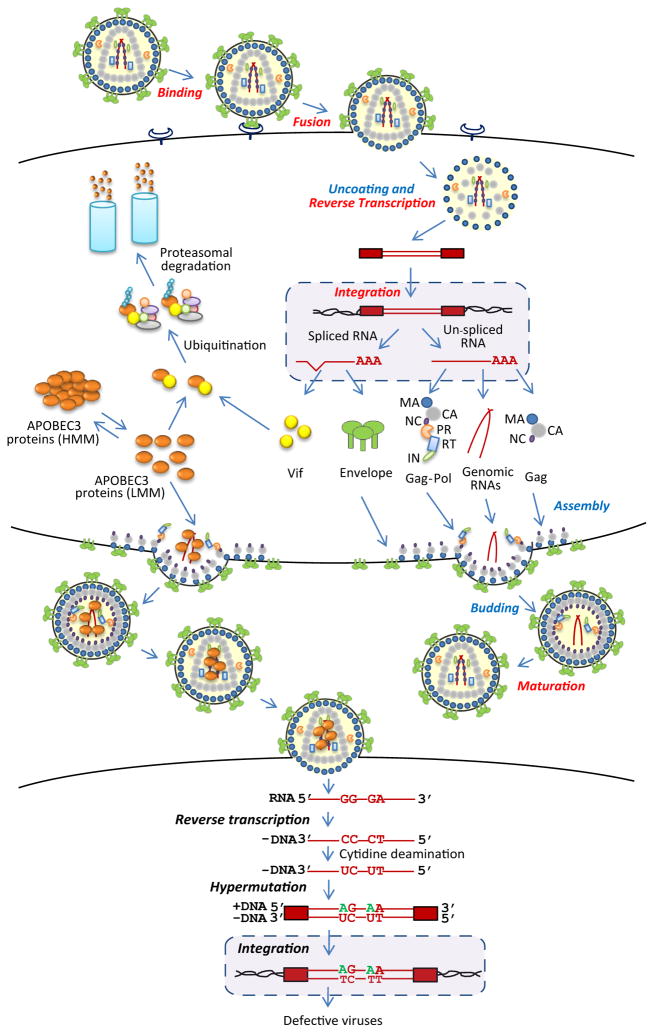
Figure 1.
HIV-1 replication and inhibition by APOBEC3 proteins. HIV-1 binds to cell surface receptors, triggering fusion of the viral and cellular membranes. Uncoating of the viral nucleoprotein complex, along with reverse transcription, results in the formation of double-stranded DNA, which is integrated into the host cell DNA to form a provirus. The proviral genome is transcribed into spliced and unspliced RNAs and translated to form viral structural proteins group antigen (Gag), Gag-polymerase (Gag-pol), and envelope (Env), along with the accessory proteins, including the virion infectivity factor (Vif). The viral genomic RNA and structural proteins are assembled and released from the plasma membrane to form immature viral particles; processing of the viral proteins by protease (PR) into their mature components, which include matrix (MA), capsid (CA), nucleocapsid (NC), PR, reverse transcriptase (RT), and integrase (IN), leads to the formation of the mature virion. Virion binding, fusion, reverse transcription, integration, and maturation (shown in red) are the targets of currently available antiviral drugs. APOBEC3 proteins exist in the cytoplasm in low molecular mass (LMM) complexes or RNA associated high molecular mass (HMM) complexes. In the absence of Vif, APOBEC3 proteins are incorporated into virion particles; Vif associates with the APOBEC3 proteins and targets them for ubiquitination and proteasomal degradation. In the infected cell, APOBEC3 proteins inhibit reverse transcription, catalyze cytidine deamination of the minus-strand DNA and G-to-A hypermutation, and inhibit viral DNA integration.
Current antiviral therapies
Most currently available antiviral drugs target the pol-encoded retroviral enzymes PR, RT, and integrase (IN); in addition, inhibitors that target HIV-1 envelope-receptor interactions or the subsequent fusion step have also been recently approved. Nucleoside analog and nonnucleoside RT inhibitors were the first successful anti-HIV-1 drugs. However, error-prone viral replication and frequent recombination coupled with high viral loads in patients results in the generation of extensive viral variation; consequently, drug-resistant variants of HIV-1 are rapidly selected in response to single drug therapy. For example, treatment of pregnant mothers with a single dose of nevirapine at the time of birth results in the selection of a drug-resistant HIV-1 variant in over 35% of patients [4]. The development of potent inhibitors of HIV-1 PR made it possible to administer highly active antiviral therapy (HAART), which typically consists of two RT inhibitors and a PR inhibitor [5, 6]. Although HAART can effectively control HIV-1 replication and maintain the plasma viral loads below the detection limits of sensitive assays (<50 RNA copies/ml) [7], patients often fail HAART therapy and develop resistance to all approved antiviral drugs. The recent approval of potent and specific inhibitors of HIV-1 IN [8] and viral entry [9] has made it possible to treat patients with multi-drug-resistant viruses and has greatly improved the management of highly treated patients. As expected, drug resistance to IN and entry inhibitors has been observed in treated patients [10, 11], and it is anticipated that new and potent antiviral drugs will be needed.
New targets for drug development
Recent elucidation of the interactions between HIV-1 and host restriction factors has provided fresh opportunities for development of novel antiviral drugs. Restriction factors are host proteins that constitute an intracellular innate immunity and inhibit the replication of a broad spectrum of pathogens. Successful pathogens have developed mechanisms to counteract these host defenses in order to establish infection. These host restriction factors include apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (APOBEC3) proteins which inhibit viral replication in the absence of Vif [12–15], TRIM5α proteins which inhibit viral replication by destabilizing the viral nucleoprotein complex [16, 17], and the recently described tetherin/BST-2/CD317 proteins that inhibit HIV-1 virion release in the absence of the virally encoded Vpu protein [18, 19]. HIV-1 expresses Vif and Vpu to evade inhibition by APOBEC3 proteins and tetherin/BST-2/CD317, respectively; similarly, HIV-1 has evolved its capsid to evade inhibition by human TRIM5α. Interactions between HIV-1 proteins and all of these host restriction factors are of tremendous interest since they provide new insights into the interplay between the virus and the host cell. In addition, interactions between restriction factors and viral proteins can be viewed as potential targets of antiviral drug development, because their inhibition could allow the host defense mechanisms to control viral replication. In this review, we will summarize the current knowledge of the structure of APOBEC3 proteins, the mechanisms by which these proteins inhibit HIV-1 replication, the structural determinants of APOBEC3/Vif interactions, and recent efforts to target these interactions for antiviral drug development.
APOBEC3 proteins
HIV-1 Vif is essential for viral replication in certain nonpermissive T cell lines and primary cell types but is dispensable in other permissive T cell lines [20–23]. Recent groundbreaking studies have identified a dominant restriction factor, APOBEC3G (A3G), which is responsible for the nonpermissive phenotype [14]. Subsequent studies identified APOBEC3F (A3F) as another potent host restriction factor that can suppress the replication of HIV-1 vif-deleted viruses (HIV-1Δvif) [12, 13, 15].
A3G and A3F (A3G/A3F) are members of a family of seven genes that encode cytidine deaminases (Figure 2) [24, 25]. These proteins contain one (A3A, A3C, and A3H) or two (A3B, A3DE, A3F, and A3G) zinc-binding catalytic domains with the consensus sequence -(H/C)-X-E-X23–28-P-C-X2–4-C [25–27]. A3G/A3F display the strongest anti-HIV-1 activities and appear to be the most important for control of HIV-1 replication. The A3G/A3F N-terminal catalytic domains are essential for RNA binding and virion incorporation [28–31], whereas the C-terminal domains are primarily associated with enzymatic activity and substrate sequence specificity [29, 32, 33]. A3G and A3F preferentially target CC and TC dinucleotides for cytidine deamination, respectively (the deaminated cytidine is underlined) [12, 34]. A3G and A3F genes are expressed in HIV-1 target cells of infection, namely activated CD4+ T cells, macrophages, and dendritic cells, and are induced by interferon [35–39]. A3G is a component of high molecular mass (HMM) complexes in association with RNA in activated CD4+ T cells and low molecular mass (LMM) complexes in resting CD4+ T cells [40]. It has been postulated that one normal function of A3G/A3F is to suppress retrotransposition of endogenous retroelements [41]. It is not known whether A3G/A3F carry out other cellular functions, but APOBEC3-deficient mice develop normally to adulthood, suggesting that they are not essential [42].
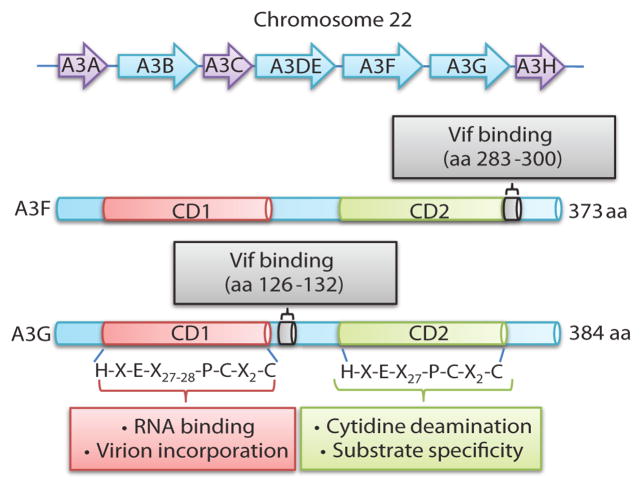
Figure 2.
Human APOBEC3 gene cluster and schematic structures of A3F and A3G proteins. The top part of the figure shows the 7 members of the human APOBEC3 gene family on chromosome 22. The bottom part of the figure schematically shows the catalytic domains (CD1 and CD2) and Vif-binding regions of the A3F and A3G proteins.
Mechanisms of antiviral activity
The primary mechanism by which A3G/A3F inhibit HIV-1Δvif replication requires their expression in virus-producer cells and their incorporation into virions [43–47]. During reverse transcription in the target cells, the virion-incorporated A3G/A3F deaminate cytidines to uridines in the viral minus-strand DNA (Figure 1). Subsequent incorporation of adenines instead of guanines in the plus strand results in extensive G-to-A hypermutation and inactivation of the viral genome [48–53].
Although G-to-A hypermutation resulting in lethal mutagenesis was initially believed to be the sole mechanism of viral inhibition, the A3G/A3F proteins also inhibit viral replication through other mechanisms [54]. Several investigators have observed that A3G/A3F proteins also inhibit viral DNA synthesis [55–63], and the mechanism of inhibition may involve interfering with tRNA primer annealing, initiation and elongation of DNA synthesis, and minus- and plus-strand DNA transfers [55–57, 59–62, 64, 65]. When A3G is expressed at levels similar to those present in CD4+ T cells, cytidine deaminase activity is essential for its antiviral activity [64, 66–68]. However, comparison of similar levels of A3G and A3F catalytic site mutants suggests that, unlike A3G, the A3F cytidine deaminase activity is not absolutely required for its ability to inhibit viral replication [62 and our unpublished observations].
The decrease in viral DNA synthesis is not sufficient to explain the overall reduction in viral infectivity. We and others have reported that expression of physiological levels of A3G also results in inhibition of viral DNA integration and provirus formation [64, 65]. This inhibition of integration was associated with aberrant structures of viral DNA ends, which were presumably poor substrates for the integration reaction [64]. Interestingly, the cytidine deaminase activity of A3G is also required for the inhibition of provirus formation [64].
It was postulated that cytidine deamination of viral DNA synthesis results in the degradation of viral DNA through the action of uracil DNA glycosylase and adenine-purine endonuclease [48, 50]. While one study supports this hypothesis [69], others have reported that inhibition of UNG did not have any impact on the amounts of viral DNA present in infected cells or viral replication in primary CD4+ T cells and macrophages [64, 67, 70, 71]. In addition to the effects of A3G/A3F in virus-producer cells, it has also been reported that A3G inhibits replication of HIV-1 in the resting CD4+ T cells that are the target cells of infection [40]. Two recent studies failed to confirm these effects [72, 73], while another study supported a role for A3G/A3F inhibition of HIV-1 in target cells [74]. Additional studies are needed to explore the potential role of A3G/A3F in infected cells.
HIV-1 Vif-mediated degradation of A3G/A3F
Vif suppresses A3G/A3F antiviral activity by targeting these proteins for polyubiquitination and proteasomal degradation [44–46, 75–78]. HIV-1 Vif is a 192 amino acid protein that contains an 144SLQYLA149 motif that binds to the Elongin C and B proteins and an 108Hx5Cx17–18Cx3–5H139 motif that binds to Cullin 5 (Figure 3) [79–82]. Vif binds to A3G/A3F and forms an E3 ubiquitin ligase complex consisting of Cullin 5, Elongin B, Elongin C, and RING finger protein 1, which results in A3G/A3F polyubquitination and degradation [77]. As a consequence, A3G/A3F are not packaged into virions, and HIV-1 replication is spared from A3G/A3F-mediated inhibition. An understanding of the interactions between Vif, A3G/A3F, and the proteasomal degradation pathway is essential for developing novel drugs for therapeutic intervention. Therapeutic agents that induce the expression of A3G/A3F could be developed, provided that the increased expression of A3G/A3F is not deleterious to the cells. Alternatively, the interactions between Vif and the E3 ubiquitin ligase components could be targeted for development of new antiviral drugs, but care must be taken to avoid inhibition of the proteasomal pathway, which could reduce cell viability. An ideal strategy could be to develop inhibitors that target the Vif-A3G/A3F interactions, since such inhibitors are much more likely to specifically disrupt viral replication without affecting normal cellular functions.
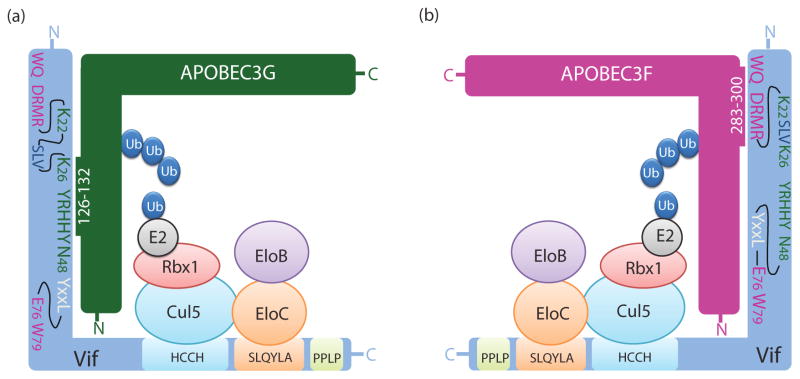
Figure 3.
Vif interactions with A3G, A3F, and the E2 ubiquitin ligase complex. (a and b) Amino acids in Vif that are involved in interaction with A3G but do not influence interaction with A3F are shown in dark green. Amino acids in Vif that are involved in interaction with A3F but do not influence interaction with A3G are shown in fuchsia. Only amino acids that reduced activity to <50% against A3G/A3F, but retained >80% activity against the other APOBEC3 protein are shown. The 69YXXL72 motif (white) interacts with both A3G and A3F and is shown in white. The 23SLV25 motif (dark blue) does not affect binding to A3G or A3F but is involved in A3G/A3F degradation. The E3 ubiquitin ligase complex consists of Cullin 5 (Cul5), Elongin B (EloB), Elongin C (EloC), RING finger protein 1 (Rbx1), and an E2 ubiquitin-conjugating enzyme (E2). The HCCH motif in Vif (108Hx5Cx17–18Cx3–5H139) binds to Cul5 and the 144SLQYLA149 motif binds to EloC and EloB. Ubiquitin (Ub) is ligated to A3G and A3F by the E3 ubiquitin ligase. The 161PPLP164 motif is involved in Vif oligomerization. A3G amino acids 126–132 are involved in interaction with Vif (a) whereas A3F determinants that interact with Vif are located to amino acids 283–300 (b).
Structural determinants of HIV-1 Vif that interact with A3G/A3F
To date, the structure of only the C-terminal domain of A3G has been determined [83–86], and because full-length A3G/A3F and HIV-1 Vif have proven difficult to express and purify in large, soluble quantities [30, 87–89], structural information with regard to the Vif-A3G/A3F interactions is not currently available. Therefore, studies reported so far have relied on functional and biochemical analysis of Vif and A3G/A3F mutants to identify the structural determinants of their interactions. The mutational analyses of key residues are summarized in Table 1.
Table 1.
Key residues in HIV-1 Vif that are important for activity against A3G/A3Fa
| Vif amino acid | Amino acid substitutionsb | A3G/A3F affected | References |
|---|---|---|---|
| W11 | A | A3F | [92, 93] |
| Q12 | A | A3F | [93] |
| 14DRMR17 | single A substitutions or14AAAA17 | A3F | [93] |
| K22 | E, D | A3G | [91, 101, 102] |
| 23SLV25 | V25S, AAAc | A3F only (V25S) or A3G and A3F | [101, 102] |
| K26 | A, D, R | A3G | [101, 102] |
| 40YRHHY44 | single A substitutions or 40AAAAA44, H42N, H43N | A3G | [93, 124] |
| E45 | G | A3G | [91] |
| N48 | A | A3G | [93] |
| 69YxxL72 | Y69Ad, L72Ad, L72Sc | A3G and A3F | [99, 100, 103] |
| E76 | A | A3F | [99, 100] |
| W79 | A | A3F | [92, 99, 100] |
Abbreviations: HIV-1, human immunodeficiency virus type 1; Vif, virion infectivity factor; A3G, APOBEC3G; A3F, APOBEC3F
For the amino acids listed, at least one substitution caused Vif activity to be reduced by more than 50% against A3G or A3F, but at least 80% activity was maintained towards the other APOBEC3 protein in infectivity assays.
Vif activity against both A3G and A3F was affected in infectivity assays, but the ability to bind A3G/A3F was maintained.
Vif activity against both A3G and A3F was affected, but the ability to bind Cullin 5 was maintained.
Initially, Vif deletion mutants were analyzed for their interaction with A3G [44, 90]. However, most deletion mutants failed to bind to A3G, suggesting that the deletions affected the overall conformation of Vif, resulting in a loss of A3G binding. Analysis of Vif variants in viruses isolated from patients identified several Vif mutants that were defective in their ability to counteract APOBEC3 proteins [91]. Importantly, some Vif mutants (K22E, Y40H, and E45G) were deficient in blocking A3G but not A3F, indicating that the overall conformation and function of the Vif mutants was retained. Mutational analysis of conserved tryptophans also showed that W11A and W79A mutants were deficient in overcoming A3F- but not A3G-mediated antiviral activity [92].
To identify structural determinants of Vif that interact with A3G/A3F, we performed extensive mutational analysis of the first 60 amino acids of Vif, and analyzed the effects of single- and double-alanine substitution mutations on anti-A3G/A3F activity and binding in co-immunoprecipitation assays [93]. These studies identified two distinct domains of the Vif-A3G/A3F interactions. The 14DRMR17 region, and to a lesser extent W11A and Q12A, are important for binding to A3F but not A3G. The 40YRHHY44 region and N48 are important for blocking the antiviral activity of A3G but not A3F. An intriguing observation is that while the 14DRMR17 region is not involved in binding to wild-type A3G, substitution of the DRMR amino acids with SERQ or SEMQ (equivalent residues in African green monkey simian immunodeficiency virus Vif) allows the Vif mutant to induce degradation of D128K-A3G, an A3G mutant that is resistant to HIV-1 Vif [93–98]. Wild-type Vif binds to the D128K-A3G but does not induce its degradation [93, 95]. These results suggest that a secondary step after Vif-A3G binding is essential to induce A3G degradation.
Other mutational analyses have shown that E76A and W79A mutants were defective in overcoming the antiviral activity of A3F but not A3G [99, 100]. Two recent studies analyzed the role of residues 22–26 and Y30 in interactions with APOBEC3 proteins [101, 102]. These studies show that K22 and K26 are important for degradation of A3G but not A3F; Y30 affects interactions with both A3G and A3F, suggesting that mutations at this site may adversely affect the overall conformation and function of Vif. A triple mutant in which 23SLV25 were replaced with alanines was able to bind to A3G and A3F but did not block their antiviral activity, suggesting that these residues affect a subsequent step that is essential for proteasomal degradation.
In addition to the amino acids in Vif that selectively block the antiviral activity of A3G or A3F, several amino acid substitutions affect the antiviral activity of both A3G and A3F [91, 93, 99–103]. These mutations could potentially affect the overall conformation of Vif and indirectly affect interactions with A3G and A3F, Cullin 5, or Elongin B and Elongin C. Based on mutagenesis of amino acids in the region from 53–79, the 69YXXL72 motif of Vif, specifically Y69 and L72, was shown to be important for blocking the antiviral activity of both A3G and A3F [99, 100, 103]. Interestingly, Y69A and L72A mutants were shown to interact with Cullin 5 in co-immunoprecipitation assays, strongly implying that the mutations did not induce conformational alterations in Vif [103].
In addition to the Vif-A3G and Vif-A3F interactions, Vif oligomerization could also be targeted for development of small molecule inhibitors [104]. The 161PPLP164 motif of Vif is important for Vif oligomerization and degradation of A3G [105]. Thus, small molecules that inhibit Vif oligomerization could be developed as antiviral drugs and block Vif’s anti-A3G/A3F activity.
Structural determinants of A3G/A3F that interact with Vif
Structural determinants of A3G/A3F that interact with Vif have been identified using functional and biochemical analysis of A3G/A3F mutants [75, 106–109]. Initially, deletion mutagenesis of A3G showed that amino acids 54–124 were important for Vif interaction [75], while another study identified amino acids 105–156 as being critical for the Vif interaction [106]. Analysis of an N-terminal deletion mutant of A3F showed that the C-terminal half of the protein was sufficient for Vif binding and Vif-induced degradation [107]. One drawback of deletion analysis is that removing large portions of the A3G/A3F proteins could potentially lead to alterations in the protein conformation and non-specific interactions with Vif. Alanine-scanning mutational analysis of amino acids near the D128 residue involved in Vif resistance identified amino acids 128–130 as being critical for the Vif interaction [108]. We performed analysis of functional A3G/A3F chimeras and identified amino acids 126–132 of A3G as the determinant involved in Vif interaction [109]. The same study showed that amino acids 283–300 in the C-terminal domain of A3F contained the A3F-Vif interaction determinant (Figure 2) [109].
Inhibitors that block Vif-induced degradation of A3G
The Vif-A3G and Vif-A3F interactions are believed to be important targets for antiviral drug development. Several studies of HIV-1Δvif viruses have shown that their replication is significantly delayed in nonpermissive cells [20–23], implying that interfering with the Vif-A3G and Vif-A3F interactions should strongly suppress viral replication. One recent study showed that Vif-deficient HIV-1 could be suppressed in nonpermissive cells for more than five weeks before replication could be detected [110]. Interestingly, replication-competent mutants evolved Vif-independent mechanisms for A3G resistance but were still susceptible to A3F [110]. A large proportion of proviruses in infected patients are hypermutated [91, 111–113] and have G-to-A mutations in RT and protease [114]. Hypermutation has the potential to increase diversity in viral populations, and one theoretical concern is that partial inhibition of Vif-A3G/A3F interactions could result in more rapid selection of viral variants resistant to other antiviral agents. One study reported that replication of Vif-defective variants can accelerate the selection of some drug resistant mutants [115]. On the other hand, another recent analysis concluded that A3G-induced hypermutation of the viral genome is unlikely to contribute to the viral mutation rate and faster evolution of drug-resistant HIV-1 [116, 117]. Additional studies are needed to determine whether partial inhibition of the Vif-A3G interaction can contribute to an increase in viral diversity and selection of drug resistant variants.
Thus far, only a few studies have reported attempts to find inhibitors that block Vif-A3G interactions. First, proline-rich peptides that contain the PPLP motif were shown to inhibit Vif oligomerization and suppress HIV-1 replication in nonpermissive cells [104, 118]. In another study, a membrane-permeable zinc chelator, N,N,N′,N′-tetrakis-(2-pyridylmethyl) ethylenediamine (TPEN), inhibited Vif-Cullin 5 binding and prevented the formation of the E3 ubiquitin ligase, thereby suppressing A3G degradation [119]. While peptide inhibitors can provide valuable insights into protein-protein interactions and their function, their development as antiviral drugs is hampered by their low stability and poor bioavailability (reviewed in [120]). Additionally, in vitro studies have shown TPEN induces apoptosis in human cells [121, 122] and considering that it acts as a zinc chelator, it is likely to inhibit cellular functions. Clearly, it would be desirable to develop small molecule inhibitors that specifically antagonize Vif and/or Vif-A3G/A3F interactions.
A high-throughput assay was previously developed and used to screen > 106 compounds that potentially inhibited any step in HIV-1 replication; since the assay was performed in the nonpermissive MT-2 cell line, Vif and Vif-A3G/A3F interactions were also targeted [123]. Although the screen resulted in the identification of 5567 compounds, secondary screens to identify compounds that target Vif or Vif-A3G/A3F interactions have not been reported.
An in vitro Vif-A3G binding assay using GST-tagged Vif and HIS-tagged A3G has been recently described [124]. A Fluorescence Resonance Energy Transfer (FRET) assay that measures interactions between GST-Vif (labeled with anti-GST-Europium) and a biotinylated A3G peptide (amino acids 110–148: labeled with streptavidin-allophycocyanin) has also been validated [124]. These assays are currently being used for high-throughput screening and identification of candidate molecules that target the Vif-A3G interaction.
Finally, an exciting recent development is the identification of a small molecule inhibitor of Vif-mediated degradation of A3G. Recently, a cell-based assay was used to screen a library of approximately 30,000 compounds that suppressed Vif-mediated degradation of A3G fused to yellow fluorescent protein (A3G-YFP) [125]. The primary screen identified 537 potential candidates; 66 candidates survived a secondary screen that eliminated false positives resulting from inherent fluorescence, increased levels of transfection, or nonspecific increased protein expression. RN-18, a compound that contains a 2-(4-nitrophenylthio)-N-phenylbenzamide moiety, was selected as the most promising compound that specifically targets Vif-mediated A3G degradation (IC50 ~ 4.5 – 10 μM in nonpermissive CEM and H9 cells). Although a thorough analysis of the cytotoxicity of the compound was not reported, RN-18 does not exhibit cytotoxic effects at 50 – 100μM concentrations. Further analysis revealed that RN-18 enhanced the levels of intracellular A3G and A3F and virion incorporation of A3G. Interestingly, RN-18 did not affect Vif-A3G/A3F binding but increased the degradation of Vif in the presence of A3G/A3F. Thus, although it is too early to determine whether RN-18 constitutes a promising lead compound, these studies validate Vif-A3G/A3F interactions as potential targets for development of novel antiviral drugs.
Conclusions
The results obtained so far indicate that Vif-A3G and Vif-A3F interactions involve distinct structural determinants in both proteins, providing two potential targets for development of antiviral therapeutics. Inhibition of either Vif-A3G or Vif-A3F interaction should allow the host APOBEC3 protein to carry out their natural activity and inhibit HIV-1 replication. Structural studies that elucidate these interactions will undoubtedly provide novel insights and could facilitate rational drug design. Such compounds could provide a novel class of antiviral agents and provide valuable tools for successful management of HIV-1 infections.
Acknowledgments
We especially thank Wei-Shau Hu, Eric Freed and Narasimhan J. Venkatachari for valuable critical comments during manuscript preparation. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has also been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robb ML. Failure of the Merck HIV vaccine: an uncertain step forward. Lancet. 2008;372:1857–1858. doi: 10.1016/S0140-6736(08)61593-7. [DOI] [PubMed] [Google Scholar]
- 3.Fischl MA, et al. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N Engl J Med. 1987;317:185–191. doi: 10.1056/NEJM198707233170401. [DOI] [PubMed] [Google Scholar]
- 4.Arrive E, et al. Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol. 2007;36:1009–1021. doi: 10.1093/ije/dym104. [DOI] [PubMed] [Google Scholar]
- 5.Sax PE, Gathe JC., Jr Beyond efficacy: the impact of combination antiretroviral therapy on quality of life. AIDS Patient Care STDS. 2005;19:563–576. doi: 10.1089/apc.2005.19.563. [DOI] [PubMed] [Google Scholar]
- 6.Vella S, et al. Saquinavir/zidovudine combination in patients with advanced HIV infection and no prior antiretroviral therapy: CD4+ lymphocyte/plasma RNA changes, and emergence of HIV strains with reduced phenotypic sensitivity. Antiviral Res. 1996;29:91–93. doi: 10.1016/0166-3542(95)00926-4. [DOI] [PubMed] [Google Scholar]
- 7.Hoen B, et al. Highly active antiretroviral treatment initiated early in the course of symptomatic primary HIV-1 infection: results of the ANRS 053 trial. J Infect Dis. 1999;180:1342–1346. doi: 10.1086/315002. [DOI] [PubMed] [Google Scholar]
- 8.Egbertson MS, et al. A potent and orally active HIV-1 integrase inhibitor. Bioorg Med Chem Lett. 2007;17:1392–1398. doi: 10.1016/j.bmcl.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 9.MacArthur RD, Novak RM. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis. 2008;47:236–241. doi: 10.1086/589289. [DOI] [PubMed] [Google Scholar]
- 10.Moore JP, Kuritzkes DR. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr Opin HIV AIDS. 2009;4:118–124. doi: 10.1097/COH.0b013e3283223d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks C, Gulick RM. Raltegravir: the first HIV type 1 integrase inhibitor. Clin Infect Dis. 2009;48:931–939. doi: 10.1086/597290. [DOI] [PubMed] [Google Scholar]
- 12.Liddament MT, et al. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Wiegand HL, et al. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. Embo J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 15.Zheng YH, et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 17.Sayah DM, et al. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 18.Neil SJ, et al. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 19.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strebel K, et al. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 21.Fisher AG, et al. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 22.Gabuzda DH, et al. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Schwedler U, et al. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conticello SG, et al. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 25.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 26.Huthoff H, Malim MH. Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins. Virology. 2005;334:147–153. doi: 10.1016/j.virol.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 27.Dang Y, et al. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gooch BD, Cullen BR. Functional domain organization of human APOBEC3G. Virology. 2008;379:118–124. doi: 10.1016/j.virol.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro F, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Iwatani Y, et al. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friew YN, et al. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hache G, et al. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. The Journal of biological chemistry. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 33.Li J, et al. Functional domains of APOBEC3G required for antiviral activity. J Cell Biochem. 2004;92:560–572. doi: 10.1002/jcb.20082. [DOI] [PubMed] [Google Scholar]
- 34.Langlois MA, et al. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic acids research. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng G, et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen K, et al. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koning FA, et al. Defining APOBEC3 Expression Patterns in Human Tissues and Hematopoietic Cell Subsets. J Virol. 2009 doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stopak KS, et al. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 39.Wang FX, et al. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008;89:722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- 40.Chiu YL, et al. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 41.Chiu YL, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikl MC, et al. Mice deficient in APOBEC2 and APOBEC3. Mol Cell Biol. 2005;25:7270–7277. doi: 10.1128/MCB.25.16.7270-7277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariani R, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 44.Marin M, et al. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 45.Sheehy AM, et al. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 46.Stopak K, et al. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 47.Svarovskaia ES, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 50.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 51.Lecossier D, et al. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science (New York, N Y) 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 52.Suspene R, et al. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 54.Shindo K, et al. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, et al. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007;365:92–100. doi: 10.1016/j.virol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 56.Guo F, et al. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo F, et al. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J Virol. 2007;81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwatani Y, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bishop KN, et al. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS pathogens. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson JL, Hope TJ. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology. 2008;375:1–12. doi: 10.1016/j.virol.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li XY, et al. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. The Journal of biological chemistry. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 62.Holmes RK, et al. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 63.Newman EN, et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 64.Mbisa JL, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luo K, et al. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyagi E, et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schumacher AJ, et al. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, et al. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 69.Yang B, et al. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J Biol Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 70.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langlois MA, Neuberger MS. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J Virol. 2008;82:4660–4664. doi: 10.1128/JVI.02469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamata M, et al. Reassessing the role of APOBEC3G in human immunodeficiency virus type 1 infection of quiescent CD4+ T-cells. PLoS Pathog. 2009;5:e1000342. doi: 10.1371/journal.ppat.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santoni de Sio FR, Trono D. APOBEC3G-depleted resting CD4+ T cells remain refractory to HIV1 infection. PLoS One. 2009;4:e6571. doi: 10.1371/journal.pone.0006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vetter ML, D’Aquila RT. Cytoplasmic APOBEC3G Restricts Incoming Vif-positive HIV-1 and Increases 2-LTR Circle Formation In Activated T Helper Subtype Cells. J Virol. 2009 doi: 10.1128/JVI.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conticello SG, et al. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 76.Mehle A, et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 77.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 78.Liu B, et al. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J Virol. 2005;79:9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehle A, et al. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Y, et al. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luo K, et al. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A. 2005;102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mehle A, et al. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 83.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 84.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. Embo J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harjes E, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stanley BJ, et al. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol. 2008;82:8656–8663. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Auclair JR, et al. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–284. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reingewertz TH, et al. The C-terminal domain of the HIV-1 Vif protein is natively unfolded in its unbound state. Protein Eng Des Sel. 2009;22:281–287. doi: 10.1093/protein/gzp004. [DOI] [PubMed] [Google Scholar]
- 90.Kao S, et al. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simon V, et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian C, et al. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J Virol. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schrofelbauer B, et al. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bogerd HP, et al. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mangeat B, et al. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 98.Schrofelbauer B, et al. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He Z, et al. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 100.Yamashita T, et al. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 2008;10:1142–1149. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 101.Chen G, et al. A patch of positively charged amino acids surrounding the HIV-1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol. 2009 doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dang Y, et al. Identification of a novel WxSLVK motif in the N-terminus of HIV and SIV Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009 doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pery E, et al. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang B, et al. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J Biol Chem. 2003;278:6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Donahue JP, et al. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang L, et al. Function analysis of sequences in human APOBEC3G involved in Vif-mediated degradation. Virology. 2008;370:113–121. doi: 10.1016/j.virol.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 107.Zhang W, et al. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS One. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Russell RA, et al. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hache G, et al. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gandhi SK, et al. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Land AM, et al. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Piantadosi A, et al. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kieffer TL, et al. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mulder LC, et al. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jern P, et al. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Russell RA, et al. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller JH, et al. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiao Z, et al. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. Faseb J. 2007;21:217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
- 120.Adessi C, Soto C. Converting a peptide into a drug: strategies to improve stability and bioavailability. Curr Med Chem. 2002;9:963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 121.Martin SJ, et al. Programmed cell death (apoptosis) in lymphoid and myeloid cell lines during zinc deficiency. Clin Exp Immunol. 1991;83:338–343. doi: 10.1111/j.1365-2249.1991.tb05639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Treves S, et al. Apoptosis is dependent on intracellular zinc and independent of intracellular calcium in lymphocytes. Exp Cell Res. 1994;211:339–343. doi: 10.1006/excr.1994.1096. [DOI] [PubMed] [Google Scholar]
- 123.Cao J, et al. High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob Agents Chemother. 2005;49:3833–3841. doi: 10.1128/AAC.49.9.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mehle A, et al. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J Virol. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nathans R, et al. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]