Abstract
A demethylated derivative of curcumin (DC; 67.8% bisdemethylcurcumin, 20.7% demethylmonodemethoxycurcumin, 5.86% bisdemethoxycurcumin, 2.58% demethylcurcumin) was prepared by using a 95% extract of curcumin (C95; 72.2% curcumin, 18.8% monodemethoxycurcumin, 4.5% bisdemethoxycurcumin). DC increased glutathione and reduced reactive oxygen species (ROS) in HT4 neuronal cells. In a model of glutamate-induced death of HT4, DC was more effective than C95 in neuroprotection. The protective effects of DC were retained even when DC was withdrawn from culture media after pretreatment. DC treatment, unlike an equal dose of C95, completely spared glutamate-induced loss of cellular GSH. Both DC and C95 prevented glutamate-induced elevation of cellular ROS but failed to attenuate glutamate-induced elevation of intracellular calcium. In human microvascular endothelial cells (HMECs) challenged with TNF-α, GeneChip analysis revealed that only a subcluster of 23 TNF-α–inducible genes were uniquely sensitive to C95. In sharp contrast, 1,065 TNF-α–inducible genes were sensitive to DC but not to C95, suggesting that DC was more effective in antagonizing the effects of TNF-α on HMECs. Functional analysis identified that the genes uniquely sensitive to DC belonged in four functional categories: cytokine-receptor interaction, focal adhesion, cell adhesion, and apoptosis. Real-time PCR as well as ELISA studies demonstrated that TNF-α–inducible CXCL10 and CXCL11 expression was sensitive to DC but not to C95. Flow-cytometry studies recognized ICAM-1 and VCAM-1 as TNF-α–inducible adhesion molecules that were uniquely sensitive to DC. Taken together, DC exhibited promising neuroprotective and antiinflammatory properties that must be characterized in vivo. Antioxid. Redox Signal. 11, 449–468.
Introduction
Curcumin, a diferuloylmethane, is the principal curcuminoid of the Indian curry spice turmeric. A member of the curcuminoid family of compounds, curcumin is a yellow phenolic pigment obtained from the powdered rhizome of Curcuma longa Linn (89). Curcuma longa, a perennial herb, is a member of the Zingiberaceae (ginger) family. With growing emphasis on desirable diet behaviors and herb-based nutraceuticals in health preservation and disease prevention, curcumin has received rapidly soaring attention (3, 37, 45, 46, 50, 89). Curcuminoids are abundantly found in turmeric preparations, which have been commonly used in Ayurveda, the original Indian system of health care that was practiced around 1900 BC (1). The current literature supports that curcumin regulates numerous transcription factors, cytokines, protein kinases, adhesion molecules, redox status, and enzymes that have been linked to inflammation. Inflammation is a major driver of several chronic disease processes including neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune, and neoplastic illnesses (1). Oral curcumin is well tolerated and, despite its limited absorption, has biologic activity in some patients with pancreatic cancer (17). In phase I clinical studies, curcumin with doses up to 3,600–8,000 mg daily for 4 months did not result in discernible toxicities, except mild nausea and diarrhea (33).
Polyphenolic phytochemicals are common in the diet and have been suggested to have a wide range of beneficial health effects, some of which have been proven in humans (66, 104). The polyphenolic structure of curcumin [(1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadien-3,5-dione; Fig. 1] is responsible for a number of its beneficial health effects (7, 31, 81). In this study, the number of phenolic groups in curcumin was doubled by a demethylation process that resulted in a derivative with two diphenols or a pyrocatechol structure (Fig. 2). Neuroprotective and antiinflammatory properties of the demethylated curcumin (DC) derivative were compared in vitro.
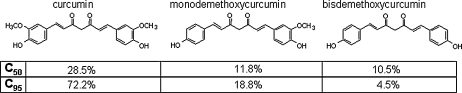
FIG. 1.
Composition of 50% (C50) and 95% (C95) curcuminoid extracts.
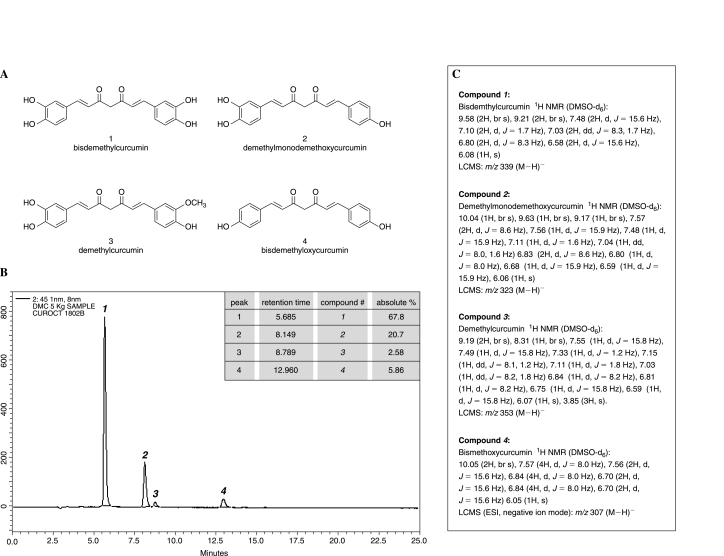
FIG. 2.
Composition of the demethylated curcumin preparation (DC). (A) major components (compounds 1–4) of DC. (B) HPLC chromatogram of compounds shown in (A), and the relative abundance (inset) of the four compounds in DC. (C) NMR characterization of compounds 1–4 in DC.
Materials & Methods
Materials
Glutamic acid monosodium salt and dimethylsulfoxide were purchased from Sigma (St. Louis, MO). For cell culture, Dulbecco's modified Eagle medium, MCDB-131 growth medium, fetal calf serum, l-glutamine, penicillin, and streptomycin were procured from Invitrogen Corporation (Carlsbad, CA). C50 and C95 were obtained from Laila Impex (Vijayawada, India). Culture dishes were obtained from Nunc, Denmark.
Synthesis of demethylated curcuminoids
The 95% curcumin extract (C95), as shown in Fig. 1, was used as the substrate. To an ice-cold solution of C95 (55 g) in ethyl acetate (2.5 L), was slowly added aluminum chloride (150 g), followed by the drop-wise addition of pyridine (350 ml) for 15 min, and the reaction mixture was heated under reflux for 24 h. After cooling the reaction mixture to 10°C, cold dilute HCl (20%) was added to decompose the aluminum chloride complex. The mixture was extracted with ethyl acetate (5 × 1.0 L). The combined ethyl acetate layer was washed with water, brine, and dried over anhydrous sodium sulfate. The solvent was filtered and evaporated under a vacuum. The residue was diluted with chloroform (50 ml) and kept at ambient temperature. After 10 h, the solid was filtered and dried under a vacuum to give the product as a yellow powder (21 g). The mixture comprised 67.8% bisdemethylcurcumin, 20.7% demethylmonodemethoxycurcumin, 2.58% demethylcurcumin, and 5.86% bisdemethoxycurcumin, as shown in Fig. 2.
Separation and characterization of demethylated curcuminoids
Enriched demethylated curcuminoids were extracted in methanol and separated on a Phenomenex C18 (4.6 × 250 mm) column and detected at 451 nm by using a Shimadzu LC 2010 CHT isocratic (A:B, 52:48; pump A, 0.1% vol/vol orthophosphoric acid in water; pump B, acetonitrile; flow rate, 1 ml/min) HPLC system. Analysis of the compounds (Fig. 2) formed by using a Bruker (Berlin, Germany) AV400 Ultrashield NMR by using deuterated dimethylsulfoxide (DMSO-d6) as solvent.
Cell culture
Mouse hippocampal HT4 neural cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 95% air and 5% CO2, as described previously (40–42, 78, 79). Human microvascular endothelial cells (HMEC-1s) were cultured under standard culture conditions in MCDB-131 growth medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 M l-glutamine, as described previously (72).
Glutamate treatment
Immediately before experiments, the culture medium was replaced with fresh medium supplemented with serum and antibiotics. Glutamate (10 mM) was added to the medium as an aqueous solution (40–42, 78, 79). No change in the medium pH was observed in response to the addition of glutamate.
Curcumin treatment
Stock solutions (×103 of working concentration) of DC, C50, or C95 were prepared in dimethylsulfoxide (DMSO). Respective controls were treated with equal volume (0.1%, vol/vol) of DMSO. In cases of pre-treatment, DC or C95 was added to the culture dishes 8 h before exposure to glutamate, followed by replacement of the culture media without any curcuminoid added to it.
Cell viability
The viability of cells in culture was assessed by measuring the leakage of lactate dehydrogenase (LDH) (29) from cells to media 18–24 h after glutamate treatment by using the in vitro toxicology assay kit from Sigma Chemical Co. (St. Louis, MO). The protocol was described in detail in previous reports (29). In brief, LDH leakage was determined by using the following equation:
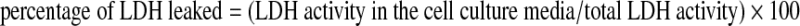 |
where total LDH activity = LDH activity in cell monolayer + LDH activity of detached cells + LDH activity in the cell-culture media (42, 43, 79).
Glutathione assay
Reduced (GSH) glutathione was detected from HT4 cell acid lysates by using HPLC coulometric electrode array detector (CoulArray Detector, model 5600 with 12 channels; ESA Inc., Chelmsford, MA), as described previously (42, 43). The CoulArray detector uses multiple channels set at specific redox potentials, as described (75). Data were collected by using channels set at 600, 700, and 800 mV. The samples were snap-frozen and stored in liquid nitrogen until HPLC assay. Sample preparation, composition of the mobile phase, and specification of the column used were previously reported (77, 79).
Reactive oxygen species (ROS)
Detection of ROS was performed by using dichlorodihydrofluorescein diacetate (H2DCF-DA) (Molecular Probes, Invitrogen). After 8 h of glutamate exposure, the cells were washed with PBS, centrifuged (500 g, 5 min), resuspended in PBS, and incubated with 10 μM H2DCF-DA for 20 min at 37°C. To detect cellular fluorescence, the fluorochromeloaded cells were excited by using a 488-nm argon-ion laser in a flow cytometer. The dichlorofluorocein (DCF) emission was recorded at 530 nm. Data were collected by using a flowcytometer from at least 10,000 cells.
Determination of intracellular Ca2+
Intracellular Ca2+ levels were measured by using cellpermeant acetomethoxyl ester of calcium green-1 (Molecular Probes), as described previously (93). After different treatments, cells were washed 3 times with PBS. Cells were detached from the monolayer by using trypsin, and centrifuged (500 g, 5 min). After another wash, the cells were resuspended in PBS and loaded with the acetomethoxyl ester of calcium green-1 (1 μM) for 20 min at room temperature. For the detection of intracellular fluorescence, the fluorochrome-loaded cells were excited by using a 488-nm argon-ion laser in a flow cytometer. Emission was recorded at 530 nm. Data were collected from at least 10,000 cells per sample.
GeneChip™ probe array analysis
To identify sets of gene differentially expressed in TNF-α–treated endothelial cells pretreated or not with DC or C95, we used the GeneChip approach, as described by our group (68, 70, 71, 73, 74, 99). Total RNA was extracted by using the RNeasy kit. The quality of RNA thus obtained was examined by using the Agilent 2100 Bioanalyzer. Targets were prepared for microarray hybridization according to previously described protocols (68, 70, 71, 73, 74). To assess the quality of hybridization, samples were hybridized for 16 h at 45°C to GeneChip test arrays. Next, the qualified samples were hybridized to Affymetrix Human Genome arrays (HGU133 2.0) for the screening of >22,000 genes and ESTs. The arrays were washed, stained with streptavidin-phycoerythrin, and scanned with the GeneArray scanner (Affymetrix) in our own facilities, as described earlier (68, 70, 71, 73, 74).
GeneChip data analyses
Data acquisition and image processing were performed by using GCOS (Gene Chip Operating Software, Affymetrix). Raw data were collected and analyzed by using a Stratagene ArrayAssist Expression Software v5.1 (Stratagene). Additional processing of data was performed by using dChip software (v 1.3, Harvard University) (68, 70, 71, 73, 74). Data normalization and background corrections were performed by using MAS 5.0. Probe sets with signal intensity ≤100 were excluded. Additionally, probe sets that showed absent call in any of the four groups were also excluded. Genes that were more than twofold upregulated in the TNF-α–treated group compared with the TNF-α–untreated control group were selected and subjected to hierarchical clustering. Major clusters of genes that were upregulated after TNF-α treatment but were downregulated uniquely by DC or C95 were selected as DC- or C95-sensitive genes. These subclusters were subjected to further functional analysis by using DAVID (Database for Annotation, Visualization and Integrated Discovery NIAID, NIH). Major known pathways (KEGG, Kyoto Encyclopedia of Genes and Genomes) identified in each of the cluster have been illustrated as pathway maps, marking the candidate genes with asterisks. In addition, selected differentially expressed candidate genes were verified by using quantitative real-time PCR and quantitative methods for protein analysis.
Quantitative gene expression analysis
mRNAs were quantified by real-time PCR assay by using the double-stranded DNA-binding dye SYBR Green-I, as described previously (68, 70, 71, 73, 74). Primer sets for CXCL10 (catalog no. PPH00765E) and CXCL11 (catalog no. PPH00506A) were purchased from SuperArray Bioscience Corporation (Frederick, MD).
The primer sets used for β-actin were
h β-actin F: 5′ GTA CCA CTG GCA TCG TGA TGG ACT 3′
h β-actin R: 5′ CCG CTC ATT GCC AAT GGT GAT 3′
Measurement of CXCL10 and CXCL11 protein
Human microvascular endothelial cells (HMECs) were seeded in six-well plates (0.5 × 106/well). DC or C95 was added to the cells for 24 h, after which cells were challenged with TNF-α for an additional 24 h. CXCL10 or CXCL11 levels in the medium were determined by using commercially available ELISA kits (R & D Systems, Minneapolis, MN). To normalize the data, total protein content was measured from cell lysates by using the BCA protein assay kit.
Determination of ICAM-1 and VCAM-1 surface expression
Cell seeding and treatment were exactly the same as described earlier for CXCL10/11 protein analyses. After TNF-α treatment, cells were washed twice with Dulbecco's PBS (D-PBS), pH 7.4, and incubated with phycoerythrin (PE)-labeled mouse anti-human CD106 (VCAM-1) antibody or mouse anti-human CD54 (ICAM-1) (BD Bioscience, San Jose, CA), for 30 min at 4°C in the dark. After incubation, cells were washed twice with D-PBS and finally resuspended in fresh D-PBS. Expression of ICAM-1 was immediately assayed by using a flow cytometer. Appropriate isotypic controls were used for background fluorescence control for the ICAM-1 and VCAM-1 assays.
Flow cytometric analysis
The fluorescence and light-scattering properties (forward scatter and side scatter) of the cells were determined by using an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI). Signals from cells labeled with the PE-conjugated ICAM-1 or VCAM-1 antibodies were collected on channel FL2 (585 ± 20 nm) after excitation with a 488-nm solid-state laser on a gated population of cells. In each sample, at least 10,000 gated viable cells were examined. A logarithmic scale was used to measure both background and endothelial cell fluorescence. Background fluorescence was then subtracted from endothelial cell fluorescence, allowing linear comparisons of ICAM-1 or VCAM-1 expression in samples.
Statistical analyses
Data are reported as mean ± SD of at least three experiments. Comparisons between multiple groups were made by using analysis of variance (ANOVA). A value of p < 0.05 was considered statistically significant.
Results
Curcumin is a biphenolic compound (Fig. 1) with hydroxyl groups at the ortho position on the two aromatic rings that are connected by a β-diketone bridge, containing two double bonds (dienone), which can undergo Michael addition, critical for some of the effects of curcumin (101). Commercial-grade curcumin is known to contain ~77% curcumin, 17% demethoxycurcumin, and 3% bisdemethoxycurcumin) (35). This is consistent with the C95 curcumin extract used in the current study (Fig. 1). The curcumin demethylation process adopted in this study resulted in a product that was rich in tetrahydroxycurcumin and its tetrahydroderivative, tetrahydrotetrahydroxycurcumin. The demethylcurcumin (DC) preparation was mostly made up of bisdemethylcurcumin, a dicatechol. Demethylmonodemethoxycurcumin represented another significant component of DC. Minor components included demethylcurcumin and bisdemethoxycurcumin (Fig. 2). The components of DC are all known to be natural products. Compounds 1, 2 and 4 (Fig. 2) are present in Curcuma longa as minor natural products (38, 39). Compound 3 has been isolated from Curcuma domestica (59). The major compound 1 has been also detected as a metabolite of curcumin in mouse and human liver microsomal preparations (91).
Glutamate challenge of the murine HT hippocampal neural cell line, lacking the intrinsic excitotoxicity pathway, represents a useful model to characterize redox-sensitive pathways involved in neurotoxicity (40–42, 78, 79). In this study, we observed that a concentration as low as 500 ng/ml DC, but not C50 or C95, completely protected HT4 cells challenged by an excessive 10 mM glutamate. No such protection by C50 or C95 was noted, even at twice the said concentration (Fig. 3). These observations suggest a higher neuroprotective property of DC compared with curcumin. Additional experiments led to the observation that even if HT4 cells were pretreated with DC and DC was then removed from the culture media by media replacement, the neuroprotective effects of DC remained. Such effect was not observed with C95 curcumin, even when used at twice the concentration (1,000 ng/ml). At 5,000 ng/ml, however, C95 curcumin did protect the neuronal cells challenged with glutamate. These results indicate that DC was more potent than C95 curcumin in protecting HT4 neuronal cells against glutamate-induced toxicity (Fig. 4).
FIG. 3.
Demethylcurcumin, but not C50 or C95 curcumin, protected HT4 neuronal cells against glutamate-induced death. HT4 cells were seeded in six-well plates (0.1 × 106/well). Demethylcurcumin (DC), C50, or C95 was added to the cells for 8 h before glutamate (10 mM) exposure. Results are expressed as mean ± SD, p < 0.05; *compared with glutamate-treated.
FIG. 4.
Demethylcurcumin pretreatment is sufficient and an order of magnitude more potent than C95 curcumin on a concentration basis to protect HT4 neuronal cells against glutamate challenge. HT4 cells were seeded in six-well plates (0.1 × 106/well). Demethylcurcumin (DC) or C95 was added to the cells (at the indicated concentrations) for 8 h and then washed before glutamate exposure. “p” indicates that cells were pretreated at the dose shown and removed before glutamate challenge. Results are expressed as mean ± SD, p < 0.05; *compared with glutamate treated.
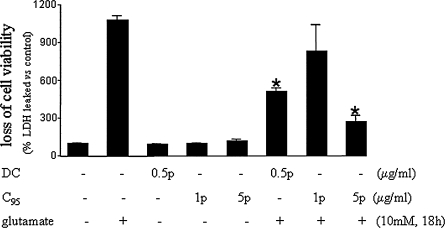
Glutamate-induced death of HT4 cells is known to be associated with oxidative stress characterized by loss of cellular GSH and elevation of cellular reactive oxygen species (ROS) (40–42, 78, 79). Therefore, we were interested to examine the effects of DC on basal cellular GSH and ROS levels. At a low concentration of 100 ng/ml, DC significantly increased the levels of cellular GSH but did not significantly influence cellular ROS, as measured by the DCF approach. At 500 ng/ml, a concentration at which DC was noted to protect HT4 against glutamate toxicity, DC significantly lowered basal cellular ROS levels (Fig. 5B). These results suggest that DC has antioxidant properties. Consistent with these findings, it was observed that at a dose of 500 ng/ml, DC completely spared glutamate-induced GSH loss in HT4 cells. Even at twice that dose, C95 curcumin failed to prevent glutamate-induced cellular GSH loss. At a dose 10 times of the dose of DC, C95 curcumin did prevent glutamate-induced GSH loss from HT4 cells, but this effect was significantly less compared with the effect of DC at 500 ng/ml (Fig. 6A). Glutamate challenge resulted in a rapid surge of cellular ROS in the HT4 cells. Both DC and C95 curcumin prevented cellular ROS accumulation in response to glutamate challenge (Fig. 6B). However, the concentration of C95 curcumin needed to generate significant effects was twice that of DC. At 500 ng/ml, curcumin was not effective in significantly lowering glutamate-induced ROS accumulation in HT4 cells (not shown). Glutamate is known to elevate intracellular calcium ion concentration in HT4 cells (93). Similar results were observed in this study. Interestingly, in concentrations at which both DC and C95 curcumin protected HT4 from glutamate, neither could prevent glutamate-induced elevation of intracellular calcium ion concentration. Thus, DC was neuroprotective against glutamate-induced toxicity despite aggravated perturbation of the cellular calcium homeostasis (Fig. 6).
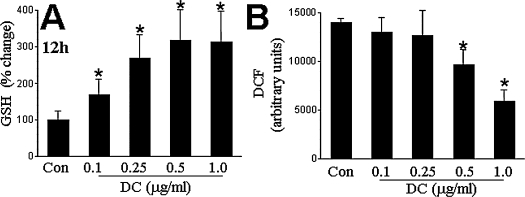
FIG. 5.
Demethylcurcumin treatment increased reduced glutathione levels and attenuated basal levels of reactive oxygen species (ROS) in HT4 neuronal cells. HT4 cells were seeded in six-well plates (0.1 × 106/well). Cells were treated with the indicated concentrations of demethylcurcumin (DC) for (A) 12 h or (B) 24 h. ROS levels were estimated as based on DCF fluorescence. Results are expressed as mean ± SD, p < 0.05; *compared with untreated control.
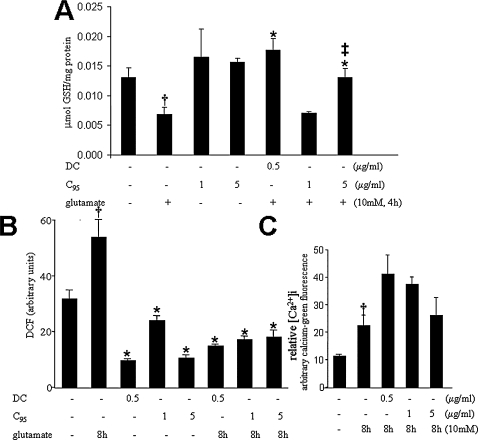
FIG. 6.
Demethylcurcumin is more potent than C95 curcumin in protecting against glutamate-induced GSH-loss (A) and elevation of cellular ROS (B). Curcuminoids do not protect against glutamateinduced intracellular Ca2+ perturbations (C). HT4 neuronal cells were treated with demethylcurcumin (DC) or C95 for 8 h and then challenged with glutamate for (A) 4 h or (B–C) 8 h. Results are expressed as mean ± SD, p < 0.05. *Compared with glutamate treated; ‡compared with DC (500 ng/ml) treated; compared with untreated control.
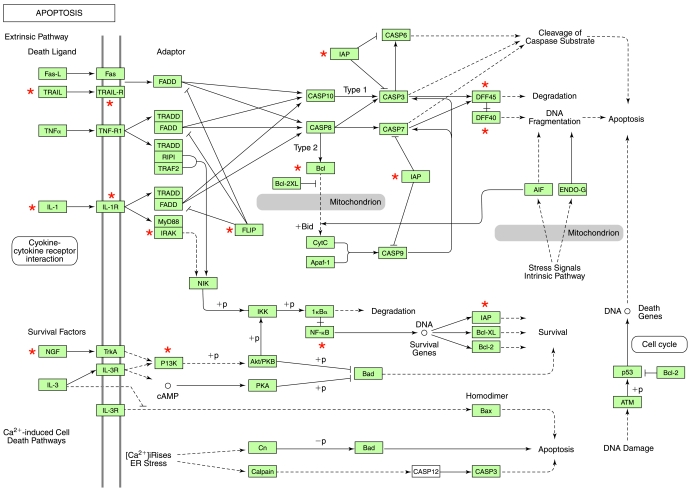
Curcuminoids have been reported to possess multifunctional bioactivities, especially the ability to inhibit proinflammatory induction (48). Specifically, curcuminoids are known to attenuate the proinflammatory effects of TNF-α (11, 44). We thus sought to compare the effects of DC and C95 curcumin in a model of TNF-α–induced gene expression in human microvascular endothelial cells (HMECs). At 0.5–1 μg/ml, both C95 curcumin and DC were not toxic to HMECs (Fig. 7). GeneChip screening of TNF-α–inducible transcriptome of HMECs identified 1,195 probe sets that were induced by a magnitude of twofold or higher (Fig. 8A). Visualization of the heat map clearly shows that globally, DC seemed to be more effective in negating the effects of TNF-α on gene induction compared with C95 curcumin (Fig. 8A). Visually, the DC column is more greenish, and the C95 curcumin column is more reddish. Subcluster analysis identified a list of 1,065 probe sets representing TNF-α–inducible genes that were differentially regulated by DC and C95 curcumin (Fig. 8B, Table 1). This observation suggests that DC and C95 curcumin function uniquely. Dominance of the greenish color in the DC column reveals that the expression of these TNF-α–inducible genes was effectively suppressed by DC but not by C95 curcumin (Fig. 8B). Conversely, the subcluster of TNF-α–inducible genes that were inhibited by C95 curcumin but not by DC only represented 23 probe sets (Fig. 8C, Table 2). Thus, overall, DC seems to be more effective in negating the effects of TNF-α on endothelial cells. DAVID-assisted functional analysis of the TNF-α–inducible pathways that were sensitive to DC but not to C95 curcumin identified four major functional categories that are also functionally related among themselves. These four categories include cytokine and cytokine-receptor interaction pathway (Fig. 9), the focal adhesion pathway (Fig. 10), the apoptosis pathway (Fig. 11) and the cell-adhesion pathway (Fig. 12). Marking of the DC-sensitive genes in these pathways provides a useful visualization of the global effects of DC, which may be used as a resource to postulate novel hypotheses characterizing the mechanism of action of DC.
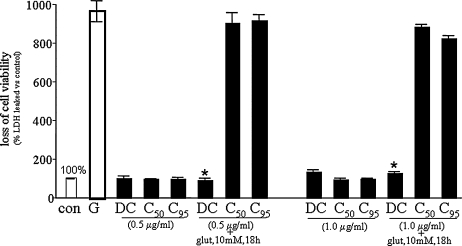
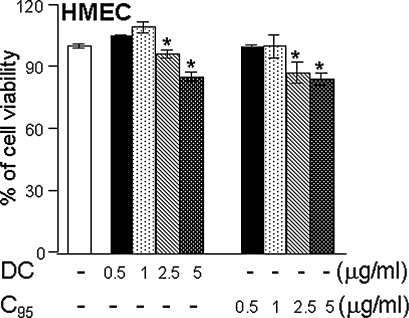
FIG. 7.
Human microvascular endothelial cell viability in response to curcuminoid treatment. Human microvascular endothelial cells (HMEC-1s) were seeded in six-well plates (0.5 × 106/well). DC or C95 was added to the cells at the indicated concentration for 24 h. Results are expressed as mean ± SD, p < 0.05, *compared with untreated control.
FIG. 8.
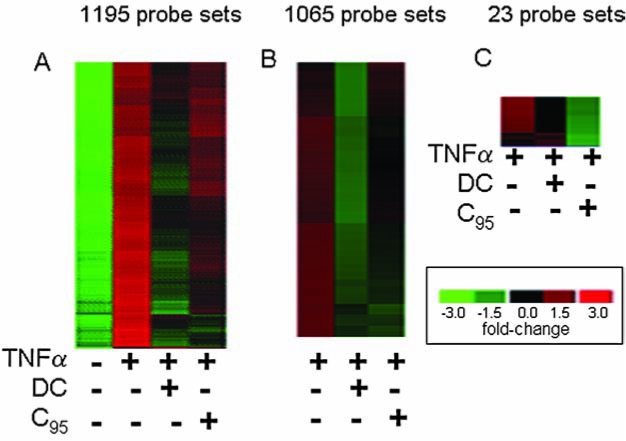
GeneChip analysis of the human microvascular endothelial cell transcriptome. Heat map illustrating how DC and C95 curcumin regulate TNF-α–inducible genes. HMEC-1 cells were treated with DC or C95 (1 μg/ml) for 24 h followed by activation with TNF-α (50 ng/ml) for 6 h. (A) All genes that showed a twofold or higher increase in response to TNF-α challenge (versus untreated control group) were subjected to hierarchical clustering. This cluster included a total of 1,195 probe sets. (B) The cluster in (A) was further subclustered to identify TNF-α–inducible genes that were specifically downregulated after DC treatment but not by C95 curcumin. This set included a total of 1,065 probe sets. Major functional pathways in this cluster were identified and are presented as Figs. 9–12. Genes were sorted based on fold change compared with an untreated control group, and the top 50 (based on highest fold change) in the TNF-α group are presented in Table 1. (C) The cluster presented in (A) was subclustered to identify TNF-α–inducible genes that were specifically downregulated after C95 treatment but not in response to DC treatment. Genes sorted based on fold change between TNF-α–treated and the untreated control groups are presented in Table 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Table 1.
Top 50 Differentially Expressed TNFα-Inducible Genes Specifically Sensitive to DC Treatment
| |
|
Fold change control vs. treatments |
|||
|---|---|---|---|---|---|
| Probe set ID | Gene title | Gene symbol | TNF alone | DC + TNF | C95 + TNF |
| 211122_s_at | Chemokine (C-X-C motif) ligand 11 | CXCL11 | 786.1 | 248 | 608.6 |
| 211506_s_at | Interleukin 8 | IL8 | 415.3 | 307.3 | 422.3 |
| 203868_s_at | Vascular cell adhesion molecule 1 | VCAM1 | 414.2 | 306.9 | 394 |
| 204533_at | Chemokine (C-X-C motif) ligand 10 | CXCL10 | 318.6 | 141.2 | 320.7 |
| 215078_at | Superoxide dismutase 2, mitochondrial | SOD2 | 202.8 | 70.5 | 84.3 |
| 202859_x_at | Interleukin 8 | IL8 | 168.3 | 153.6 | 170.8 |
| 207517_at | Laminin, gamma 2 | LAMC2 | 117.1 | 61.1 | 66.5 |
| 204439_at | interferon-induced protein 44-like | IFI44L | 84.1 | 34.3 | 102.9 |
| 228617_at | XIAP-associated factor 1 | XAF1 | 81.8 | 26.8 | 72.6 |
| 204580_at | Matrix metallopeptidase 12 (macrophage elastase) | MMP12 | 80.5 | 52 | 68.3 |
| 205599_at | TNF receptor-associated factor 1 | TRAF1 | 78 | 5 | 64 |
| 205828_at | Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 | 60.4 | 34.2 | 38.6 |
| 210229_s_at | Colony-stimulating factor 2 (granulocyte-macrophage) | CSF2 | 57.3 | 54.2 | 55 |
| 235116_at | TNF receptor-associated factor 1 | TRAF1 | 53.3 | 33.3 | 41.8 |
| 221371_at | Tumor necrosis factor (ligand) superfamily, member 18 | TNFSF18 | 48.1 | 35.4 | 45.5 |
| 219181_at | Lipase, endothelial | LIPG | 44.3 | 19.6 | 28.6 |
| 223484_at | Chromosome 15 open reading frame 48 | C15orf48 | 42.2 | 23.8 | 31.3 |
| 232593_at | likely orthologue of mouse lung-inducible neutralized-related C3HC4 RING domain Protein | LINCR | 36.9 | 17.4 | 30.1 |
| 202643_s_at | Tumor necrosis factor, α-induced protein 3 | TNFAIP3 | 35.8 | 23.3 | 31 |
| 216841_s_at | Superoxide dismutase 2, mitochondrial | SOD2 | 35.7 | 20.4 | 24.6 |
| 204475_at | Matrix metallopeptidase 1 (interstitial collagenase) | MMP1 | 34.5 | 21.4 | 24.2 |
| 216598_s_at | Chemokine (C-C motif) ligand 2 | CCL2 | 33.3 | 32.1 | 32 |
| 202644_s_at | Tumor necrosis factor, α-induced protein 3 | TNFAIP3 | 33.2 | 21.9 | 27.7 |
| 214974_x_at | Chemokine (C-X-C motif) ligand 5 | CXCL5 | 33.1 | 23.5 | 25.4 |
| 217767_at | Complement component 3 | C3 | 31.9 | 19.9 | 33.3 |
| 210538_s_at | Baculoviral IAP repeat-containing 3 | BIRC3 | 31.4 | 24.6 | 32.7 |
| 230966_at | Interleukin 4-induced 1 | IL4I1 | 30.8 | 18.7 | 24.9 |
| 205680_at | Matrix metallopeptidase 10 (stromelysin 2) | MMP10 | 30.7 | 19.4 | 26.2 |
| 218182_s_at | Claudin 1 | CLDN1 | 26.1 | 20.4 | 21.5 |
| 215485_s_at | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 28.8 | 19.8 | 26 |
| 204655_at | Chemokine (C-C motif) ligand 5 | CCL5 | 28.5 | 14.6 | 30.6 |
| 1405_i_at | Chemokine (C-C motif) ligand 5 | CCL5 | 27.9 | 17.5 | 28.7 |
| 204994_at | Myxovirus (influenza virus) resistance 2 (mouse) | MX2 | 27.1 | 11.1 | 26.2 |
| 204470_at | Chemokine (C-X-C motif) ligand 1 (melanoma growth-stimulating activity, alpha) | CXCL1 | 26.9 | 21 | 28.2 |
| 222549_at | Claudin 1 | CLDN1 | 26.1 | 22.5 | 22.5 |
| 202086_at | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | MX1 | 25.1 | 12.4 | 22.6 |
| 202376_at | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | SERPINA3 | 24.9 | 17.4 | 29.2 |
| 1555759_a_at | Chemokine (C-C motif) ligand 5 | CCL5 | 23.6 | 16.5 | 26 |
| 221085_at | Tumor necrosis factor (ligand) superfamily, member 15 | TNFSF15 | 22.3 | 10 | 15.5 |
| 203236_s_at | Lectin, galactoside-binding, soluble, 9 (galectin 9) | LGALS9 | 22.3 | 8.7 | 19.3 |
| 204933_s_at | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | TNFRSF11B | 22.3 | 15 | 19.5 |
| 207339_s_at | Lymphotoxin beta (TNF superfamily, member 3) | LTB | 22.1 | 15.1 | 15.7 |
| 228607_at | 2′ −5′-oligoadenylate synthetase 2, 69/71 kDa | OAS2 | 21.7 | 10 | 16.1 |
| 215223_s_at | Superoxide dismutase 2, mitochondrial | SOD2 | 21.3 | 18.7 | 21.4 |
| 203828_s_at | Interleukin 32 | IL32 | 20.4 | 14.8 | 17.9 |
| 221477_s_at | Hypothetical protein MGC5618 | MGC5618 | 20.3 | 16 | 17.1 |
| 202267_at | Laminin, gamma 2 | LAMC2 | 19.7 | 11.7 | 15.1 |
| 202638_s_at | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 18.4 | 14.7 | 16.9 |
| 202510_s_at | Tumor necrosis factor, α-induced protein 2 | TNFAIP2 | 18.2 | 15.3 | 17.2 |
| 223502_s_at | Tumor necrosis factor (ligand) superfamily, member 13b | TNFSF13B | 17.6 | 7.6 | 16.6 |
Data presented indicate fold change in gene expression compared with TNF-α-untreated control group. Probe set ID, Affymetrix probe identifications. Genes in bold font were verified by using quantitative PCR or protein analysis studies. Supplement for Fig. 8, cluster B.
Table 2.
Differentially Expressed TNFα-Inducible Genes Specifically Sensitive to C95 Treatment
| |
|
Fold change, control vs. treatments |
|||
|---|---|---|---|---|---|
| Probe set ID | Gene title | Gene symbol | TNF alone | DC + TNF | C95 + TNF |
| 1552277_a_at | Transmembrane protein with EGF-like and two follistatin-like domains 1 | C9orf30 | 2.6 | 2.3 | 2 |
| 1552914_a_at | CD276 molecule | CD276 | 8.9 | 5.3 | 3.1 |
| 1559064_at | Nucleoporin, 153 kDa | NUP153 | 4.4 | 4.7 | 3.7 |
| 201502_s_at | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NFKBIA | 6.8 | 6.4 | 5.9 |
| 203921_at | Carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | CHST2 | 3.4 | 2.6 | 2 |
| 205476_at | Chemokine (C-C motif) ligand 20 | CCL20 | 181.3 | 181.6 | 162.6 |
| 207937_x_at | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | FGFR1 | 2.1 | 1.5 | 0.8 |
| 208711_s_at | Cyclin D1 | CCND1 | 4.6 | 4 | 3.4 |
| 211924_s_at | Plasminogen activator, urokinase receptor | PLAUR | 4.5 | 3.9 | 3.5 |
| 216074_x_at | WW and C2 domain containing 1 | WWC1 | 41.8 | 58.4 | 8.5 |
| 219488_at | α1,4-Galactosyltransferase (globotriosylceramide synthase) | A4GALT | 3 | 2.5 | 2.1 |
| 220353_at | Family with sequence similarity 86, member C | FAM86C | 2.2 | 2.1 | 1.3 |
| 220382_s_at | Rho GTPase activating protein 28 | ARHGAP28 | 4.2 | 4.3 | 3.9 |
| 221530_s_at | Basic helix-loop-helix domain containing, class B, 3 | BHLHB3 | 5.5 | 4.7 | 3.7 |
| 226142_at | GLI pathogenesis-related 1 (glioma) | GLIPR1 | 2.2 | 2.3 | 1.9 |
| 226281_at | Delta/notch-like EGF repeat containing | DNER | 2.2 | 2 | 1.9 |
| 227340_s_at | RGM domain family, member B | RGMB | 2.4 | 2.4 | 2.3 |
| 235417_at | SPOC domain containing 1 | SPOCD1 | 4.9 | 4.3 | 3.6 |
| 238417_at | Phosphoglucomutase 2-like 1 | PGM2L1 | 2.2 | 1.8 | 1.4 |
| 238532_at | D4, zinc and double PHD fingers, family 3 | DPF3 | 3.2 | 2.3 | 1.7 |
Data presented indicate fold change in gene expression compared with TNF-α-untreated control group. Probe set ID, Affymetrix probe identifications. Supplement for Fig. 8, cluster C.
FIG. 9.
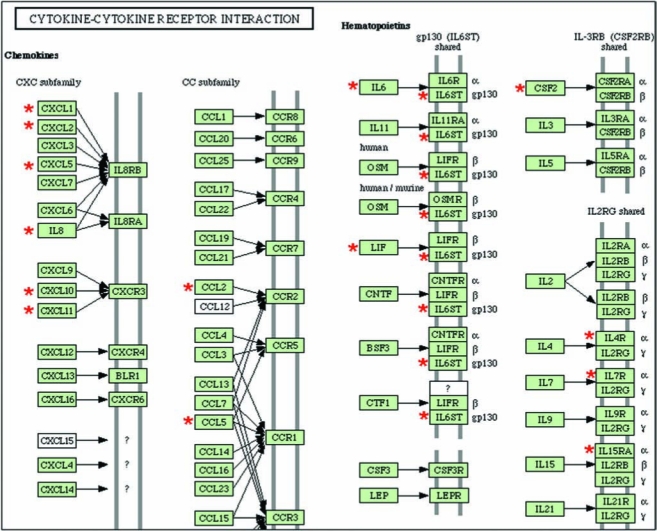
TNF- α–inducible genes that were specifically downregulated after DC treatment but not by C95 curcumin: the cytokine and cytokine-receptor interaction pathway. The genes shown in cluster Fig. 8B were subjected to functional analysis by using DAVID (NIAID, NIH). One of the major known pathways (KEGG) identified in this category was the cytokine–cytokine receptor interaction pathway as shown. The gene candidates that were differentially expressed in response to DC are marked with asterisks. DAVID, Database for Annotation, Visualization, and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
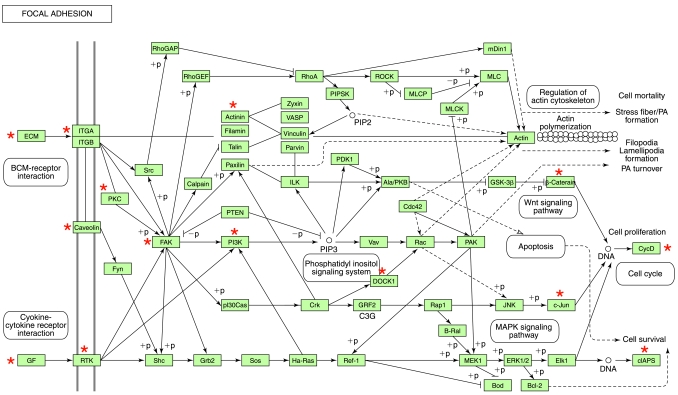
FIG. 10.
TNF- α–inducible genes that were specifically downregulated after DC treatment but not by C95 curcumin: the focal adhesion pathway. The genes shown in cluster Fig. 8B were subjected to functional analysis by using DAVID (NIAID, NIH). One of the major known pathways (KEGG) identified in this category was the focal adhesion pathway, as shown. The gene candidates that were differentially expressed in response to DC are marked with asterisks. DAVID, Database for Annotation, Visualization, and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 11.
TNF- α–inducible genes that were specifically downregulated after DC treatment but not by C95 curcumin: the apoptosis pathway. The genes shown in cluster Fig. 8B were subjected to functional analysis by using DAVID (NIAID, NIH). One of the major known pathways (KEGG) identified in this category was the apoptosis pathway. The gene candidates that were differentially expressed in response to DC are marked with asterisks. DAVID, Database for Annotation, Visualization, and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 12.
TNF- α–inducible genes that were specifically downregulated after DC treatment but not by C95 curcumin: the cell-adhesion pathway. The genes shown in cluster Fig. 8B were subjected to functional analysis by using DAVID (NIAID, NIH). One of the major known pathways (KEGG) identified in this category was the cell-adhesion pathway. The gene candidates that were differentially expressed in response to DC are marked with asterisks. DAVID, Database for Annotation, Visualization, and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Selected candidates from the list of DC-sensitive but C95-insensitive TNF-α–inducible genes were verified by using quantitative approaches to measure gene and protein expression.
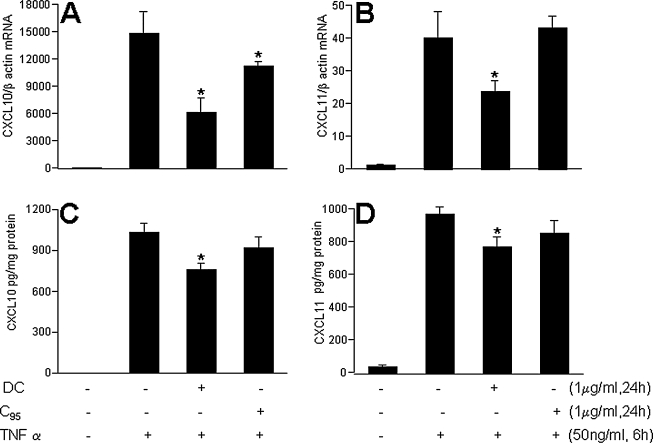
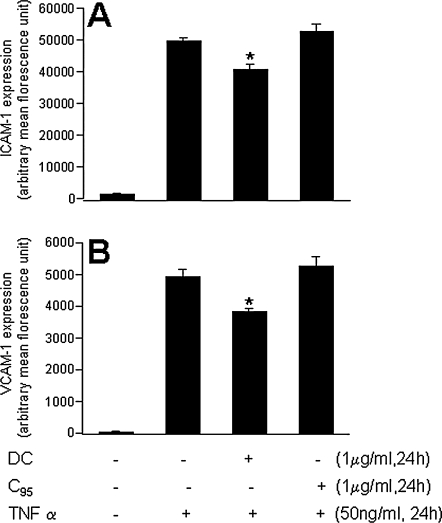
Chemokine (C-X-C motif) ligand 10 (CXCL10) or IP-10 is a small cytokine belonging to the CXC chemokine family that is also known as 10-kDa interferon-γ–induced protein. CXCL10 is secreted by several cell types in response to interferon (IFN)-γ. These cell types include monocytes, endothelial cells and fibroblasts. CXCL10 has been attributed to several roles, such as chemoattraction for monocytes/macrophages, T cells, NK cells, and dendritic cells, and promotion of T-cell adhesion to endothelial cells. In endothelial cells, induction of CXCL10 expression depends on an IFN-stimulated response element (ISRE) within the CXCL10 promoter (10). CXCL10 is known to be TNF-α inducible in other cell types (83). Consistently, we noted that both the CXCL10 gene and protein were potently induced by TNF-α in HMECs. TNF-α–inducible CXCL10 gene as well as protein expression was significantly blunted by DC but not by C95 curcumin (Fig. 13A and C). Angiostatic CXCL11 is another small cytokine belonging to the CXC chemokine family that is also called IFN-inducible T-cell α chemoattractant (I-TAC) and IFN-γ–inducible protein 9 (IP-9) (36). It is known to be present in endothelial cells, serves as a ligand for CXCR3, and is assumed to be involved in inflammatory diseases (92). TNF-α–induced marked induction of both the CXCL11 gene and the protein in HMECs. DC, but not C95 curcumin, significantly downregulated the expression of the TNF-α–induced CXCL11 gene, as well as the protein (Fig. 13B and D). TNF-α is well known to be a potent inducer of adhesion molecules and inflammatory disorders in endothelial cells (67, 109). Consistent results were obtained from our microarray studies in which the TNF-α–inducible expression of several major adhesion molecules, such as ICAM-1 and VCAM-1, was noted to be inhibited by DC. In experiments looking at the abundance of ICAM-1 and VCAM-1 in TNF-α–treated HMECs, it was observed that DC significantly attenuated inducible expression of both ICAM-1 and VCAM-1. These effects were not shared by C95 curcumin (Fig. 14). Thus, taken together, DC exhibited several desirable properties that were in contrast to curcumin rich in C95.
FIG. 13.
Demethylcurcumin, but not C95 curcumin, prevented TNF- α–induced CXCL10 and CXCL11 expression in HMEC cells. HMEC-1 cells were seeded in six-well plates (0.5 × 106/well). DC or C95 was added to the cells for 24 h followed by activation with TNF-α for 6 h (mRNA expression, A and B) or 24 h (protein expression, C and D). Results are expressed as mean ± SD, p < 0.05; *, compared with TNF-α treated.
FIG. 14.
Demthylcurcumin (DC) downregulated TNF- α–induced ICAM-1 and VCAM-1 protein expression in HMEC cells. Human microvascular endothelial cells (HMECs) were seeded in six-well plates (0.5 × 106/well). DC or C95 was added to the cells for 24 h and then activated with TNF-α for 24 h. A significant decrease in (A) ICAM-1 and (B) VCAM-1 protein expression was noted with DC treatment but not the C95 treatment. Results are expressed as mean ± SD, p < 0.05; *, compared with TNF-α treated.
Discussion
Current interest in the therapeutic properties of curcumin is fueled by a large body of literature, providing experimental data supporting a wide range of pharmacologic properties of curcumin, including chemosensitizing, radiosensitizing, wound healing, antimicrobial, antiviral, antifungical, immunomodulatory, neuroprotective, antioxidant and antiinflammatory properties (89). Pilot phase I clinical trials have shown curcumin to be safe even when consumed at a daily dose of 12 g for 3 months (24). The pleiotropic activities of curcumin seem to derive from its complex chemistry as well as its ability to influence multiple signaling pathways (30). Furthermore, curcumin is a free radical scavenger and hydrogen donor and exhibits both pro- and antioxidant activity. It also binds metals, particularly iron and copper, and can function as an iron chelator (30). In nature, curcumin exists as a family of structural analogues (Fig. 1). The potent antioxidant property of the curcumin family of compounds is viewed as a major contributor to the multifaceted functions displayed by naturally occurring curcumin (2, 55). Attempts to characterize the chemical basis of the antioxidant properties of the curcumin family of compounds have resulted in the synthesis of a number of ring-substituted analogues of curcumin. It has been consistently noted that the phenolic group is important for the antioxidant activity of curcumin (98). Curcumin possesses pro-glutathione properties (88) possibly via the Nrf2/ARE pathway (20). The current study provides first evidence that the dicatechol tetraphenolic form of curcumin has pro-glutathione antioxidant properties in vitro. Unlike curcumin, bisdimethylcurcumin is known to be ineffective in regulating TPA-induced tumor promotion on mouse skin (14). However, bis-dimethylcurcumin–rich DC clearly was superior to curcuminrich C95 in protective neuronal cells against glutamate-induced oxidative stress and death. This initial observation lays the foundation to test the neuroprotective efficacy of DC in vivo. Although curcumin has been demonstrated to be protective against stroke in vivo, it is notable that very a high dosage (100 mg/kg in rats) was required (84). This is consistent with the high dose of C95 required in this study to protect neuronal cells against glutamate-induced cytotoxicity. The comparative effects of DC and C95 observed in the current study provide the rationale to hypothesize that a lower dose of DC would be effective against stroke in vivo.
Unresolved inflammation is widely recognized as the driver of several major health disorders (9, 16, 22, 51, 52, 60, 80, 87, 97, 100). The vascular endothelium is a major target of actions of the proinflammatory cytokine TNF-α (5, 49). Anti–TNF-a strategies are therefore of therapeutic significance in conditions ranging from inflammatory vascular diseases, neuroimmune disorders, to cancer (6, 53, 82). Consistent with the observation that curcumin attenuates inflammatory responses of TNF-α–stimulated human endothelial cells, we noted that C95 treatment clearly attenuated TNF-a-induced gene expression in HMECs. Interestingly, in the same setting, an equal dose of DC prevented the change in expression of a much larger number of TNF-α–inducible genes. Such observations suggest that at a comparable dose, DC is more potent in antagonizing the effects of TNF-α on human microvascular endothelial cells. Results from the quantitative study of individual TNF-α–inducible chemokines and adhesion molecules and proteins support this contention.
Functional analysis of the microarray data identified specific TNF-a–inducible pathways that were uniquely sensitive to DC. In the category of cytokine-receptor interaction, DC-sensitive pathways were identified in the CXC, CC, and hematopoietin subfamilies. In the CXC subfamily, IL-8 and CXCR3 signaling were sensitive to DC. IL-8, a proinflammatory chemokine, induces trafficking of neutrophils across the vascular wall. It plays an important role in tumor angiogenesis, progression, and metastasis in a variety of human cancers (106). Antagonism of IL-8 is viewed as a therapeutic strategy for the management of a wide range of inflammatory disorders (65). The CXCR3 chemokine receptor was first discovered in 1996 and has been shown to play an important role in several diseases, most of which are related to inflammation. New antagonist classes are being developed to reveal the full therapeutic potential of CXCR3 (102). In the CC subfamily, inducible expression of CCL2 [formerly known as monocyte chemoattractant protein or MCP-1(103)] and CCL5 (formerly known as RANTES or regulated upon activation, normal T cell expressed and secreted) were sensitive to DC. CCL2 is known to be TNF-α inducible (32), and excessive levels of CCL2 are implicated in a broad range of inflammatory disorders (18, 90, 94). Anti-CCL5 strategies are effective in diminishing leukocyte infiltration into the central nervous system and in reducing inflammatory neurologic disease (23). Among the hematopoietins, the IL-6 pathway was DC sensitive.
Signal transduction in eukaryotic cells is a complex process mediated, normally, by the interaction of soluble extrinsic protein factors and their cognate receptors. One example of this phenomenon is the inflammatory cytokine IL-6 and the IL-6 receptor. However, the IL-6 receptor, once its ligand is bound, associates with another membrane glycoprotein, gp130 (also known as IL-6 signal transducer or IL6ST), to potentiate the cytokine response (28).
The vascular wall contains intimal endothelium and medial smooth muscle that act as contiguous tissues with tight spatial and functional coordination in response to tonic and episodic input from the bloodstream and the surrounding parenchyma. Focal adhesions lie at the convergence of integrin adhesion, signaling, and the actin cytoskeleton. Cells modify focal adhesions in response to changes in the molecular composition, two-dimensional (2D) versus three-dimensional (3D) structure, and physical forces present in their extracellular matrix environment (69, 95). Excessive focal adhesions drive numerous pathologic disorders ranging from cerebral infarction to tumor progression (12, 34, 56). Numerous key mediators of the focal adhesion pathway were identified as being specifically sensitive to DC.
Induction of apoptosis is a hallmark TNF-α response (5, 62). Although the killing of tumor cells is desirable, TNF-induced death of nontumor cells represents a major concern (21, 105, 108). The TRAIL and IL-1 death pathways were recognized as being DC sensitive. Anti-TRAIL strategies are being sought to address inflammatory vascular disorders. In particular, emerging data indicate that recombinant soluble TRAIL may act as a molecule with potential antiinflammatory activity in vascular pathophysiology (15). IL-1 has been implicated as a critical mediator of neuroimmune communication. In the brain, the functional receptor for IL-1, type 1 IL-1 receptor (IL-1R1), is localized primarily to the endothelial cells and is known to contribute to inflammatory disorders such as leukocyte infiltration into the CNS, activation of hypothalamic neurons, fever, and reduced locomotor activity in normal mice (13). Survival signals are often elicited in response to a terminal insult. Because the DC substantially attenuates the toxic insult of TNF-α on HMECs, the NGFPI3K survival pathway (61) seems to be no longer required in DC-treated TNF-a–challenged HMECs. TNF-α–induced expression of cell-adhesion molecules is implicated in a range of inflammatory vascular disorders (8, 25, 47, 57, 64, 86). Both anti–ICAM-1 and VCAM-1 strategies are in development for the treatment of inflammatory vascular disorders (26, 27, 58, 63, 76, 107). Observations made in this study reveal that DC is more effective than curcumin-rich C95 in attenuating TNF-α–induced ICAM-1 as well as VCAM-1 expression in human microvascular endothelial cells. Further studies examining the effects of DC on cell-adhesion function are therefore warranted.
Our current state of understanding of the therapeutic potential of curcuminoids recognizes the multifaceted beneficial properties of this family of compounds. Conversely, significant limitations must be addressed. The seven-carbon β-diketone linker in curcumin is known to be responsible for its instability (48). Studies addressing the stability of DC in vivo are therefore necessary. In general, plant polyphenols are widely recognized for the potential benefits to human health. However, polyphenol metabolism may pose certain metabolic challenges, such as those related to interaction with other pharmaceutical agents (54). It is important that DC be examined for such possible limitations. Although the observation that large doses of curcumin are safe in humans is encouraging (24), specific studies examining DC in vivo are necessary. Results of this study suggest that DC may have potent neuroprotective properties by acting as a pro-GSH agent. Pro-GSH approaches are known to be effective in neuroprotection in a wide range of models (4, 19, 85, 96). The significance of the neuroprotective observations reported herein will be substantially clearer if it is known whether DC, administered in vivo, crosses the blood–brain barrier. Addressing such questions about bioavailability, biotransformation, and metabolic interaction with other dietary and pharmaceutical agents has the potential to elevate DC as a highly potent nutraceutical curcuminoid to address neurodegenerative as well as inflammatory disorders.
Acknowledgments
This study was supported in part by NIH grants NS42617 and DK076566.
Abbreviations
C95, 95% extract of curcumin, commercial grade curcumin; C50, 50% extract of curcumin; CXCL10, chemokine (C-X-C motif) ligand 10; CXCL11, chemokine (C-X-C motif) ligand 11; CCL2, chemokine (C-C motif) ligand 2; CCL5, chemokine (C-C motif) ligand 5; DAVID, database for annotation, visualization, and integrated discovery; DC, demethylated derivative of curcumin; DCF, dichlorodihydrofluorescein; GSH, reduced glutathione; H2DCF-DA, dichlorodihydrofluorescein diacetate; IFN, interferon; ISRE, IFN-stimulated response element; I-TAC, IFN-inducible T-cell α chemoattractant; ICAM-1, intercellular adhesion molecule 1; IP, IFN-γ–inducible protein; KEGG, Kyoto encyclopedia of genes and genomes; LDH, lactate dehydrogenase; MCP, monocyte chemoattractant protein; NGF, nerve growth factor; PI3K, phosphatidyl inositol 3-kinase; ROS, reactive oxygen species; TNF-α, tumor necrosis factor alpha; TRAIL, TNF-related apoptosis-inducing ligand; VCAM-1, vascular cell adhesion molecule-1.
Disclosure Statement
No competing financial interests exist.
References
- 1.Aggarwal BB. Sundaram C. Malani N. Ichikawa H. Curcumin: the Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Ak T. Gulcin I. Antioxidant and radical scavenging properties of curcumin. Chem Biol Interact. 2008;174:27–37. doi: 10.1016/j.cbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Anand P. Kunnumakkara AB. Newman RA. Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MF. Nilsson M. Eriksson PS. Sims NR. Glutathione monoethyl ester provides neuroprotection in a rat model of stroke. Neurosci Lett. 2004;354:163–165. doi: 10.1016/j.neulet.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 5.Asija A. Peterson SJ. Stec DE. Abraham NG. Targeting endothelial cells with heme oxygenase-1 gene using VE-cadherin promoter attenuates hyperglycemia-mediated cell injury and apoptosis. Antioxid Redox Signal. 2007;9:2065–2074. doi: 10.1089/ars.2007.1804. [DOI] [PubMed] [Google Scholar]
- 6.Avouac J. Allanore Y. Cardiovascular risk in rheumatoid arthritis: effects of anti-TNF drugs. Expert Opin Pharmacother. 2008;9:1121–1128. doi: 10.1517/14656566.9.7.1121. [DOI] [PubMed] [Google Scholar]
- 7.Bachmeier B. Nerlich AG. Iancu CM. Cilli M. Schleicher E. Vene R. Dell'Eva R. Jochum M. Albini A. Pfeffer U. The chemopreventive polyphenol curcumin prevents hematogenous breast cancer metastases in immunodeficient mice. Cell Physiol Biochem. 2007;19:137–152. doi: 10.1159/000099202. [DOI] [PubMed] [Google Scholar]
- 8.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 9.Bringardner BD. Baran CP. Eubank TD. Marsh CB. The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxid Redox Signal. 2008;10:287–301. doi: 10.1089/ars.2007.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buttmann M. Berberich-Siebelt F. Serfling E. Rieckmann P. Interferon-beta is a potent inducer of interferon regulatory factor-1/2-dependent IP-10/CXCL10 expression in primary human endothelial cells. J Vasc Res. 2007;44:51–60. doi: 10.1159/000097977. [DOI] [PubMed] [Google Scholar]
- 11.Chan MM. Inhibition of tumor necrosis factor by curcumin, a phytochemical. Biochem Pharmacol. 1995;49:1551–1556. doi: 10.1016/0006-2952(95)00171-u. [DOI] [PubMed] [Google Scholar]
- 12.Cheung PF. Wong CK. Ip WK. Lam CW. FAK-mediated activation of ERK for eosinophil migration: a novel mechanism for infection-induced allergic inflammation. Int Immunol. 2008;20:353–363. doi: 10.1093/intimm/dxm146. [DOI] [PubMed] [Google Scholar]
- 13.Ching S. Zhang H. Belevych N. He L. Lai W. Pu XA. Jaeger LB. Chen Q. Quan N. Endothelial-specific knockdown of interleukin-1 (IL-1) type 1 receptor differentially alters CNS responses to IL-1 depending on its route of administration. J Neurosci. 2007;27:10476–10486. doi: 10.1523/JNEUROSCI.3357-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conney AH. Enzyme induction and dietary chemicals as approaches to cancer chemoprevention: the Seventh De-Witt S. Goodman Lecture. Cancer Res. 2003;63:7005–7031. [PubMed] [Google Scholar]
- 15.Corallini F. Rimondi E. Secchiero P. TRAIL and osteoprotegerin: a role in endothelial physiopathology? Front Biosci. 2008;13:135–147. doi: 10.2741/2665. [DOI] [PubMed] [Google Scholar]
- 16.Csiszar A. Wang M. Lakatta EG. Ungvari ZI. Inflammation and endothelial dysfunction during aging: role of NF-κB. J Appl Physiol. 2008;105:1333–1341. doi: 10.1152/japplphysiol.90470.2008. Epub 2008 Jul 3 Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhillon N. Aggarwal BB. Newman RA. Wolff RA. Kunnumakkara AB. Abbruzzese JL. Ng CS. Badmaev V. Kurzrock R. Phase II Trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 18.Dogan RN. Elhofy A. Karpus WJ. Production of CCL2 by central nervous system cells regulates development of murine experimental autoimmune encephalomyelitis through the recruitment of TNF- and iNOS-expressing macrophages and myeloid dendritic cells. J Immunol. 2008;180:7376–7384. doi: 10.4049/jimmunol.180.11.7376. [DOI] [PubMed] [Google Scholar]
- 19.Dringen R. Hirrlinger J. Glutathione pathways in the brain. Biol Chem. 2003;384:505–516. doi: 10.1515/BC.2003.059. [DOI] [PubMed] [Google Scholar]
- 20.Farombi EO. Shrotriya S. Na HK. Kim SH. Surh YJ. Curcumin attenuates dimethylnitrosamine-induced liver injury in rats through Nrf2-mediated induction of heme oxygenase-1. Food Chem Toxicol. 2008;46:1279–1287. doi: 10.1016/j.fct.2007.09.095. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Ruiz C. Fernandez-Checa JC. Redox regulation of hepatocyte apoptosis. J Gastroenterol Hepatol. 2007;22(suppl 1):S38–S42. doi: 10.1111/j.1440-1746.2006.04644.x. [DOI] [PubMed] [Google Scholar]
- 22.Gelderman KA. Hultqvist M. Olsson LM. Bauer K. Pizzolla A. Olofsson P. Holmdahl R. Rheumatoid arthritis: the role of reactive oxygen species in disease development and therapeutic strategies. Antioxid Redox Signal. 2007;9:1541–1567. doi: 10.1089/ars.2007.1569. [DOI] [PubMed] [Google Scholar]
- 23.Glass WG. Hickey MJ. Hardison JL. Liu MT. Manning JE. Lane TE. Antibody targeting of the CC chemokine ligand 5 results in diminished leukocyte infiltration into the central nervous system and reduced neurologic disease in a viral model of multiple sclerosis. J Immunol. 2004;172:4018–4025. doi: 10.4049/jimmunol.172.7.4018. [DOI] [PubMed] [Google Scholar]
- 24.Goel A. Kunnumakkara AB. Aggarwal BB. Curcumin as “Curecumin”: from kitchen to clinic. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Gorbunov NV. Das DK. Goswami SK. Gurusamy N. Atkins JL. Spatial coordination of cell-adhesion molecules and redox cycling of iron in the microvascular inflammatory response to pulmonary injury. Antioxid Redox Signal. 2007;9:483–495. doi: 10.1089/ars.2006.1296. [DOI] [PubMed] [Google Scholar]
- 26.Gorcyznski RM. Chung S. Fu XM. Levy G. Sullivan B. Chen Z. Manipulation of skin graft rejection in alloimmune mice by anti-VCAM-1:VLA-4 but not anti-ICAM-1:LFA-1 monoclonal antibodies. Transpl Immunol. 1995;3:55–61. doi: 10.1016/0966-3274(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 27.Gosk S. Moos T. Gottstein C. Bendas G. VCAM-1 directed immunoliposomes selectively target tumor vasculature in vivo. Biochim Biophys Acta. 2008;1778:854–863. doi: 10.1016/j.bbamem.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Gottardo L. De Cosmo S. Zhang YY. Powers C. Prudente S. Marescotti MC. Trischitta V. Avogaro A. Doria A. A polymorphism at the IL6ST (gp130) locus is associated with traits of the metabolic syndrome. Obesity (Silver Spring) 2008;16:205–210. doi: 10.1038/oby.2007.28. [DOI] [PubMed] [Google Scholar]
- 29.Han D. Sen CK. Roy S. Kobayashi MS. Tritschler HJ. Packer L. Protection against glutamate-induced cytotoxicity in C6 glial cells by thiol antioxidants. Am J Physiol. 1997;273:R1771–R1778. doi: 10.1152/ajpregu.1997.273.5.R1771. [DOI] [PubMed] [Google Scholar]
- 30.Hatcher H. Planalp R. Cho J. Torti FM. Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayeshi R. Mutingwende I. Mavengere W. Masiyanise V. Mukanganyama S. The inhibition of human glutathione S-transferases activity by plant polyphenolic compounds ellagic acid and curcumin. Food Chem Toxicol. 2007;45:286–295. doi: 10.1016/j.fct.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Ho AW. Wong CK. Lam CW. Tumor necrosis factoralpha up-regulates the expression of CCL2 and adhesion molecules of human proximal tubular epithelial cells through MAPK signaling pathways. Immunobiology. 2008;213:533–544. doi: 10.1016/j.imbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Hsu CH. Cheng AL. Clinical studies with curcumin. Adv Exp Med Biol. 2007;595:471–480. doi: 10.1007/978-0-387-46401-5_21. [DOI] [PubMed] [Google Scholar]
- 34.Huang J. Upadhyay UM. Tamargo RJ. Inflammation in stroke and focal cerebral ischemia. Surg Neurol. 2006;66:232–245. doi: 10.1016/j.surneu.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Huang MT. Ma W. Lu YP. Chang RL. Fisher C. Manchand PS. Newmark HL. Conney AH. Effects of curcumin, demethoxycurcumin, bisdemethoxycurcumin and tetrahydrocurcumin on 12-O-tetradecanoylphorbol-13-acetate-induced tumor promotion. Carcinogenesis. 1995;16:2493–2497. doi: 10.1093/carcin/16.10.2493. [DOI] [PubMed] [Google Scholar]
- 36.Indraccolo S. Pfeffer U. Minuzzo S. Esposito G. Roni V. Mandruzzato S. Ferrari N. Anfosso L. Dell'Eva R. Noonan DM. Chieco-Bianchi L. Albini A. Amadori A. Identification of genes selectively regulated by IFNs in endothelial cells. J Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 37.Jacob A. Wu R. Zhou M. Wang P. Mechanism of the anti-inflammatory effect of curcumin: PPAR-gamma activation. PPAR Res. 2007;2007:89369. doi: 10.1155/2007/89369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H. Somogyi A. Jacobsen NE. Timmermann BN. Gang DR. Analysis of curcuminoids by positive and negative electrospray ionization and tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1001–1012. doi: 10.1002/rcm.2401. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H. Timmermann BN. Gang DR. Use of liquid chromatography-electrospray ionization tandem mass spectrometry to identify diarylheptanoids in turmeric (Curcuma longa L.) rhizome. J Chromatogr A. 2006;1111:21–31. doi: 10.1016/j.chroma.2006.01.103. [DOI] [PubMed] [Google Scholar]
- 40.Khanna S. Roy S. Parinandi NL. Maurer M. Sen CK. Characterization of the potent neuroprotective properties of the natural vitamin E alpha-tocotrienol. J Neurochem. 2006;98:1474–1486. doi: 10.1111/j.1471-4159.2006.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khanna S. Roy S. Park HA. Sen CK. Regulation of c-Src activity in glutamate-induced neurodegeneration. J Biol Chem. 2007;282:23482–23490. doi: 10.1074/jbc.M611269200. [DOI] [PubMed] [Google Scholar]
- 42.Khanna S. Roy S. Ryu H. Bahadduri P. Swaan PW. Ratan RR. Sen CK. Molecular basis of vitamin E action: tocotrienol modulates 12-lipoxygenase, a key mediator of glutamate-induced neurodegeneration. J Biol Chem. 2003;278:43508–43515. doi: 10.1074/jbc.M307075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanna S. Roy S. Slivka A. Craft TK. Chaki S. Rink C. Notestine MA. DeVries AC. Parinandi NL. Sen CK. Neuroprotective properties of the natural vitamin E alphatocotrienol. Stroke. 2005;36:2258–2264. doi: 10.1161/01.STR.0000181082.70763.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YS. Ahn Y. Hong MH. Joo SY. Kim KH. Sohn IS. Park HW. Hong YJ. Kim JH. Kim W. Jeong MH. Cho JG. Park JC. Kang JC. Curcumin attenuates inflammatory responses of TNF-alpha-stimulated human endothelial cells. J Cardiovasc Pharmacol. 2007;50:41–49. doi: 10.1097/FJC.0b013e31805559b9. [DOI] [PubMed] [Google Scholar]
- 45.Kunnumakkara AB. Anand P. Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. Epub 2008 May 13. Review. [DOI] [PubMed] [Google Scholar]
- 46.Kurup VP. Barrios CS. Immunomodulatory effects of curcumin in allergy. Mol Nutr Food Res. 2008;52:1031–1039. doi: 10.1002/mnfr.200700293. [DOI] [PubMed] [Google Scholar]
- 47.Laurindo FR. Fernandes DC. Amanso AM. Lopes LR. Santos CX. Novel role of protein disulfide isomerase in the regulation of NADPH oxidase activity: pathophysiological implications in vascular diseases. Antioxid Redox Signal. 2008;10:1101–1113. doi: 10.1089/ars.2007.2011. [DOI] [PubMed] [Google Scholar]
- 48.Liang G. Yang S. Zhou H. Shao L. Huang K. Xiao J. Huang Z. Li X. Synthesis, crystal structure and anti-inflammatory properties of curcumin analogues. Eur J Med Chem. 2008 Feb 3; doi: 10.1016/j.ejmech.2008.01.031. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 49.Liu RM. Oxidative stress, plasminogen activator inhibitor 1, and lung fibrosis. Antioxid Redox Signal. 2008;10:303–319. doi: 10.1089/ars.2007.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Lazaro M. Anticancer and carcinogenic properties of curcumin: considerations for its clinical development as a cancer chemopreventive and chemotherapeutic agent. Mol Nutr Food Res. 2008;52(suppl 1):S103–S127. doi: 10.1002/mnfr.200700238. [DOI] [PubMed] [Google Scholar]
- 51.Mantovani A. Marchesi F. Portal C. Allavena P. Sica A. Linking inflammation reactions to cancer: novel targets for therapeutic strategies. Adv Exp Med Biol. 2008;610:112–127. doi: 10.1007/978-0-387-73898-7_9. [DOI] [PubMed] [Google Scholar]
- 52.Mashaly HA. Provencio JJ. Inflammation as a link between brain injury and heart damage: the model of subarachnoid hemorrhage. Cleve Clin J Med. 2008;75(suppl 2):S26–S30. doi: 10.3949/ccjm.75.suppl_2.s26. [DOI] [PubMed] [Google Scholar]
- 53.Mata M. Hao S. Fink DJ. Gene therapy directed at the neuroimmune component of chronic pain with particular attention to the role of TNF alpha. Neurosci Lett. 2008;437:209–213. doi: 10.1016/j.neulet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mennen LI. Walker R. Bennetau-Pelissero C. Scalbert A. Risks and safety of polyphenol consumption. Am J Clin Nutr. 2005;81:326S–329S. doi: 10.1093/ajcn/81.1.326S. [DOI] [PubMed] [Google Scholar]
- 55.Menon VP. Sudheer AR. Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol. 2007;595:105–125. doi: 10.1007/978-0-387-46401-5_3. [DOI] [PubMed] [Google Scholar]
- 56.Mon NN. Ito S. Senga T. Hamaguchi M. FAK signaling in neoplastic disorders: a linkage between inflammation and cancer. Ann N Y Acad Sci. 2006;1086:199–212. doi: 10.1196/annals.1377.019. [DOI] [PubMed] [Google Scholar]
- 57.Monteiro HP. Arai RJ. Travassos LR. Protein tyrosine phosphorylation and protein tyrosine nitration in redox signaling. Antioxid Redox Signal. 2008;10:843–889. doi: 10.1089/ars.2007.1853. [DOI] [PubMed] [Google Scholar]
- 58.Muro S. Garnacho C. Champion JA. Leferovich J. Gajewski C. Schuchman EH. Mitragotri S. Muzykantov VR. Control of endothelial targeting and intracellular delivery of therapeutic enzymes by modulating the size and shape of ICAM-1-targeted carriers. Mol Ther. 2008;16:1450–1458. doi: 10.1038/mt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakayama R. Tamura Y. Yamanaka H. Kikuzaki H. Nakatani N. Curcuminoid pigments from Curcuma domestica. Phytochemistry. 1993;33:501–502. [Google Scholar]
- 60.Navab M. Gharavi N. Watson AD. Inflammation and metabolic disorders. Curr Opin Clin Nutr Metab Care. 2008;11:459–464. doi: 10.1097/MCO.0b013e32830460c2. [DOI] [PubMed] [Google Scholar]
- 61.Nico B. Mangieri D. Benagiano V. Crivellato E. Ribatti D. Nerve growth factor as an angiogenic factor. Microvasc Res. 2008;75:135–141. doi: 10.1016/j.mvr.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Nishikawa T. Araki E. Impact of mitochondrial ROS production in the pathogenesis of diabetes mellitus and its complications. Antioxid Redox Signal. 2007;9:343–353. doi: 10.1089/ars.2006.1458. [DOI] [PubMed] [Google Scholar]
- 63.Ozer K. Siemionow M. Combination of anti-ICAM-1 and anti-LFA-1 monoclonal antibody therapy prolongs allograft survival in rat hind-limb transplants. J Reconstr Microsurg. 2001;17:511–517. doi: 10.1055/s-2001-17753. discussion 518. [DOI] [PubMed] [Google Scholar]
- 64.Papatheodorou L. Weiss N. Vascular oxidant stress and inflammation in hyperhomocysteinemia. Antioxid Redox Signal. 2007;9:1941–1958. doi: 10.1089/ars.2007.1750. [DOI] [PubMed] [Google Scholar]
- 65.Pease JE. Sabroe I. The role of interleukin-8 and its receptors in inflammatory lung disease: implications for therapy. Am J Respir Med. 2002;1:19–25. doi: 10.1007/BF03257159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Putics A. Vegh EM. Csermely P. Soti C. Resveratrol induces the heat-shock response and protects human cells from severe heat stress. Antioxid Redox Signal. 2008;10:65–75. doi: 10.1089/ars.2007.1866. [DOI] [PubMed] [Google Scholar]
- 67.Rajan S. Ye J. Bai S. Huang F. Guo YL. NF-kappaB, but not p38 MAP Kinase, is required for TNF-alpha-induced expression of cell adhesion molecules in endothelial cells. J Cell Biochem. 2008;105:477–486. doi: 10.1002/jcb.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rink C. Roy S. Khanna S. Rink T. Bagchi D. Sen CK. Transcriptome of the subcutaneous adipose tissue in response to oral supplementation of type 2 Leprdb obese diabetic mice with niacin-bound chromium. Physiol Genomics. 2006;27:370–379. doi: 10.1152/physiolgenomics.00071.2006. [DOI] [PubMed] [Google Scholar]
- 69.Romer LH. Birukov KG. Garcia JG. Focal adhesions: paradigm for a signaling nexus. Circ Res. 2006;98:606–616. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- 70.Roy S. Khanna S. Kuhn DE. Rink C. Williams WT. Zweier JL. Sen CK. Transcriptome analysis of the ischemiareperfused remodeling myocardium: temporal changes in inflammation and extracellular matrix. Physiol Genomics. 2006;25:364–374. doi: 10.1152/physiolgenomics.00013.2006. [DOI] [PubMed] [Google Scholar]
- 71.Roy S. Khanna S. Rink C. Biswas S. Sen CK. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics. 2008;34:162–184. doi: 10.1152/physiolgenomics.00045.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy S. Khanna S. Shah H. Rink C. Phillips C. Preuss H. Subbaraju GV. Trimurtulu G. Krishnaraju AV. Bagchi M. Bagchi D. Sen CK. Human genome screen to identify the genetic basis of the anti-inflammatory effects of Boswellia in microvascular endothelial cells. DNA Cell Biol. 2005;24:244–255. doi: 10.1089/dna.2005.24.244. [DOI] [PubMed] [Google Scholar]
- 73.Roy S. Patel D. Khanna S. Gordillo GM. Biswas S. Friedman A. Sen CK. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci U S A. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roy S. Shah H. Rink C. Khanna S. Bagchi D. Bagchi M. Sen CK. Transcriptome of primary adipocytes from obese women in response to a novel hydroxycitric acid-based dietary supplement. DNA Cell Biol. 2007;26:627–639. doi: 10.1089/dna.2007.0617. [DOI] [PubMed] [Google Scholar]
- 75.Roy S. Venojarvi M. Khanna S. Sen CK. Simultaneous detection of tocopherols and tocotrienols in biological samples using HPLC-coulometric electrode array. Methods Enzymol. 2002;352:326–332. doi: 10.1016/s0076-6879(02)52029-2. [DOI] [PubMed] [Google Scholar]
- 76.Seguin C. Abid MR. Spokes KC. Schoots IG. Brkovic A. Sirois MG. Aird WC. Priming effect of homocysteine on inducible vascular cell adhesion molecule-1 expression in endothelial cells. Biomed Pharmacother. 2008;62:395–400. doi: 10.1016/j.biopha.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen CK. Khanna S. Babior BM. Hunt TK. Ellison EC. Roy S. Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J Biol Chem. 2002;277:33284–33290. doi: 10.1074/jbc.M203391200. [DOI] [PubMed] [Google Scholar]
- 78.Sen CK. Khanna S. Roy S. Tocotrienol: the natural vitamin E to defend the nervous system? Ann N Y Acad Sci. 2004;1031:127–142. doi: 10.1196/annals.1331.013. [DOI] [PubMed] [Google Scholar]
- 79.Sen CK. Khanna S. Roy S. Packer L. Molecular basis of vitamin E action: tocotrienol potently inhibits glutamateinduced pp60(c-Src) kinase activation and death of HT4 neuronal cells. J Biol Chem. 2000;275:13049–13055. doi: 10.1074/jbc.275.17.13049. [DOI] [PubMed] [Google Scholar]
- 80.Sethi G. Sung B. Aggarwal BB. TNF: a master switch for inflammation to cancer. Front Biosci. 2008;13:5094–5107. doi: 10.2741/3066. [DOI] [PubMed] [Google Scholar]
- 81.Shankar S. Srivastava RK. Bax and Bak genes are essential for maximum apoptotic response by curcumin, a polyphenolic compound and cancer chemopreventive agent derived from turmeric, Curcuma longa. Carcinogenesis. 2007;28:1277–1286. doi: 10.1093/carcin/bgm024. [DOI] [PubMed] [Google Scholar]
- 82.Shealy DJ. Visvanathan S. Anti-TNF antibodies: lessons from the past, roadmap for the future. Handb Exp Pharmacol. 2008;181:101–129. doi: 10.1007/978-3-540-73259-4_5. Review. [DOI] [PubMed] [Google Scholar]
- 83.Sheng WS. Hu S. Ni HT. Rowen TN. Lokensgard JR. Peterson PK. TNF-alpha-induced chemokine production and apoptosis in human neural precursor cells. J Leukoc Biol. 2005;78:1233–1241. doi: 10.1189/jlb.0405221. [DOI] [PubMed] [Google Scholar]
- 84.Shukla PK. Khanna VK. Ali MM. Khan MY. Srimal RC. Anti-ischemic effect of curcumin in rat brain. Neurochem Res. 2008;33:1036–1043. doi: 10.1007/s11064-007-9547-y. [DOI] [PubMed] [Google Scholar]
- 85.Sims NR. Nilsson M. Muyderman H. Mitochondrial glutathione: a modulator of brain cell death. J Bioenerg Biomembr. 2004;36:329–333. doi: 10.1023/B:JOBB.0000041763.63958.e7. [DOI] [PubMed] [Google Scholar]
- 86.Sluiter W. Pietersma A. Lamers JM. Koster JF. Leukocyte adhesion molecules on the vascular endothelium: their role in the pathogenesis of cardiovascular disease and the mechanisms underlying their expression. J Cardiovasc Pharmacol. 1993;22(suppl 4):S37–S44. [PubMed] [Google Scholar]
- 87.Smith PF. Inflammation in Parkinson's disease: an update. Curr Opin Invest Drugs. 2008;9:478–484. [PubMed] [Google Scholar]
- 88.Strasser EM. Wessner B. Manhart N. Roth E. The relationship between the anti-inflammatory effects of curcumin and cellular glutathione content in myelomonocytic cells. Biochem Pharmacol. 2005;70:552–559. doi: 10.1016/j.bcp.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 89.Strimpakos AS. Sharma RA. Curcumin: preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid Redox Signal. 2008;10:511–545. doi: 10.1089/ars.2007.1769. [DOI] [PubMed] [Google Scholar]
- 90.Talvani A. Rocha MO. Barcelos LS. Gomes YM. Ribeiro AL. Teixeira MM. Elevated concentrations of CCL2 and tumor necrosis factor-alpha in chagasic cardiomyopathy. Clin Infect Dis. 2004;38:943–950. doi: 10.1086/381892. [DOI] [PubMed] [Google Scholar]
- 91.Tamvakopoulos C. Dimas K. Sofianos ZD. Hatziantoniou S. Han Z. Liu ZL. Wyche JH. Pantazis P. Metabolism and anticancer activity of the curcumin analogue, dimethoxycurcumin. Clin Cancer Res. 2007;13:1269–1277. doi: 10.1158/1078-0432.CCR-06-1839. [DOI] [PubMed] [Google Scholar]
- 92.Tensen CP. Flier J. Rampersad SS. Sampat-Sardjoepersad S. Scheper RJ. Boorsma DM. Willemze R. Genomic organization, sequence and transcriptional regulation of the human CXCL 11(1) gene. Biochim Biophys Acta. 1999;1446:167–172. doi: 10.1016/s0167-4781(99)00084-6. [DOI] [PubMed] [Google Scholar]
- 93.Tirosh O. Sen CK. Roy S. Packer L. Cellular and mitochondrial changes in glutamate-induced HT4 neuronal cell death. Neuroscience. 2000;97:531–541. doi: 10.1016/s0306-4522(00)00028-2. [DOI] [PubMed] [Google Scholar]
- 94.Toft-Hansen H. Buist R. Sun XJ. Schellenberg A. Peeling J. Owens T. Metalloproteinases control brain inflammation induced by pertussis toxin in mice overexpressing the chemokine CCL2 in the central nervous system. J Immunol. 2006;177:7242–7249. doi: 10.4049/jimmunol.177.10.7242. [DOI] [PubMed] [Google Scholar]
- 95.Ushio-Fukai M. VEGF signaling through NADPH oxidasederived ROS. Antioxid Redox Signal. 2007;9:731–739. doi: 10.1089/ars.2007.1556. [DOI] [PubMed] [Google Scholar]
- 96.Vargas MR. Pehar M. Cassina P. Beckman JS. Barbeito L. Increased glutathione biosynthesis by Nrf2 activation in astrocytes prevents p75NTR-dependent motor neuron apoptosis. J Neurochem. 2006;97:687–696. doi: 10.1111/j.1471-4159.2006.03742.x. [DOI] [PubMed] [Google Scholar]
- 97.Vasto S. Carruba G. Lio D. Colonna-Romano G. Di Bona D. Candore G. Caruso C. Inflammation, ageing and cancer. Mech Ageing Dev. 2008 Jul 10; doi: 10.1016/j.mad.2008.06.003. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 98.Venkatesan P. Rao MN. Structure-activity relationships for the inhibition of lipid peroxidation and the scavenging of free radicals by synthetic symmetrical curcumin analogues. J Pharm Pharmacol. 2000;52:1123–1128. doi: 10.1211/0022357001774886. [DOI] [PubMed] [Google Scholar]
- 99.Verducci JS. Melfi VF. Lin S. Wang Z. Roy S. Sen CK. Microarray analysis of gene expression: considerations in data mining and statistical treatment. Physiol Genomics. 2006;25:355–363. doi: 10.1152/physiolgenomics.00314.2004. [DOI] [PubMed] [Google Scholar]
- 100.Watson T. Goon PK. Lip GY. Endothelial progenitor cells, endothelial dysfunction, inflammation, and oxidative stress in hypertension. Antioxid Redox Signal. 2008;10:1079–1088. doi: 10.1089/ars.2007.1998. [DOI] [PubMed] [Google Scholar]
- 101.Weber WM. Hunsaker LA. Gonzales AM. Heynekamp JJ. Orlando RA. Deck LM. Vander Jagt DL. TPA-induced up-regulation of activator protein-1 can be inhibited or enhanced by analogs of the natural product curcumin. Biochem Pharmacol. 2006;72:928–940. doi: 10.1016/j.bcp.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 102.Wijtmans M. Verzijl D. Leurs R. de Esch IJ. Smit MJ. Towards small-molecule CXCR3 ligands with clinical potential. Chem Med Chem. 2008;3:861–872. doi: 10.1002/cmdc.200700365. [DOI] [PubMed] [Google Scholar]
- 103.Xing L. Remick DG. Promoter elements responsible for antioxidant regulation of MCP-1 gene expression. Antioxid Redox Signal. 2007;9:1979–1989. doi: 10.1089/ars.2007.1921. [DOI] [PubMed] [Google Scholar]
- 104.Yang CS. Sang S. Lambert JD. Lee MJ. Bioavailability issues in studying the health effects of plant polyphenolic compounds. Mol Nutr Food Res. 2008;52(suppl 1):S139–S151. doi: 10.1002/mnfr.200700234. [DOI] [PubMed] [Google Scholar]
- 105.Yaoita H. Maruyama Y. Intervention for apoptosis in cardiomyopathy. Heart Fail Rev. 2008;13:181–191. doi: 10.1007/s10741-007-9074-6. [DOI] [PubMed] [Google Scholar]
- 106.Yuan A. Chen JJ. Yao PL. Yang PC. The role of interleukin-8 in cancer cells and microenvironment interaction. Front Biosci. 2005;10:853–865. doi: 10.2741/1579. [DOI] [PubMed] [Google Scholar]
- 107.Yuan H. Gaber MW. McColgan T. Naimark MD. Kiani MF. Merchant TE. Radiation-induced permeability and leukocyte adhesion in the rat blood-brain barrier: modulation with anti-ICAM-1 antibodies. Brain Res. 2003;969:59–69. doi: 10.1016/s0006-8993(03)02278-9. [DOI] [PubMed] [Google Scholar]
- 108.Zhou L. Opalinska J. Verma A. p38 MAP kinase regulates stem cell apoptosis in human hematopoietic failure. Cell Cycle. 2007;6:534–537. doi: 10.4161/cc.6.5.3921. [DOI] [PubMed] [Google Scholar]
- 109.Zhou Z. Connell MC. MacEwan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal. 2007;19:1238–1248. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]