Abstract
In the vertebrate nervous system, the myelin sheath allows for rapid and efficient conduction of action potentials along axons. Despite the essential function of myelin, many questions remain unanswered about the mechanisms that govern the development of myelinated axons. The fundamental properties of myelin are widely shared among vertebrates, and the zebrafish has emerged as a powerful system to study myelination in vivo. This review will highlight recent advances from genetic screens in zebrafish, including the discovery of the role of kif1b in mRNA localization in myelinating oligodendrocytes.
Introduction
Myelin is a multilayered membrane formed by the wrapping of glial cells around axons that allows for efficient conduction of action potentials in the vertebrate nervous system [1,2]. In the central nervous system (CNS), oligodendrocytes form myelin, whereas Schwann cells myelinate axons in the peripheral nervous system (PNS). Myelinating glial cells are critical for the proper function of the vertebrate nervous system, not only in allowing rapid propagation of action potentials, but also in providing axons with trophic support [2]. At the gaps between adjacent myelin segments are the nodes of Ranvier, which contain abundant voltage-gated sodium channels. These channels propagate action potentials, and the specific localization of channels at the nodes is required for saltatory conduction in myelinated axons [3,4]. The importance of myelinating glia is underscored by human disorders in which myelinated axons are disrupted, including multiple sclerosis (MS) and Charcot-Marie-Tooth disease [5,6]. Studies in human patients and mammalian model organisms have identified key genes involved in myelination [6,7], but important questions about the genetic control of glial development and differentiation remain unanswered. The goal of this review is to summarize recent advances from genetic studies of myelination in zebrafish.
Zebrafish as a model organism to study myelination
In the last two decades, the zebrafish has become a well-established model system for the study of vertebrate biology [8]. The embryos are large, externally developing, and translucent, which is advantageous for timelapse imaging, cellular transplantation, and microinjection. The ability to perform timelapse imaging is particularly advantageous for the investigation of dynamic interactions between cells that occur during glial development, and several studies have exploited this property to examine glial migration and behavior in zebrafish [9–11]. Additionally, the zebrafish is fast-growing and fecund, which facilitates genetic screens. These attributes have also allowed zebrafish embryos and larvae to be used to screen for small molecules affecting development or behavior in a whole animal [12]. In light of these experimental advantages, the zebrafish has emerged as a model to study glial cell development and myelination in vivo [9,10,13–15].
As in mammals, oligodendrocytes and Schwann cells form myelin in the zebrafish CNS and PNS, respectively [9,10,13]. Additionally, nodes of Ranvier have similar protein composition and ultrastructure in mammals and zebrafish [16,17]. Biochemical studies have described both similarities and differences between zebrafish and mammalian myelin. For example, Myelin basic protein (Mbp) and myelin proteolipid protein (PLP/DM20) are components of myelin in fish and mammals, but there are also teleost-specific myelin proteins in the CNS including 36K, Zwilling-A, and Zwilling-B [13,18–20]. Myelin Protein Zero (P0), an Ig-like protein, is present in peripheral myelin of both tetrapods and teleosts, but only in fish is this protein prominently expressed in the CNS [13,21–23]. X-ray diffraction studies have shown that the structure of myelin is fundamentally similar in zebrafish and mammals, but that the periodicity of myelin varies slightly [23]. Thus, despite these differences, the properties of vertebrate myelin are overall highly conserved.
Many genes appear to have conserved functions in glial development of zebrafish and mammals. For example, Olig1, Olig2, and Sox10 act in oligodendrocyte development in zebrafish and mouse [24,25]. Likewise, Sox 10, ErbB2/ErbB3, Oct6, and Krox20 act in Schwann cells of both species [7,9,14,26]. These similarities provide evidence that genes identified by genetic approaches in zebrafish will also have essential functions in glial development and myelination in mammals.
Mutational screens for genes with essential function in glial development and myelination
In the mid-1990s, large-scale genetic screens in zebrafish identified mutations in more than 1000 genes that disrupt the morphology of the embryo and early larva [27,28]. These mutations fueled advances in many areas, including the genetic control of axis formation, gastrulation, and organogenesis. Because these initial screens identified mutants with readily observable morphological defects, many of the mutations disrupt global patterning of the embryo or otherwise affect the development of numerous different cell types. Nonetheless, examination of mbp expression in 39 previously identified mutations identified four mutants in which axons were present but mbp expression was reduced in the posterior lateral line nerve (PLLn, a prominent sensory nerve in zebrafish; [29]). Further analysis of mutations in the aldh1a2 gene supported a role for retinoic acid signaling at an early step of development essential for the eventual differentiation of myelinating glia [29].
In another approach, two complementary screens were performed to find new mutations that affect the development of myelinated axons. In the first of these, Pogoda et al. [14] screened more than 1850 clutches of F3 larvae for mutations with specific defects in the expression of mbp mRNA. This screen identified 13 mutations in 10 genes that disrupted mbp expression in the CNS, in the PNS, or in both [14]. A second smaller screen expanded the scope of phenotypes examined by assaying sodium channel (NaCh) expression at nodes of Ranvier in myelinated axons [17]. It is important to note that neither screen was pursued to saturation, so that continued screening will surely yield mutations in genes that have not yet been defined. Most of the mutants have normal morphology, and were therefore not uncovered in previous large-scale screens. The absence of obvious morphological defects suggests that the mutated genes have relatively specific functions in the development of myelinating glia and the associated axons; this also implies that the screens overlooked genes with interesting but more general functions in myelination and other processes. As summarized in Table 1, the available phenotypic studies indicate that the mutated genes act at many different steps in the development of myelinated axons, including glial fate specification, axon outgrowth, Schwann cell migration, and organization of the nodes of Ranvier. In addition, at least two of the genes have been associated with human disease, suggesting that analysis of these zebrafish mutants may yield important insights into diseases that disrupt myelinated axons.
Table 1.
Mutated genes identified in screens for abnormal mbp expression and NaCh localization
| Mutant allele | Gene (if reported) |
Functions | Screen | Reference |
|---|---|---|---|---|
| st50, st61 | erbb2 | SC proliferation, migration, myelination | Both | [9] |
| st14, st48, st71 | erbb3 | SC proliferation, migration, myelination | Both | [9] |
| st20 | foxa2 | Development of midline OL precursors | mbp | [30] |
| st25, st53 | nsf | Forming NaCh clusters at nodes | mbp | [16] |
| st60 | aII-spectrin | Stabilizing NaCh clusters at nodes | NaCh | [17] |
| st23 | kbp* | Axon outgrowth in PNS and CNS | mbp | [31] |
| st43 | kif1b* | Axon outgrowth, CNS: mRNA localization | mbp | [32] |
| st49, st63 | Unpubl. | SC development | mbp | [14] |
| st51 | Unpubl. | Development of hindbrain OL | mbp | [14] |
| st64 | Unpubl. | SC development | mbp | [14] |
| st67 | Unpubl. | Myelin morphogenesis | NaCh | Unpubl. |
SC: Schwann cell; OL: oligodendrocyte; NaCh: sodium channel
Associated with human disease
Defining the functions of mutated genes at the molecular and cellular level
Recent studies have provided insight into the cellular and biochemical functions of several of the genes defined by the mutational screens discussed above [9,16,17,30–32]. In the following section, we discuss three examples of mutational studies in zebrafish that have identified genes that function in glial development and myelination, and describe how these studies have contributed new insights.
ErbB signaling
ErbB2/ErbB3 receptors and their Neuregulin ligands are critical for many steps in Schwann cell development including proliferation, ensheathment, and myelination [33]. Accordingly, the mbp and NaCh screens recovered mutations in erbb2 and erbb3 [9,14]. As in mammals [33], erbb2 and erbb3 mutant zebrafish lack Schwann cells in peripheral nerves. Phenotypic studies of the mutants showed that ErbB signaling is required for Schwann cell proliferation in vivo, supporting previous work in mammals [34]. Time-lapse imaging of the erbb mutants and embryos treated with ErbB inhibitors demonstrated that ErbB signaling is essential for directed migration of Schwann cells in growing nerves [9]. Thus, analysis of these mutants not only underscored the fundamental similarities between zebrafish and mammalian PNS myelination, but also provided new insight into a role for ErbB signaling during Schwann cell migration.
αII-spectrin
In myelinated axons, the cytoskeletal protein αII-spectrin is localized at paranodes, sites of axoglial adhesion immediately adjacent to the nodes of Ranvier [35,36]. The screen for mutants with aberrant sodium channel clustering at nodes of Ranvier uncovered a mutant allele of αII-spectrin, providing an opportunity to examine its function in vivo [17]. Mutational studies showed that αII-spectrin is essential to stabilize nascent sodium channel clusters in developing axons and to assemble mature nodes of the correct dimensions. These functional studies, together with previous biochemical analyses showing that αII-spectrin forms a complex with βII-spectrin and Ankyin B [35], have added important understanding to role of these cytoskeletal proteins in the organization of myelinated axons.
Kif1b
In the CNS of mammals and zebrafish, mRNAs encoding Mbp and a small number of other structural components of myelin are localized to processes of myelinating oligodendrocytes (Figure 1 [13,37]). Analyses in cultured cells have implicated microtubules and kinesin motors in the transport of mbp mRNA [38–40], but genes required for endogenous mRNA localization in vivo had not been identified until recently. Furthermore, the role of this specific mRNA localization process in oligodendrocyte differentiation was not understood.
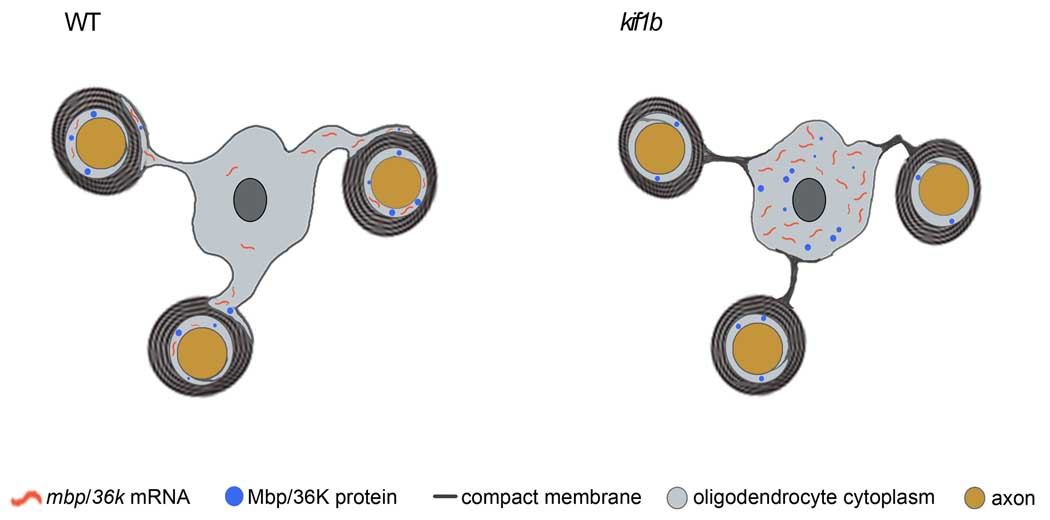
Figure 1. A model of defects in the CNS of kif1b zebrafish mutants.
In WT zebrafish, axons (orange) are surrounded by compact myelin. Mbp and 36K mRNA and protein are localized to the uncompacted region of the myelinating process (gray), but are largely excluded from the cell body. In kif1b mutant zebrafish, mbp and 36k mRNA are not transported into myelinating process, although proteins are localized to these regions. Mbp and 36K protein are aberrantly localized in oligodendrocyte cell body, coincident with mislocalization of compacted, myelin-like membrane. Adapted from [32].
Lyons et al. recently reported that mutations in kif1b, which encodes a member of the kinesin motor superfamily, cause defects in mbp mRNA localization in oligodendrocytes and disruptions of the outgrowth on long axons in the spinal cord and PNS [32]. Analysis of genetic chimeras indicated that kif1b is required in neurons for axon outgrowth, perhaps to transport key cargo to the distal region of rapidly growing axons.
In the CNS, kif1b mutants display a striking phenotype: mbp mRNA is present in oligodendrocyte cell bodies, but not in myelinating processes [32]. Analysis of genetic chimeras indicated that kif1b is required autonomously in oligodendrocytes for proper localization of mbp mRNA. Kif1b mutant oligodendrocytes form myelinating processes, and Mbp protein is present in these processes despite the lack of mbp mRNA. The mRNA for another myelin protein, 36K, is also localized to oligodendrocyte processes in wildtype zebrafish [19], but not in kif1b mutants. These results indicate that Kif1b is essential to localize specific mRNAs within myelinating processes. Future studies will examine if the Kif1b motor acts directly to transport mRNA cargo into myelinating processes.
The finding that mbp mRNA is mislocalized in kif1b mutants provided the opportunity to investigate the role of mRNA localization in the differentiation of oligodendrocytes. Ultrastructural analyses showed that myelin forms in the CNS of kif1b mutants, although the number of myelinated axons and the thickness of the myelin are somewhat reduced [32]. The most striking observation from the ultrastructural studies was that that kif1b mutants possess marked ectopic myelin-like membrane compaction throughout the anterior spinal cord. Long stretches of compact myelin-like membranes were observed surrounding neuronal cell bodies and in processes that did not ensheath axons. Mbp and 36K protein are largely restricted to myelin in wild-type oligodendrocytes, but these proteins are present throughout the oligodendrocytes of kif1b mutants at stages when ectopic myelin-like membrane is evident [32]. These studies suggest that mislocalized myelin proteins contribute to aberrant membrane compaction in kif1b mutants. According to this model (Figure 1), the role of mRNA localization in oligodendrocytes is to restrict the localization of Mbp, 36K and some other myelin proteins to the myelin itself, thereby preventing the deleterious action of these proteins in other parts of the cell. An additional implication is that the subset of localized mRNAs encodes the proteins that bring about myelin membrane compaction. Interestingly, in a recent genome-wide association study, KIF1B was associated with susceptibility to MS [41]. Thus, the discovery of a function for Kif1b in oligodendrocytes may shed some light on its possible roles in MS, and the mutant zebrafish may represent a useful tool to explore the pathophysiology of MS in the future.
Conclusions
Mutational screens in zebrafish have begun to contribute to the genetic dissection of myelination in vertebrates. The cellular and molecular analysis of zebrafish mutations has revealed new insight into the functions of genes essential for glial development and myelination. Additional discoveries are expected, as existing mutants are characterized and new screens recover mutations in genes that have not yet been identified. Additionally, zebrafish myelination mutations could be utilized in future small molecule screens for chemicals that rescue or exacerbate phenotypes. These studies may further our understanding of pathways involved in diseases of myelinated axons and speed the development of human treatments.
Acknowledgements
We thank David Lyons and members of our laboratory for critical reading of this manuscript. KRM is supported by a postdoctoral award from the NMSS (FG 1719-A-1), and our research has been supported by grants from the NIH, NMSS, and MDA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kelly R. Monk, Email: kmonk@stanford.edu.
William S. Talbot, Email: wtalbot@stanford.edu.
References
- 1.Sherman DL, Brophy PJ. Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci. 2005;6:683–690. doi: 10.1038/nrn1743. [DOI] [PubMed] [Google Scholar]
- 2.Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- 3.Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nat Rev Neurosci. 2003;4:968–980. doi: 10.1038/nrn1253. [DOI] [PubMed] [Google Scholar]
- 4.Salzer JL. Polarized domains of myelinated axons. Neuron. 2003;40:297–318. doi: 10.1016/s0896-6273(03)00628-7. [DOI] [PubMed] [Google Scholar]
- 5.Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 2008;31:247–269. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- 6.Suter U, Scherer SS. Disease mechanisms in inherited neuropathies. Nature Reviews Neuroscience. 2003;4:714–726. doi: 10.1038/nrn1196. [DOI] [PubMed] [Google Scholar]
- 7.Jessen KR, Mirsky RM. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 8.Grunwald DJ, Eisen JS. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 9.Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for Schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- 11.Gilmour DT, Maischein HM, Nüsslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 12.Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006;39:255–261. doi: 10.1016/j.ymeth.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- 14.Pogoda HM, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, Bhatt DH, Franzini-Armstrong C, Dominguez C, Arana N, et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol. 2006;298:118–131. doi: 10.1016/j.ydbio.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Buckley CE, Goldsmith P, Franklin RJ. Zebrafish myelination: a transparent model for remyelination? Dis Model Mech. 2008;1:221–228. doi: 10.1242/dmm.001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woods IG, Lyons DA, Voas MG, Pogoda HM, Talbot WS. nsf is essential for organization of myelinated axons in zebrafish. Curr Biol. 2006;16:636–648. doi: 10.1016/j.cub.2006.02.067. [DOI] [PubMed] [Google Scholar]
- 17.Voas MG, Lyons DA, Naylor SG, Arana N, Rasband MN, Talbot WS. alphaII-spectrin is essential for assembly of the nodes of Ranvier in myelinated axons. Curr Biol. 2007;17:562–568. doi: 10.1016/j.cub.2007.01.071. [DOI] [PubMed] [Google Scholar]
- 18.Schweitzer J, Becker T, Schachner M, Nave KA, Werner H. Evolution of myelin proteolipid proteins: gene duplication in teleosts and expression pattern divergence. Mol Cell Neurosci. 2006;31:161–177. doi: 10.1016/j.mcn.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Morris JK, Willard BB, Yin X, Jeserich G, Kinter M, Trapp BD. The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia. 2004;45:378–391. doi: 10.1002/glia.10338. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer K, Brösamle C. Zwilling-A and -B, two related myelin proteins of teleosts, which originate from a single bicistronic transcript. Mol Biol Evol. 2009;263:495–499. doi: 10.1093/molbev/msn298. [DOI] [PubMed] [Google Scholar]
- 21.Waehneldt TV. Phylogeny of myelin proteins. Ann N Y Acad Sci. 1990;605:15–28. doi: 10.1111/j.1749-6632.1990.tb42377.x. [DOI] [PubMed] [Google Scholar]
- 22.Schweigreiter R, Roots BI, Bandtlow CE, Gould RM. Understanding myelination through studying its evolution. Int Rev Neurobiol. 2006;73:219–273. doi: 10.1016/S0074-7742(06)73007-0. [DOI] [PubMed] [Google Scholar]
- 23.Avila RL, Tevlin BR, Lees JPB, Inouye H, Kirschner DA. Myelin structure and composition in zebrafish. Neurochem Res. 2007;32:197–209. doi: 10.1007/s11064-006-9136-5. [DOI] [PubMed] [Google Scholar]
- 24.Park HC, Shin J, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2004;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive Myelin Basic Protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levavasseur F, Mandemakers W, Visser P, Broos L, Grosveld F, Zivkovic D, Meijer D. Comparison of sequence and function of the Oct-6 genes in zebrafish, chicken and mouse. Mech Dev. 1998;74:89–98. doi: 10.1016/s0925-4773(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 27.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 28.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Kazakova N, Li H, Mora A, Jessen KR, Mirsky R, Richardson WD, Smith HK. A screen for mutations in zebrafish that affect myelin gene expression in Schwann cells and oligodendrocytes. Dev Biol. 2006;297:1–13. doi: 10.1016/j.ydbio.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 30.Norton WH, Mangoli M, Lele Z, Pogoda HM, Diamond B, Mercurio S, Russell C, Teraoka H, Stickney HL, Rauch GJ, et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphé neurons and cranial motoneurones. Development. 2005;132:645–658. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyons DA, Naylor SG, Mercurio S, Dominguez C, Talbot WS. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development. 2008;135:599–608. doi: 10.1242/dev.012377. [DOI] [PubMed] [Google Scholar]
- 32.Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet. 2009 doi: 10.1038/ng.376. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nave KA, Salzer JL. Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006;16:492–500. doi: 10.1016/j.conb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa Y, Schafer DP, Horresh I, Bar V, Hales K, Yang Y, Susuki K, Peles E, Stankewich MC, Rasband MN. Spectrins and ankyrinB constitute a specialized paranodal cytoskeleton. J Neurosci. 2006;26:5230–5239. doi: 10.1523/JNEUROSCI.0425-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Fresco GP, Sousa AD, Pillai AM, Moy SS, Crawley JN, Tessarollo L, Dupree JL, Bhat MA. Disruption of axo-glial junctions causes cytoskeletal disorganization and degeneration of Purkinje neuron axons. Proc Natl Acad Sci U S A. 2006;103:5137–5142. doi: 10.1073/pnas.0601082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colman DR, Kreibich G, Frey AB, Sabatini DD. Synthesis and incorporation of myelin polypeptides into CNS myelin. J Cell Biol. 1982;95:598–608. doi: 10.1083/jcb.95.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carson JH, Worboys K, Ainger K, Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Song J, Carson JH, Barbarese E, Li FY, Duncan ID. RNA transport in oligodendrocytes from the taiep mutant rat. Mol Cell Neurosci. 2003;24:926–938. doi: 10.1016/s1044-7431(03)00254-9. [DOI] [PubMed] [Google Scholar]
- 40.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, Link J, Lundström W, Greiner E, Dessa Sadovnick A, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]