Abstract
BTG/TOB factors are a family of antiproliferative proteins whose expression is altered in numerous cancers. They have been implicated in cell differentiation, development and apoptosis. Although proposed to affect transcriptional regulation, these factors interact with CAF1, a subunit of the main eukaryotic deadenylase, and with poly(A)-binding-proteins, strongly suggesting a role in post-transcriptional regulation of gene expression. The recent determination of the structures of BTG2, TOB1 N-terminal domain (TOB1N138) and TOB1N138–Caf1 complexes support a role for BTG/TOB proteins in mRNA deadenylation, a function corroborated by recently published functional characterizations. We highlight possible molecular mechanisms by which BTG/TOB proteins influence deadenylation and discuss the need for a better understanding of BTG/TOB physiological functions.
The BTG/TOB family of antiproliferative proteins
The BTG/TOB (http://www.genenames.org/data/hgnc_data.php?hgnc_id=1130B-cell translocation gene/http://www.genenames.org/data/hgnc_data.php?hgnc_id=11979transducer of ERBB2) family comprises a group of antiproliferative proteins first identified in mammals. Ectopic expression of several members of this protein family resulted in inhibition of cell proliferation [1-5]. Consistent with their growth inhibitory properties, BTG/TOB expression is reduced in several cancer tissues, pointing to a role for the BTG/TOB family in tumor suppression [6, 7]. Moreover, suppression of mammary carcinoma cell growth by retinoic acid is paralleled by the induction of BTG2, a mediator of retinoic acid receptor signaling [8].
In 19991, the first members of the BTG/TOB family were cloned were rat and mouse BTG2 (termed Pc3 and Tis21, respectively); they were isolated in screens designed to identify primary response genes induced by growth factors and tumor promoters [9, 10]. Human BTG1 was identified the following year in a chromosomal translocation observed in a lymphocytic leukemia [1; the TOB1 was found through its ability to bind to the growth factor receptor p185erbB2 [2]. These proteins share a common N-terminal domain (named the BTG/TOB or APRO domain) with no similarity to any known motifs, and thus constitute a new family of proteins involved in the control of cell proliferation [11].
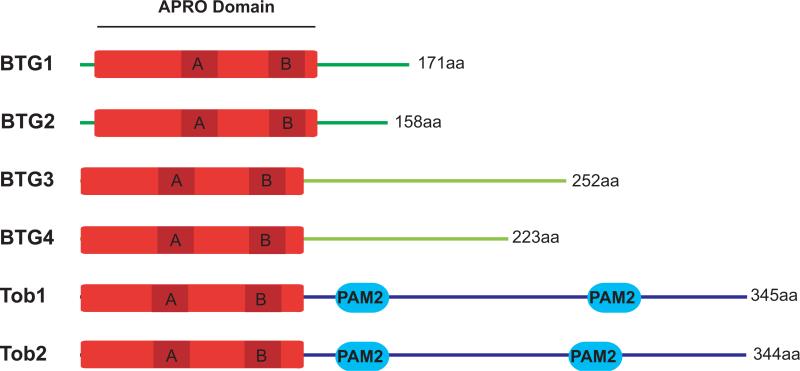
The APRO domain contains approximately 120 amino acids and includes two short conserved motifs, box A and box B, which define the BTG/TOB family. Related proteins have subsequently been identified in several vertebrate and invertebrate animal species, including six BTG/TOB proteins in humans (Figure 1). Whereas the N-terminal APRO domain is conserved, the C-terminal moiety of these proteins is not conserved equally among the family members, allowing their classification into three distinct subfamilies: the BTG1/BTG2 subfamily, the BTG3/BTG4 subfamily, and the TOB subfamily. Until recently, the phylogenetic distribution of APRO domain-containing proteins was thought to be exclusively restricted to metazoan animals. With the availability of an ever-increasing number of genome sequences, BLAST searches reveal that divergent members are also present in some fungi and amoeba [12].
Figure 1.
Organization of the six human BTG/TOB proteins. The presence of an APRO domain (red) at the N-terminus characterizes the BTG/TOB family. Two more conserved sequences, box A and box B (dark red), constitute signature motifs defining the family. The C-terminal parts of the proteins are less conserved and possess no other known domains except for PAM2 motifs found in the TOB members.
In metazoans, BTG/TOB proteins have been implicated in different cellular processes including embryonic development, cellular differentiation, and apoptosis (for review see [11, 13-16]), in addition to their role in cell proliferation and cancer suppression. Their expression levels can fluctuate during the cell cycle and can be induced by diverse stimuli such as growth factors, tumor promoters or genotoxic stresses. Furthermore, these proteins are substrates for post-translational modifications including phosphorylation by mitogen-activated protein kinases (MAPKs) [17-19] and ubiquitylation, which target them for degradation resulting in short half-lives [20]. The analysis of BTG/TOB biological functions has been complicated by the pleiotropic roles attributed to these proteins, the tight regulation of their expression levels, and the identification of numerous protein partners. For example, BTG1 and BTG2 interact with PRMT1 (http://www.genenames.org/data/hgnc_data.php?hgnc_id=5187protein arginine methyltransferase 1) and HOXB9 [21, 22] whereas Tob1 binds p185erbB2 and SMAD proteins [2, 23, 24]. The presence of transcription factors among the BTG/TOB partners suggests that the BTG/TOB proteins may be involved in regulating mRNA production and emerging evidence indicates that BTG/TOB proteins affect chromatin modification and/or modulate the activity of transcriptional activators and repressors [22, 25-28]. Nonetheless, the molecular mechanism by which BTG/TOB proteins exert their tumor suppression and antiproliferative function remains unclear.
Among the BTG/TOB protein partners, http://www.genenames.org/data/hgnc_data.php?hgnc_id=1910chromatin assembly factor 1 (CAF1) is of special interest as it can interact with all BTG/TOB proteins tested [4, 29, 30]. CAF1 is a subunit of the CCR4–NOT complex, long considered to be a transcription factor. However, during the past decade, it has been well documented that CAF1 and its partner CCR4 possess deadenylase activities in vitro and that both factors have a predominant role in mRNA poly(A) tail shortening in vivo [31-39]. These observations suggested that BTG/TOB proteins could be involved in mRNA deadenylation. This hypothesis was recently investigated and these results, combined with the available structural data, provide compelling evidence for an important role of BTG/TOB factors in controlling deadenylation.
Deadenylation: a key step to control gene expression
Eukaryotic mRNAs are synthesized with long poly(A) tails at their 3’ end that are covered by poly(A)-binding-proteins (PABP). Interaction between PABP and the eukaryotic initiation factor eIF4F cap-binding complex (composed of eIF4E, eIF4G and eIF4A) is important for efficient translation initiation. This situation is particularly well illustrated during early embryonic development where gene expression is regulated in the absence of embryonic RNA transcription by variation of maternal mRNA poly(A) tail length [40]. The poly(A) tail also helps protect mRNA from degradation and its progressive shortening by deadenylase is the first step observed in several eukaryotic mRNA decay pathways (reviewed in [41, 42]). The observation that deadenylation rates correlate with mRNA half-lives indicates a central role for poly(A) removal in controlling mRNA degradation [43]. Consistently, destabilizing sequences such as AU-rich elements in the 3’ UTR of mRNAs [44], as well as microRNAs, which promote mRNA decay through partially base-pairing with sequences in the 3’UTR of their target mRNAs [45, 46], have been shown to control the deadenylation of target mRNAs. Once the poly(A) tail of the mRNA is reduced to a few residues, degradation of its body is initiated either via decapping followed by rapid 5’ to 3’ digestion by the exonuclease Xrn1 or alternatively via 3’ to 5’ digestion by the exosome complex (Figure 2) [47]. Overall, deadenylation plays a critical role in the post-transcriptional regulation of gene expression, allowing the rapid shut-off of target mRNA expression through its combined effect on protein translation initiation and mRNA degradation.
Figure 2.
Major mRNA degradation pathways. Deadenylation is the first event observed during most major mRNA decay processes. Deadenylation is biphasic with the first step catalyzed by the PAN2–PAN3 complex (blue–purple); the second step is mediated by CCR4-CAF1 (blue–aqua). After deadenylation, the mRNA is either decapped (yellow) and then rapidly degraded in the 5’ to 3’ direction by Xrn1 (aqua), or alternatively, the body of the mRNA can be degraded in the 3’ to 5’ direction by the exosome (blue). In specific cell types, deadenylated mRNAs can be stored as translationally inactive molecules (not shown).
Several enzymes which catalyze poly(A) removal have been characterized in eukaryotes [42]. Two universally conserved heterodimeric complexes, namely the PAN2–PAN3 and CCR4-CAF1 complexes, have been ascribed a central role in eukaryotic mRNA poly(A) shortening. Additional deadenylases, such as poly(A)-specific ribonuclease (PARN) and nocturnin, are present in only some species and might perform specific functions [48, 49]. Only the PAN2 subunit of the PAN2–PAN3 complex is endowed with nuclease activity; PAN3 is an essential auxiliary factor. By contrast, both subunits of the CCR4–CAF1 complex possess deadenylase activities. In mammals, the situation is further complicated by the existence of two CAF1 paralogs (CAF1, also called CAF1A; and POP2, also called CAF1B of CALIF; these proteins are hereafter collectively named CAF1). As a similar situation exists for CCR4 (CCR4A and CCR4B, hereafter named CCR4), in theory 4 different heterodimeric deadenylase complexes could be produced in mammals. The CCR4–CAF1 deadenylases are further associated in large assemblies with NOT proteins, whichare important for deadenylation in vivo but whose molecular functions remain elusive [34, 36]. Transcriptional pulse-chase strategies have revealed that deadenylation is biphasic in mammalian cells; the first step is a synchronous poly(A) shortening catalyzed by PAN2–PAN3 followed by a second more rapid and heterogeneous step mediated by CCR4-CAF1 [38]. The mechanism mediating the transition between the two phases is not understood. A similar biphasic deadenylation process has been proposed for yeast [50]. Altogether, a precisely coordinated choreography of deadenylases appears to play a major role in controlling poly(A) tail length and thereby mRNA stability and translation. To decipher this process, it is important to understand the function(s) of partners that directly interact with the deadenylases.
BTG/TOB physical links to deadenylation
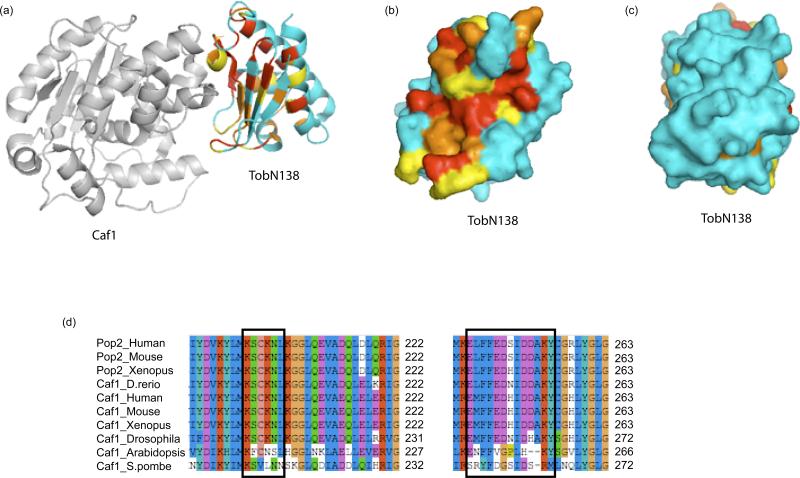
Recently, the resolution of the structures of human and mouse BTG2 [51] and, importantly, of a complex consisting of the human TOB APRO domain (residues 1-138, hereafter denoted TOBN138) associated with human CAF1A[12] provided important information about the structural organization of the BTG/TOB proteins and their interaction with the CAF1 deadenylase. The crystal structures of human and mouse BTG2 at 2.3 Å and 2.2 Å, respectively, reveal that the APRO domain (residues 7-130) forms a single globular domain. It is composed of five α-helices and four β-strands, forming one anti-parallel β-sheet, arranged in a new topology (Figure 3a). Superimposing the crystal structure backbone of the APRO domain of human TOB1 in the TOBN138–CAF1 complex and that of BTG2 reveals a high degree of similarity between the two structures, suggesting that this fold is characteristic of the APRO domain of the BTG/TOB protein family and that this structure is not significantly rearranged upon CAF1 binding. The crystal structure of the TAOBN138–CAF1 complex reveals the surface contact between the two proteins and demonstrates that key residues from both box A and box B motifs of TOB1 are involved in the TOB1–CAF1 interaction. This observation validates mutagenesis results which showed that alterations of the equivalent residues prevented BTG-CAF1 interaction [30, 52]. Moreover, plotting the residues conserved among the BTG/TOB members on the TOBN138 structure reveals a major conserved surface (figure 3b) that corresponds to the CAF1 interaction site whereas other exposed regions of the APRO domain are not well conserved (Figure 3c). These structural data indicate that interaction with the CAF1 deadenylase is of central importance for the function of BTG/TOB proteins. Consistently, co-expression of TOB1 with a CAF1 point mutant which is unable to interact with TOB1 abrogated the antiproliferative effect of TOB1 expression [12]. Reciprocally, residues on the CAF1 surface which are involved in the interaction with TOBN138 are well conserved through higher eukaryote CAF1 homologs, but are more variable in organisms that are not known to encode APRO domain-containing proteins in their genome (Figure 3d). A comparison of the structure of CAF1 in the TOBN138-CAF1 complex with the structures of its yeast Pop2p orthologs [53, 54] indicates that the association with BTG/TOB proteins does not appreciably affect the structure of the deadenylase active site nor does it block its access to the poly(A) tail, suggesting that the CAF1 catalytic capacities are unlikely to be strongly modified. Collectively, the structures of human and mouse BTG2, together with that of the TOBN138-CAF1 complex, suggest that the APRO domain is a universally conserved adaptor which binds the CAF1 deadenylase without significant effects on its catalytic deadenylation activity.
Figure 3.
The APRO domain is a conserved adaptor binding the Caf1 deadenylase.
(a) Structure of the CAF1-TOBN138 complex shown as ribbon diagram (PDB entry 2D5R, [12]). CAF1 is shown in gray. For TOBN138, invariant residues are shown in red, well-conserved residues in orange, moderately conserved residues in yellow and non-conserved residues in blue. The degree of conservation is taken from a ClustalW multiple sequence alignment of vertebrate APRO domains (shown in supplemental data). All structural images were prepared using PyMOL (www.pymol.org).
(b) Surface representation of TOBN138 showing the surface in contact with CAF1. The coloring scheme is as in (a).
(c) Surface representation of TOBN138 showing the opposite face as compared to (b). The coloring scheme is as in (a).
(d) The CAF1 regions interacting with TOB1 are conserved only in orthologs from species encoding APRO domains. Clustal W sequence alignments of two regions of CAF1 are shown. Residues found to contact TOBN138 [12] are indicated in black boxes. Note that these residues are not conserved in Arabidopsis and S. pombe, which do not encode APRO-containing proteins in their genomes.
It is noteworthy that interaction with CAF1 is not the sole structural feature linking TOB factors to deadenylation. Indeed, an interaction between TOB1 and the NOT1 subunit of the large CCR4-NOT assembly has been described recently [55]. This interaction involves a C-terminal region of TOB1 that is not conserved between the BTG/TOB proteins. Thus this property is probably not a universal feature of the BTG/TOB family. Curiously, this interaction involves both the N-terminal and C-terminal domains of NOT1, but it is not clear whether this interaction is direct or indirect.
Of particular interest, two PAM2 motifs[SC1] have been identified in the TOB protein sequences (Figure 1). PAM2 motifs are peptide sequences found in numerous proteins, which have been shown to be necessary for their interaction with the C-terminal domain of PABP proteins[SC2] [56]. The physical interaction between TOB proteins and PABP has been confirmed by GST-pull down experiments using wild type and mutant proteins [57-60]. The BTG members of the family do not contain PAM2 sequences, nor have they been reported to interact directly with PABP. Such an interaction remains possible, however, especially if it is dependent upon specific protein modification(s).
BTG/TOB functional links to deadenylation
The identification of CAF1 as a conserved partner of BTG/TOB factors, together with the direct interaction of TOB proteins with PABP, prompted several groups to look for a role(s) in mRNA decay. Using a Tet-off transcriptional pulse-chase approach, Ezzedine et al [59]showed that ectopic co-expression of TOB1 activated deadenylation of two different reporters: a stable β-globin mRNA reporter that is degraded by the basic mRNA decay pathway, and a β-globin reporter containing a premature termination codon (PTC) that is degraded by the nonsense-mediated mRNA decay. Thus TOB1 can activate mRNA deadenylation independently of the decay pathways. Interestingly, this effect was abrogated when TOB1 mutant proteins lacking functional PAM2 motifs, and thus unable to interact with PABP, were used. This finding demonstrated that the ability of TOB1 to activate mRNA deadenylation was dependent upon its ability to bind PABP. Previous experiments showed that recruitment of the CAF1–CCCR4 deadenylase onto specific mRNAs either by specific RNA binding proteins [61, 62] or in tethering experiments [63] was sufficient to activate deadenylation and degradation of the cognate mRNA. Thus it is possible that TOB1 activates deadenylation by recruiting CAF1-CCR4 to poly(A)mRNA through its interaction with PABP.
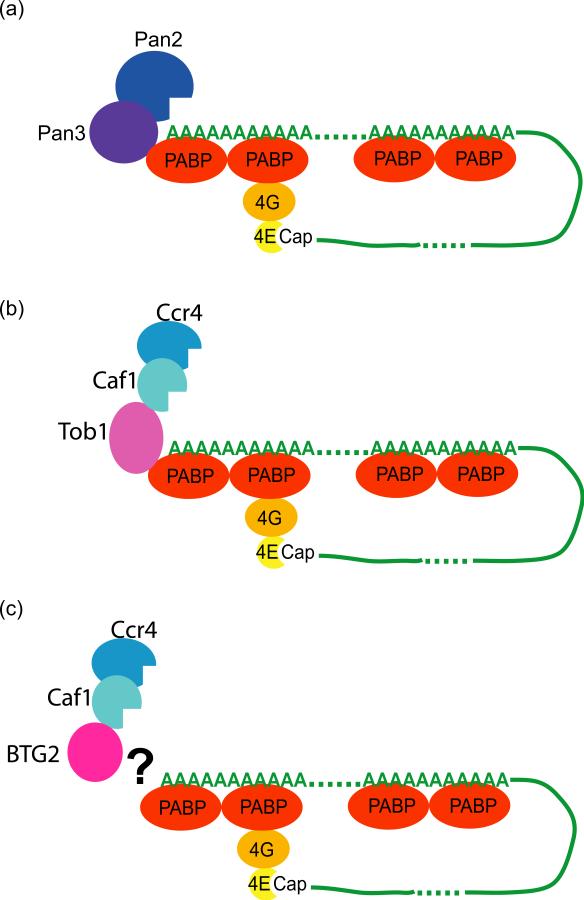
It is worth noting that the translation termination factor eRF3 as well as the PAN3 subunit of the PAN2–PAN3 deadenylase complex both contain PAM2 motifs and also interact with PABP [60]. Titration experiments have shown that either PAN3 or TOB1 can compete with eRF3 to form a complex with PABP [60]. This observation led Funakoshi et al. to propose a model in which deadenylation induction would be coupled to translation: following translation termination, the PAM2-mediated interaction between PABP and eRF3 would somehow be replaced by a PAN3–PABP interaction, initiating the first phase of deadenylation (Figure 4a) [60]. In the presence of TOB, this initial step would be bypassed (Figure 4b). However, as a direct connection between translation and deadenylation remains controversial, validation of this model requires further experimentation.
Figure 4.
Models of recruitment of deadenylases onto mRNAs.
(a) a translation-dependent exchange of eRF3 for the PAM2-containing PAN3 (purple) factor on PABP (red) allows the recruitment of the PAN2 deadenylase (blue) and initiation of deadenylation.
(b) Expression of the PAM2-containing TOB1 (pink) recruits the CCR4-CAF1 (aqua–blue) deadenylases onto mRNA possibly bypassing PAN2 and activating deadenylation.
(c) Expression of BTG2 (pink) activates CAF1-dependent deadenylation by an unknown mechanism.
Interestingly, ectopic expression of BTG2 also activates deadenylation of a β-globin mRNA reporter in transcriptional pulse-chase experiments [52]. This effect was independent of the expression system and the particular reporter used, as it was also observed with endogenous mRNAs in cells stably expressing ectopic BTG2. Moreover, this activation strictly required an active CAF1 deadenylase. Thus, althoug the BTG members of the BTG/TOB family do not possess PAM2 motifs and are not known to interact with PABP or with other proteins present in mRNA–protein complexes (mRNPs; e.g., cap binding factors), they are nevertheless able to activate CAF1-dependent deadenylation. The mechanisms by which BTG2 and TOB1 activate deadenylation are thus unlikely to be identical; instead they are likely to be variations on a common theme. In the case of BTG2, the precise mechanism remains to be elucidated (Figure 4c).
In vitro nuclease assays performed with either recombinant CAF1 deadenylase or with in vivo immunoprecipitated CAF1 complexes generated conflicting results. In one report [51], addition of recombinant BTG2 slowed down the deadenylase activity of recombinant CAF1. Similarly, co-immunoprecipitation of FLAG-TOB[SC4] with GFP-CAF1 resulted in slower nuclease activity of the complex [55], suggesting that interaction of BTG/TOB factors with CAF1 would reduce deadenylation. These in vitro results contradict the observations made in vivo in transcriptional pulse-chase experiments and the structural data indicating that interaction of an APRO domain does not affect the CAF1 structure. Consistent with the latter remarks, another group reported that addition of TOBN138 had no effect on the nuclease activity of recombinant CAF1 deadenylase [12]. It remains unknown why conflicting results were observed for the effects of BTG2 or TOB1 on CAF1 activity. As CAF1 nuclease activity can be affected, for example by divalent metal ions [12, 64], it is possible that the discrepancy results from the conditions used for the in vitro assays. Another important parameter is that in vivo, RNA substrates are associated with other factors to form ribonucleoparticles (RNPs)[SC5] and thus their accessibility differs from that under in vitro conditions. In agreement with this hypothesis, addition of recombinant PABP stimulated nuclease activity of an immunoprecipitated CAF1–TOB complex in vitro [60], lending further suppor to the hypothesis that TOB directs CAF1 to its mRNA substrates via the poly(A)–PABP complex.
Concluding remarks and future perspectives
Recent results indicate unequivocally that some members of the BTG/TOB family function in mRNA deadenylation. As the residues involved in the interaction with the CAF1 deadenylase are strikingly conserved among the members of this protein family, it would not be unexpected for all members of the BTG/TOB family to also target this step in mRNA decay. It would be interesting to know whether all BTG/TOB members activate deadenylation, or if some members of this family might have the opposite effect and inhibit the deadenylation process. At this stage, it is worth remembering that BTG/TOB factors are targets of multiple signaling pathways that induce their production, affect their stability and might control their activity and/or localization. Combined with the recent data, these findings suggest that the BTG/TOB proteins relay these signals to influence protein translation and/or mRNA stability.
Several questions related to the function of BTG/TOB proteins in deadenylation remain open. The first concerns the functional consequences of BTG/TOB-activated deadenylation. As the poly(A) tail is required for both protein translation and mRNA stability, one can wonder whether BTG/TOB induced deadenylation always promotes degradation of the target transcripts, as observed in ectopic expression studies in cultured cells, or if in some cases deadenylation causes a block in translation and storage of the corresponding transcripts, for example in structures named P-bodies (also known as GW-bodies or Dcp-bodies), which are a site for storage of non-translating mRNPs [65, 66]. In the latter case, BTG/TOB proteins could have more robust affect at the protein level than at the mRNA level. In agreement with this hypothesis is the observation that human TOB2 colocalizes with P-bodies in tissue culture cells [59]. This finding might suggest a role for the TOB proteins in facilitating the exit of mRNPs from the translation pool to the nontranslated pool residing in P-bodies. It will be interesting to see if BTG/TOB factors are recruited to P-bodies together with the CCR4–CAF1 deadenylase or if these factors might have a function in P-bodies that is independent of deadenylases.
Another important issue concerns the specificity of the mRNAs targeted by BTG and TOB factors. The proposed mechanism of TOB action suggests that they should act on every mRNP harboring PABP, as observed in the case of ectopic expression [59]. Similarly, ectopically expressed BTG2 targets every mRNA tested [52]. However, it remains unclear whether this is the case under physiological conditions. One could imagine that other mRNP-associated factors might interact with BTG/TOB proteins and provide specificity to their action. Alternatively, activation of BTG/TOB, for example by kinases [17-19], could provide spatially restricted activity without providing specificity towards particular mRNP substrates.By contrast, if BTG/TOB activity displays no mRNA specificity, one could imagine that BTG/TOB-mediated deadenylation could be prevented by other factors.
Whatever the exact mechanism of BTG/TOB action is, the observation that these proteins target deadenylation can easily be reconciled with their antiproliferative activity. Preventing translation and promoting degradation of all, or a specific subset, of the cellular mRNAs would undoubtedly have a negative impact on cell proliferation. The observation that TOB2 undergoes cell cycle-dependent by BTG/TOB contributes to their antiproliferative properties [59]. These observations might also explain the pleiotropic functions of BTG/TOB proteins in development, stress responses and apoptosis. Indeed, a predisposition to develop cancer was reported for Tob1 and Btg3 knock-out mice [6, 67]. This phenotype can be easily explained by the antiproliferative properties of the proteins. Unexpectedly, Btg/Tob knock-out mice display bone morphogenesis phenotypes: the Btg2 mutant showed abnormalities in vertebral patterning [68], Btg3 inactivation resulted in increased ectopic bone formation [69], Tob1 deletion increased bone mass [23], and in contrast, Tob2 knock-out decreased bone mass [70]. The relation of bone formation to deadenylation is unclear at present. Additional experiments will be required to test whether BTG/TOB factors preferentially control deadenylation of specific mRNAs involved in bone morphogenesis. Alternatively, other activities proposed for these proteins, for example in transcription, could be required to produce the observed physiological effects. Nevertheless, recent structural and functional studies have provided a strong framework supporting the role of BTG/TOB factors in deadenylation. Deciphering the mechanisms involved will provide new insight into the physiological roles of this interesting and pleiotropic protein family in regulating gene expression.
Supplementary Material
Acknowledgements
We apologize to our colleagues whose contributions were omitted due to space limitation. We thank M. Graille for suggestion for structure drawing and J. Lever for critical reading of the manuscript. The work in the laboratory of Bertrand Séraphin is supported by La Ligue contre le Cancer (Equipe labellisée 2008) and the CNRS, Agence Nationale de la Recherche (ANR-07-BLAN-0093), and the work by the laboratory of Ann-Bin Shyu is supported by National Institutes of Health (RO1GM 046454) and by the Houston Endowment, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rouault JP, et al. BTG1, a member of a new family of antiproliferative genes. Embo J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda S, et al. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- 3.Yoshida Y, et al. ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene. 1998;16:2687–2693. doi: 10.1038/sj.onc.1201805. [DOI] [PubMed] [Google Scholar]
- 4.Ikematsu N, et al. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- 5.Buanne P, et al. Cloning of PC3B, a novel member of the PC3/BTG/TOB family of growth inhibitory genes, highly expressed in the olfactory epithelium. Genomics. 2000;68:253–263. doi: 10.1006/geno.2000.6288. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida Y, et al. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–1206. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiko AD, et al. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donato LJ, et al. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 2007;67:609–615. doi: 10.1158/0008-5472.CAN-06-0989. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury A, et al. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci U S A. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher BS, et al. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem. 1991;266:14511–14518. [PubMed] [Google Scholar]
- 11.Matsuda S, et al. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 12.Horiuchi M, et al. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J Biol Chem. 2009;284:13244–13255. doi: 10.1074/jbc.M809250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol. 2001;187:155–165. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- 14.Duriez C, et al. BTG2, its family and its tutor. Bull Cancer. 2004;91:E242–253. [PubMed] [Google Scholar]
- 15.Lim IK. TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol. 2006;132:417–426. doi: 10.1007/s00432-006-0080-1. [DOI] [PubMed] [Google Scholar]
- 16.Jia S, Meng A. Tob genes in development and homeostasis. Dev Dyn. 2007;236:913–921. doi: 10.1002/dvdy.21092. [DOI] [PubMed] [Google Scholar]
- 17.Maekawa M, et al. Identification of the Anti-proliferative protein Tob as a MAPK substrate. J Biol Chem. 2002;277:37783–37787. doi: 10.1074/jbc.M204506200. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, et al. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 2002;16:1356–1370. doi: 10.1101/gad.962802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong JW, et al. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J Biol Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- 20.Sasajima H, et al. Antiproliferative proteins of the BTG/Tob family are degraded by the ubiquitin-proteasome system. Eur J Biochem. 2002;269:3596–3604. doi: 10.1046/j.1432-1033.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin WJ, et al. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 22.Prevot D, et al. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida Y, et al. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–1097. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida Y, et al. Tob proteins enhance inhibitory Smad-receptor interactions to repress BMP signaling. Mech Dev. 2003;120:629–637. doi: 10.1016/s0925-4773(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 25.Guardavaccaro D, et al. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol. 2000;20:1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prevot D, et al. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- 27.Berthet C, et al. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells. 2002;7:29–39. doi: 10.1046/j.1356-9597.2001.00497.x. [DOI] [PubMed] [Google Scholar]
- 28.Ou YH, et al. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. Embo J. 2007;26:3968–3980. doi: 10.1038/sj.emboj.7601825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdan JA, et al. Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J. 1998;336(Pt 2):471–481. doi: 10.1042/bj3360471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouault JP, et al. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem. 1998;273:22563–22569. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- 31.Daugeron MC, et al. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucker M, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, et al. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. Embo J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker M, et al. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. Embo J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanathan P, et al. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 36.Temme C, et al. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bianchin C, et al. Conservation of the deadenylase activity of proteins of the Caf1 family in human. Rna. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 39.Schwede A, et al. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter JD. Breaking the code of polyadenylation-induced translation. Cell. 2008;132:335–337. doi: 10.1016/j.cell.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Meyer S, et al. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 42.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 43.Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. Rna. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 45.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, et al. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garneau NL, et al. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 48.Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 49.Wu M, et al. Structural insight into poly(A) binding and catalytic mechanism of human PARN. Embo J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 2008;36:6872–6881. doi: 10.1093/nar/gkn825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mauxion F, et al. The BTG2 protein is a general activator of mRNA deadenylation. Embo J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonstrup AT, et al. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007;35:3153–3164. doi: 10.1093/nar/gkm178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thore S, et al. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyasaka T, et al. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci. 2008;99:755–761. doi: 10.1111/j.1349-7006.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy G, et al. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol. 2002;22:3769–3782. doi: 10.1128/MCB.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okochi K, et al. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells. 2005;10:151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 58.Lim NS, et al. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J Biol Chem. 2006;281:14376–14382. doi: 10.1074/jbc.M600307200. [DOI] [PubMed] [Google Scholar]
- 59.Ezzeddine N, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funakoshi Y, et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldstrohm AC, et al. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 62.Zaessinger S, et al. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 63.Finoux AL, Seraphin B. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J Biol Chem. 2006;281:25940–25947. doi: 10.1074/jbc.M600132200. [DOI] [PubMed] [Google Scholar]
- 64.Liu WF, Yan YB. Biophysical and biochemical characterization of recombinant human Pop2 deadenylase. Protein Expr Purif. 2008;60:46–52. doi: 10.1016/j.pep.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Eulalio A, et al. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 66.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 67.Yoneda M, et al. Deficiency of antiproliferative family protein Ana correlates with development of lung adenocarcinoma. Cancer Sci. 2008 doi: 10.1111/j.1349-7006.2008.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park S, et al. B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/smad signaling. Mol Cell Biol. 2004;24:10256–10262. doi: 10.1128/MCB.24.23.10256-10262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyai K, et al. ANA deficiency enhances bone morphogenetic protein-induced ectopic bone formation via transcriptional events. J Biol Chem. 2009;284:10593–10600. doi: 10.1074/jbc.M807677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ajima R, et al. Osteoporotic bone formation in mice lacking tob2; involvement of Tob2 in RANK ligand expression and osteoclasts differentiation. FEBS Lett. 2008;582:1313–1318. doi: 10.1016/j.febslet.2008.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.