Abstract
Mitochondrial 12S rRNA 1555A>G mutation is one of the important causes of aminoglycoside-induced and nonsyndromic deafness. Our previous investigations showed that the A1555G mutation was a primary factor underlying the development of deafness but was insufficient to produce deafness phenotype. However, it has been proposed that mitochondrial haplotypes modulate the phenotypic manifestation of the 1555A>G mutation. Here, we performed systematic and extended mutational screening of 12S rRNA gene in a cohort of 1742 hearing-impaired Han Chinese pediatric subjects from Zhejiang Province, China. Among these, 69 subjects with aminoglycoside-induced and nonsyndromic deafness harbored the homoplasmic 1555A>G mutation. These translated to a frequency of ~3.96% for the 1555A>G mutation in this hearing impaired population. Clinical and genetic characterizations of 69 Chinese families carrying the 1555A>G mutation exhibited a wide range of penetrance and expressivity of hearing impairment. The average penetrances of deafness were 29.5% and 17.6%, respectively, when aminoglycoside-induced hearing loss was included or excluded. Furthermore, the average age-of-onset for deafness without aminoglycoside exposure ranged from 5 and 30 years old, with the average of 14.5 years. Their mitochondrial genomes exhibited distinct sets of polymorphisms belonging to ten Eastern Asian haplogroups A, B, C, D, F, G, M, N, R and Y, respectively. These indicated that the 1555A>G mutation occurred through recurrent origins and founder events. The haplogroup D accounted for 40.6% of the patient’s mtDNA samples but only 25.8% of the Chinese control mtDNA samples. Strikingly, these Chinese families carrying mitochondrial haplogroup B exhibited higher penetrance and expressivity of hearing loss. In addition, the mitochondrial haplogroup specific variants: 15927G>A of haplogroup B5b, 12338T>C of haplogroup F2, 7444G>A of haplogroup B4, 5802T>C, 10454T>C, 12224C>T and 11696G>A of D4 haplogroup, 5821G>A of haplogroup C, 14693A>G of haplogroups Y2 and F, and b of Y2 may enhance the penetrace of hearing loss in these Chinese families. Moreover, the absence of mutation in nuclear modifier gene TRMU suggested that TRMU may not be a modifier for the phenotypic expression of the 1555A>G mutation in these Chinese families. These observations suggested that mitochondrial haplotypes modulate the variable penetrance and expressivity of deafness among these Chinese families.
INTRODUCTION
Deafness is one of the most common human health problems, affecting one in 700–1000 newborns (Nance and Sweeney 1975; Morton 1991). Deafness can be caused by gene alterations and environmental factors including ototoxic drugs such as aminoglycoside antibiotics. Mutations in mitochondrial DNA (mtDNA) are one of the important causes of sensorineural hearing loss in some countries (Fischel-Ghosian 2005; Guan 2005). Of these, the 12S rRNA 1555A>G mutation has been found to be responsible for both aminoglycoside-induced and nonsyndromic deafness in many families worldwide (Prezant et al. 1993; Matthijs et al. 1996; Pandya et al. 1997; Estivill et al. 1998; del Castillo et a. 2003; Li et al. 2004a; 2004b; Tang et al. 2007). The administration of aminoglycosides can induce or worsen hearing loss in these subjects carrying the 1555A>G mutation. In the absence of aminoglycosides, matrilineal relatives within and among families carrying the 1555A>G mutation exhibited a considerable phenotypic variation with respect to severity, age-of-onset and penetrance of hearing loss (Prezant et al. 1993; Matthijs et al. 1996; Estivill et al. 1998; Li et al. 2004a; 2004b; Young et al. 2005; 2006; Tang et al. 2007; Chen et al. 2008). Notably, some Chinese families carrying the 1555A>G mutation exhibited very low penetrance of hearing loss (Young et al. 2005; Dai et al. 2006; Tang et al. 2007), while a large Arab-Israeli family carrying the 1555A>G mutation revealed high penetrance of hearing loss (Bykhovskaya et al. 1998). Functional characterization of cell lines derived from matrilineal relatives of a large Arab-Israeli family demonstrated that the 1555A>G mutation conferred mild mitochondrial dysfunction and sensitivity to aminoglycosides (Guan et al. 1996; 2000; 2001). These findings indicated that the 1555A>G mutation by itself is insufficient to produce the deafness phenotype. Therefore, additional modifier factors such as aminoglycosides, nuclear and mitochondrial genetic modifiers contribute to the phenotypic variability of these mtDNA mutations (Guan et al. 1996; 2000; 2001; 2006; Young et al. 2006; Chen et al. 2008; Wang et al. 2008; Qian and Guan 2009).
However, the role of mitochondrial haplogroups/variants, in the phenotypic expression of these primary deafness-associated mtDNA mutations remains poorly defined. In the present investigation, we carried out a systematic and extended mutational screening of 12S rRNA gene in a large cohort of 1742 hearing-impaired Han Chinese pediatric subjects from Zhejiang Province, Eastern China. Mutational analysis of 12S rRNA gene in these subjects identified 69 genetically unrelated individuals harboring the 1555A>G mutation. We then performed the clinical, genetic and molecular characterization of these hearing-impaired subjects carrying the 1555A>G mutation. A wide range of penetrance, severity and age-at-onset of hearing loss was observed in the matrilineal relatives within and among these Chinese families. To assess the contribution that mtDNA variants or haplogroups make toward the variable penetrance and expressivity of hearing loss in these pedigrees, we performed PCR-amplification of fragments spanning entire mtDNA and subsequent DNA sequence analysis in the matrilineal relatives of those families. These analyses showed that there were distinct sets of mtDNA variants belonging to ten Eastern Asian haplogroups in these Chinese pedigrees carrying the 1555A>G mutation. Furthermore, we evaluated the potential role of these mtDNA haplogroups and variants in the phenotypic manifestation of the 1555A>G mutation in these Chinese families. In the previous investigations, we showed that the 10A>S mutation in TRMU, a nuclear modifier gene encoding a highly conserved 5-methylaminomethyl-2-thiouridylate-methyltransferase modulated the phenotypic manifestation of the 1555A>G mutation (Guan et al. 2006). To examine whether TRMU gene plays a modifying role in the phenotypic expression of the 1555A>G mutation, we performed a mutational analysis of the TRMU gene in the hearing-impaired and normal hearing subjects of these families.
MATERIALS AND METHODS
Subjects and audiological examinations
A total of 1742 pediatric Chinese subjects, who were diagnosed with sporadic aminoglycoside-induced and nonsyndromic hearing impairment by the Otology Clinic of Wenzhou Medical College, were used for the screening of the 1555A>G mutation. A total of 69 Han Chinese pedigrees carrying the 1555A>G mutation were included in this study. A comprehensive history and physical examination for these participating subjects were performed to identify any syndromic findings, the history of the use of aminoglycosides, genetic factors related to the hearing impairment. An age-appropriate audiological examination was performed and this examination included pure-tone audiometry (PTA) and/or auditory brainstem response (ABR), immittance testing and Distortion product otoacoustic emissions (DPOAE). The PTA was calculated from the average of the audiometric thresholds at 500, 1000, 2000, 4000 and 8000 Hz. The severity of hearing impairment was classified into five grades: normal <26 Decibel (dB); mild =26–40 dB; moderate=41–70 dB; severe=71–90 dB; and profound >90 dB. The 262 control DNA were obtained from a panel of unaffected subjects from Han Chinese ancestry from the same region. Penetrance (%) was calculated as affected matrilineal relatives/total matrilineal relatives in each family. Informed consent was obtained from participants prior to their participation in the study, in accordance with the Cincinnati Children’s Hospital Medical Center Institutional Review Board and Ethnics Committee of Wenzhou Medical College.
Mutational analysis of mitochondrial genome
Genomic DNA was isolated from whole blood of participants using Puregene DNA Isolation Kits (Gentra Systems, Minneapolis, MN). The homoplasmy of the 1555A>G mutation in these subjects was determined as detailed previously (Li et al. 2004a; 2004b; Li et al. 2005). Briefly, subject’s DNA fragments spanning the 12S rRNA gene were amplified by PCR using oligodeoxynucleotides corresponding to positions 618–635 and 1988–2007 (Andrews et al. 1999). The amplified segments were digested with a restriction enzyme BsmAI (Li et al. 2004a; 2004b). Equal amounts of various digested samples were then analyzed by electrophoresis through 1.5% agarose gel. The proportions of digested and undigested PCR products were determined by the Image-Quant program after ethidium bromide staining to determine if the 1555A>G mutation is in homoplasmy in these subjects.
The entire mitochondrial genomes of 43 probands and 93 normal hearing Han Chinese control subjects were PCR amplified in 24 overlapping fragments by use of sets of the light-strand and the heavy-strand oligonucleotide primers, as described elsewhere (Rieder et al. 1998). Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the BigDye Terminator Cycle sequencing reaction kit. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920) (Andrews et al. 1999). Multiple sequence alignments were performed using the ClustalW2 program from European Bioinformatics Institute. The allele frequency of variants in CO1 and tRNASer(AGY) genes was determined by PCR amplification of fragments spanning the corresponding regions, using the genomic DNA derived from 262 Chinese controls as templates and performing subsequent sequence analysis of PCR products, as described above. These sequence results were compared with the updated consensus Cambridge sequence (Andrews et al. 1999).
Haplogroup and phylogenetic analyses
The entire mtDNA sequences of the 69 hearing-impaired Chinese probands carrying the 1555A>G mutation and 93 Han Chinese controls were assigned to the Asian mitochondrial haplogroups by using the nomenclature of mitochondrial haplogroup (Tanaka et al. 2004; Kong et al. 2006). The phylogenetic relationships between the haplogroups of 69 hearing-impaired Chinese subjects and those of 93 Chinese control subjects were determined by the bootstrapping tests (Marshall 1991). The phylogenetic tree was generated by Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0., as detailed elsewhere (Tamura et al. 2007).
Mutational analysis of TRMU gene
Nine pairs of primers for PCR-amplifying exons and their franking sequences, including splicing donor and acceptor consensus sequences of TRMU, were used for this analysis. The forward and reverse primers for PCR amplification and sequence analysis were as detailed elsewhere (Guan et al. 2006). Fragment spanning 11 exons and flanking sequence from 69 matrilineal relatives of Chinese families and 11 unrelated controls were PCR-amplified, purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the BigDye Terminator Cycle sequencing reaction kit. These sequence results were compared with the TRMU genomic sequence (GenBank accession number AF448221) (Guan et al. 2006).
Statistical analysis
Statistical analysis was carried out using the Student’s unpaired, two-tailed t-test contained in the Microsoft-Excel program. Unless indicated otherwise, a P value <0.05 was considered statistically significant.
RESULTS
Incidence of the 1555A>G mutation in a large cohort of Chinese pediatric subjects with hearing loss
We have performed a mutational screening of the 1555A>G mutation in a cohort of 1742 Han Chinese pediatric subjects, who were diagnosed as aminoglycoside-induced and nonsyndromic hearing loss by the Otology Clinic at the Wenzhou Medical College. Firstly, DNA fragments spanning the 12S rRNA gene were PCR-amplified from each affected subject. Each fragment was digested by restriction enzyme BsmAI and subsequent electrophoresis analysis. Of those, 69 genetically unrelated subjects harbored the homoplasmic 1555A>G mutation in the 12S rRNA gene (data not shown). These translate to a frequency of ~3.96% for the 1555A>G mutation in this hearing impaired Chinese pediatric population. The presence of the homoplasmic 1555A>G mutation in those subjects was further confirmed by PCR-amplification of fragments spanning the 12S rRNA gene and subsequent DNA sequence analysis (data not shown).
Clinical and genetic evaluation of Chinese families carrying the 1555A>G mutation
In the previous investigations, we have performed the clinical, genetic and molecular characterization of 26 Chinese pedigrees carrying the 1555A>G mutation (Young et al. 2005; 2006; Zhao et al. 2005; Yuan et al. 2005; Liao et al. 2007; Tang et al. 2007; Chen et al. 2008; Wang et al. 2008). In the present investigation, all available members of another 43 Han Chinese pedigrees carrying the 1555A>G mutation underwent a comprehensive history and physical examination as well as audiological examination identify any syndromic findings, the history of the use of aminoglycosides and genetic factors related to the hearing impairment. Comprehensive family medical histories of those probands and other members of these Chinese families showed no other clinical abnormalities, including diabetes, muscular diseases, visual dysfunction, and neurological disorders.
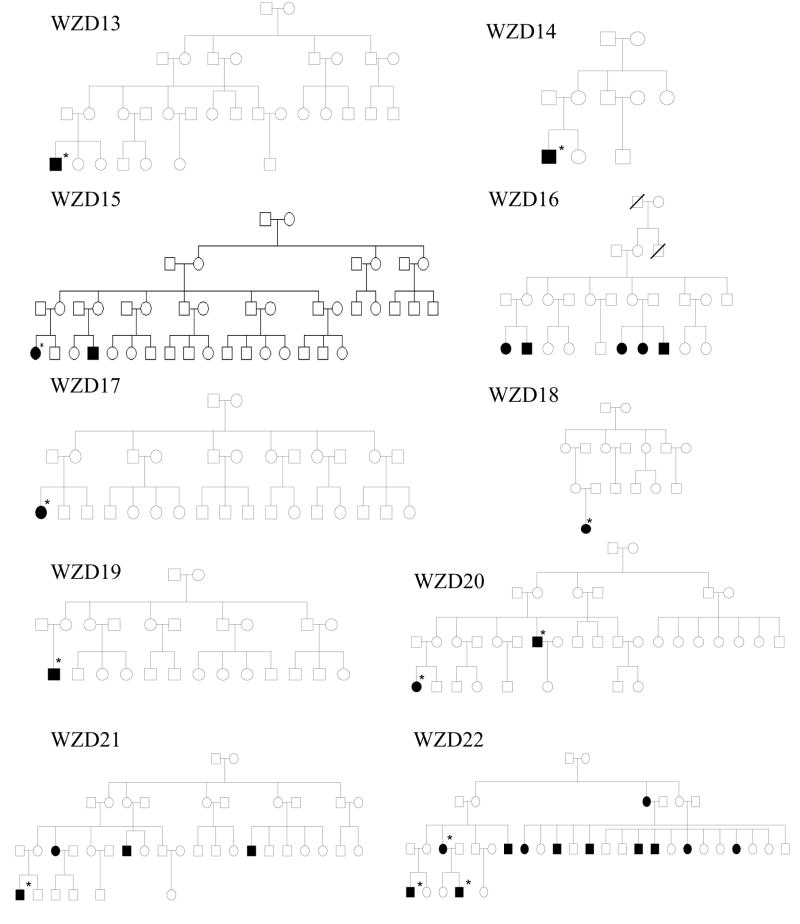
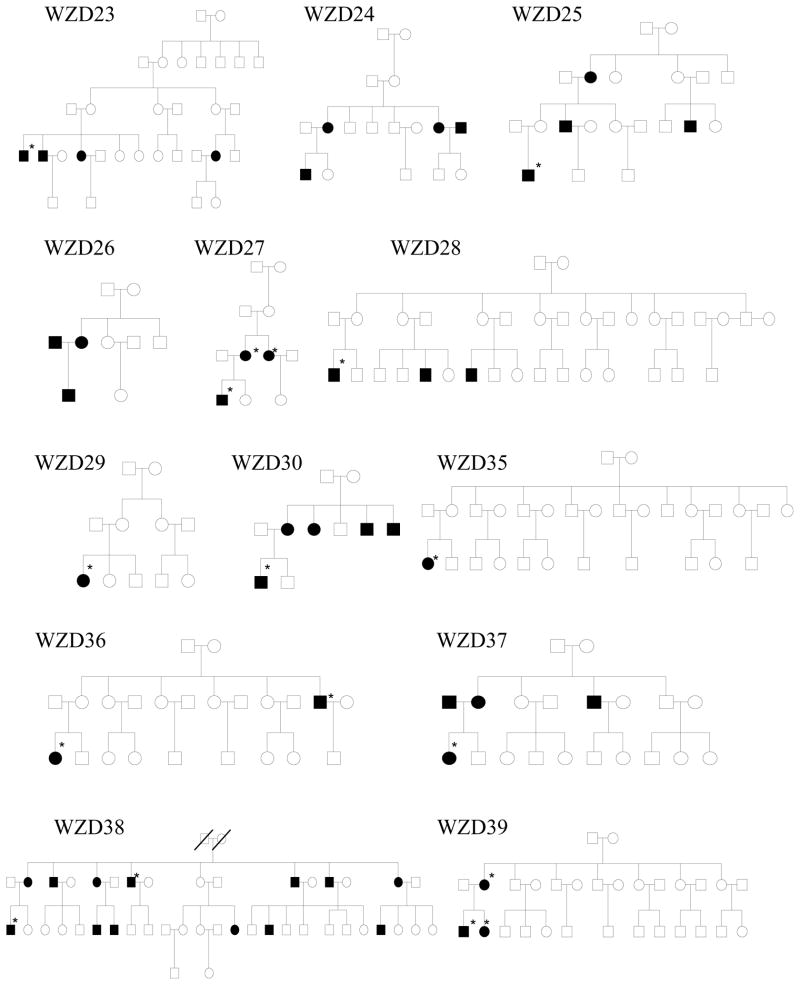
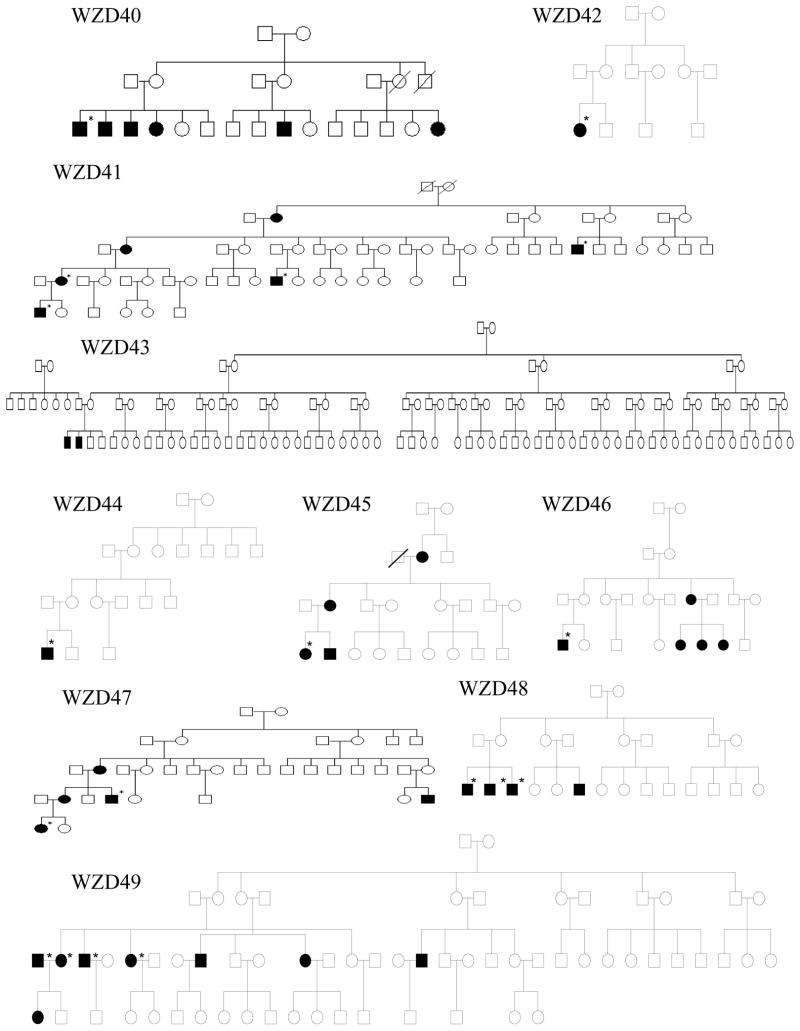
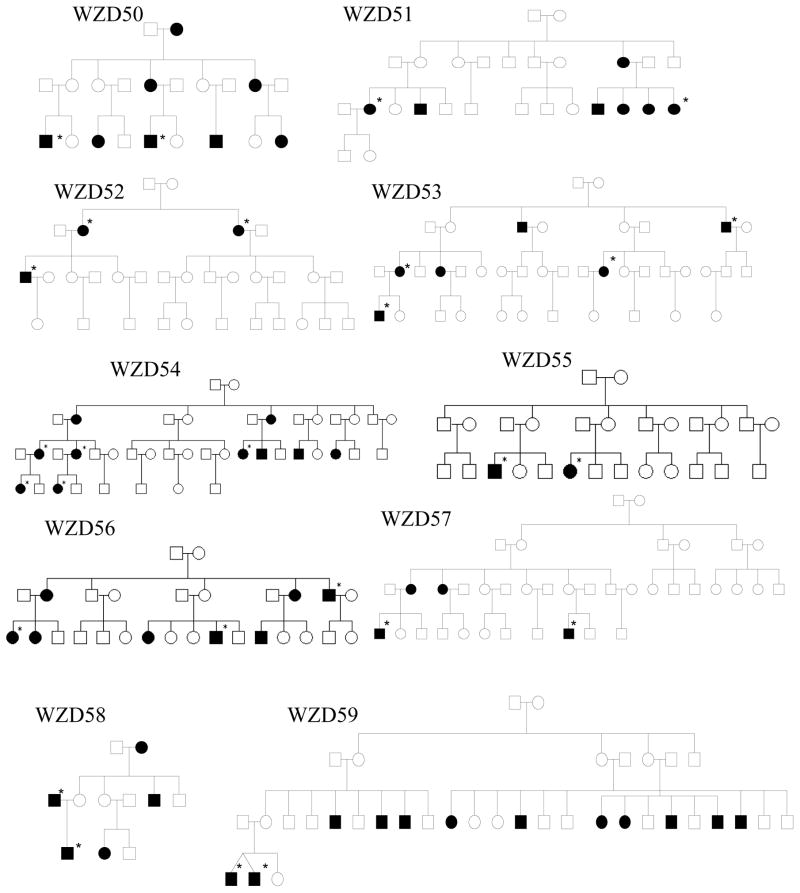
As shown in Figure 1 and Table 1, one hundred eighty-two of 786 matrilineal relatives among 43 Chinese pedigrees exhibited bilateral and sensorineural hearing impairment. All affected individuals showed the loss of high frequencies. Of these, 71 hearing-impaired matrilineal relatives, who had a history of exposure to gentamicin and/or streptomycin, had subsequent severe-to-profound hearing loss. They received a regular dose of aminoglycosides (3–5 mg/kg/dose every 8 h for gentamicin or 15–25 mg/kg/dose every 12 h for streptomycin) for various illness at the age of 3 months to 10 years old. Hearing loss usually occurred from 3 days to 3 months after drug administration. In the absence of aminoglycosides, matrilineal relatives of these Chinese families exhibited congenital to late-onset hearing impairment. Audiometric studies showed a variable severity of hearing impairment in the matrilineal relatives of these families, ranging from severe hearing impairment, to moderate hearing impairment, to mild hearing impairment, to completely normal hearing. Furthermore, there was a wide range in the age at onset of hearing impairment in these families, varying from infant to 65 years old. As shown in Table 1, the average age-at-onset (excluding the use of aminoglycosides) of each family in these 43 pedigrees varied from 5 years to 30 years old. In addition, there was a wide range of the penetrance of hearing loss among these pedigrees. In particular, the penetrances of hearing loss in these pedigrees ranged from 3.2% to 62.5%, when aminoglycoside-induced hearing loss was included. However, the average penetrances of hearing loss in these pedigrees varied from 0% to 50%, when aminoglycoside-induced hearing loss was excluded. In particular, none of matrilineal relatives in 16 out of 43 pedigrees here suffered from hearing loss, when aminoglycoside-induced hearing loss was excluded.
Figure 1. Forty-three Han Chinese pedigrees with aminoglycoside-induced and nonsyndromic hearing impairment.
Hearing impaired individuals are indicated by filled symbols. Asterisks denote individuals who had a history of exposure to aminoglycosides.
TABLE 1.
Summary of clinical and molecular data for 69 Chinese families carrying the A1555G mutation
| Pedigree | Number of matrilineal relatives | a Penetrance (including the use of drugs) (%) | Penetrance (excluding the use of drugs) (%) | Average age-at-onset (excluding the use of drugs) | Second mtDNA mutations | mtDNA haplogroup |
|---|---|---|---|---|---|---|
| WZD13 | 15 | 6.7 | 0 | None | D4 | |
| WZD14 | 6 | 16.7 | 0 | None | D4a | |
| WZD15 | 22 | 9.1 | 4.5 | 5 | None | F1a |
| WZD16 | 16 | 31.3 | 31.3 | 12 | None | N9a3 |
| WZD17 | 17 | 5.9 | 0 | None | D5b1 | |
| WZD18 | 10 | 10 | 0 | None | D5a | |
| WZD19 | 12 | 8.3 | 0 | None | D5a | |
| WZD20 | 16 | 12.5 | 0 | None | R9b | |
| WZD21 | 25 | 16 | 12 | 15 | None | D4 |
| WZD22 | 29 | 41.4 | 31 | 14 | tRNACys T5802C | D4 |
| WZD23 | 22 | 18.2 | 13.6 | 18 | None | D4 |
| WZD24 | 11 | 36.4 | 36.4 | 12 | None | D5a |
| WZD25 | 13 | 30.8 | 23.1 | 16 | None | F1a |
| WZD26 | 6 | 33.3 | 0 | None | M10a | |
| WZD27 | 7 | 42.9 | 0 | None | M7b1 | |
| WZD28 | 24 | 12.5 | 8.3 | 16 | None | N9a |
| WZD29 | 8 | 12.5 | 0 | None | D4 | |
| WZD30 | 8 | 62.5 | 50 | 11 | None | N9a |
| WZD35 | 15 | 6.7 | 0 | None | F1 | |
| WZD36 | 15 | 13.3 | 0 | None | D4h | |
| WZD37 | 30 | 30 | 20 | 16 | None | F2 |
| WZD38 | 22 | 59.1 | 50 | 11 | None | D5 |
| WZD39 | 16 | 18.8 | 0 | None | M7 | |
| WZD40 | 20 | 30 | 25 | 12 | ND5 T12338C | F2 |
| WZD41 | 37 | 16.2 | 5.4 | 30 | None | N9a |
| WZD42 | 7 | 14.3 | 0 | None | G | |
| WZD43 | 62 | 3.2 | 3.2 | 9 | None | F1a1 |
| WZD44 | 14 | 7.1 | 0 | None | B4 | |
| WZD45 | 12 | 33 | 25 | 15 | tRNACys G5821A | C |
| WZD46 | 14 | 35.7 | 28.6 | 13 | None | D5a |
| WZD47 | 25 | 20 | 12 | 14 | None | N9a3 |
| WZD48 | 15 | 26.7 | 6.7 | 18 | None | B4a1 |
| WZD49 | 30 | 23.3 | 13.3 | 10 | None | F2 |
| WZD50 | 15 | 53.3 | 46.7 | 13 | tRNASer(AGY) C12224T | D5 |
| WZD51 | 18 | 38.9 | 27.8 | 15 | tRNAGlu A14693G | Y2 |
| WZD52 | 19 | 15.8 | 0 | None | D4b | |
| WZD53 | 17 | 35.3 | 11.8 | 21 | None | F2 |
| WZD54 | 25 | 40 | 20 | 18 | None | A |
| WZD55 | 15 | 13.3 | 0 | None | D5a | |
| WZD56 | 17 | 47.1 | 29.4 | 13 | None | D5b |
| WZD57 | 20 | 20 | 10 | 20 | None | F3b |
| WZD58 | 8 | 50 | 37.5 | 13 | CO1/tRNASer(UCN) G7444A | B4b1c |
| WZD59 | 31 | 41.9 | 35.5 | 12 | CO1/tRNASer(UCN) G7444A | B4 |
| WZD31b | 12 | 66.7 | 50 | 14 | tRNAThr G15927A | B5b1 |
| WZD32 | 12 | 66.7 | 41.7 | 13 | tRNAThr G15927A | B5b1 |
| WZD33 | 27 | 51.9 | 44.4 | 16 | tRNAThr G15927A | B5b1 |
| WZD34 | 19 | 50 | 39 | 15 | tRNAThr G15927A | B5b1 |
| WZD8c | 13 | 46 | 23 | 13 | tRNACys T5802C | D4b2b |
| WZD9 | 13 | 46 | 31 | 16 | tRNAThr G15927A | B5b1 |
| WZD10 | 8 | 50 | 37.5 | 12 | ND5 T12338C | F2 |
| WZD11 d | 19 | 53 | 42 | 12 | ND4 G11696A | D4 |
| WZD12 | 12 | 58 | 25 | 11 | CO1/tRNASer(UCN) G7444A | B4c1 |
| WZD1e | 17 | 5.9 | 0 | none | B4C1C1 | |
| WZD2 | 21 | 9.5 | 4.7 | 19 | none | D4a |
| WZD3 | 16 | 12.5 | 0 | none | D5a2 | |
| WZD4 | 24 | 29.2 | 16.7 | 20 | none | F1a1 |
| WZD5 | 31 | 3.2 | 0 | none | D5a2b | |
| WZD6 | 8 | 25 | 12.5 | 19 | none | D4b2b |
| WZD7 | 30 | 10 | 3.3 | 21 | none | D5a2 |
| BJ101 f | 14 | 5 | 0 | none | F3 | |
| BJ102 | 13 | 13 | 8 | 18 | none | N9a1 |
| BJ103 | 20 | 5 | 0 | none | D4a | |
| BJ104 | 11 | 4 | 0 | none | D4b2b | |
| BJ105 g | 15 | 67 | 45 | 13 | tRNACys G5821A | C |
| BJ106 | 18 | 33 | 33 | 15 | none | M7b |
| BJ107h | 34 | 35 | 24 | 11 | tRNAThr T15908C | Y2 |
| BJ108 | 16 | 63 | 38 | 14 | tRNAGlu A14693G | F2 |
| BJ109 | 9 | 67 | 44 | 12 | tRNAArg T10454C | D4 |
| BJ110i | 17 | 59 | 5.9 | 5 | CO1/tRNASer(UCN) G7444A | D4a |
affected matrilineal relatives/total matrilineal relatives;
Analysis of the complete mitochondrial genomes
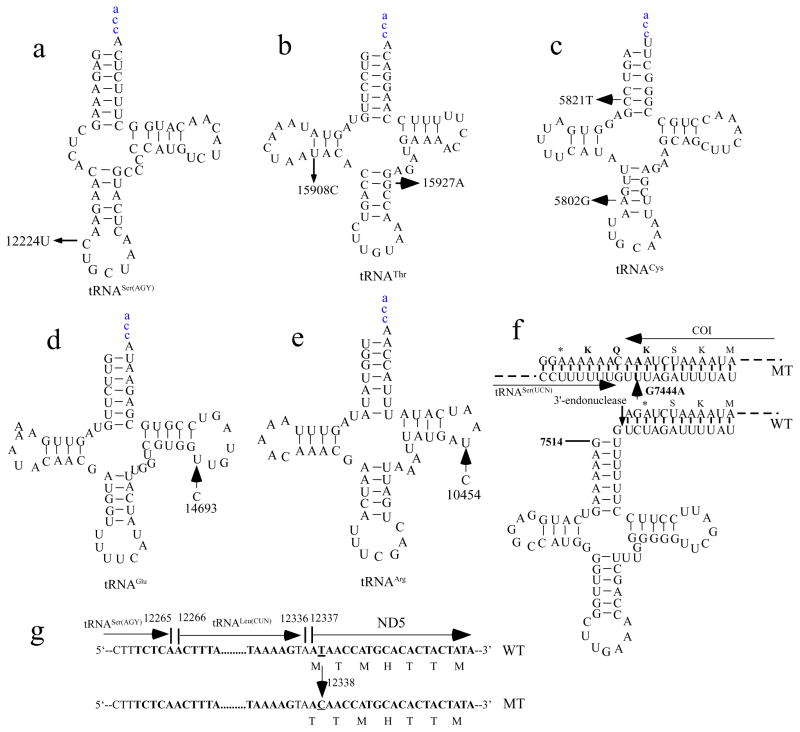
To assess the contribution that mtDNA variants or haplogroups make toward the variable penetrance and expressivity of hearing loss in these Chinese pedigrees, we performed a PCR-amplification of fragments spanning entire mtDNA and subsequent DNA sequence analysis in 43 Chinese probands carrying the 1555A>G mutation and 93 normal hearing Chinese controls. The sequence results from these Chinese subjects were aligned with the updated consensus Cambridge sequence (Andrews et al. 1999). In addition to the identical 1555A>G mutation, these probands, as shown in Table 2 (supplement data), exhibited distinct sets of mtDNA polymorphisms including a number of known and novel variants. Of the known variants (Brandon et al. 2005), five deafness-associated secondary mtDNA mutations were identified in the following probands: tRNACys 5802T>C (WZD22 pedigree), tRNACys 5821G>A (WZD45 pedigree), ND5 12338T>C (M1T) (WZD40 pedigree), tRNAGlu 14693A>G (WZD51 pedigree) and CO1/tRNASer(UCN) 7444G>A (WZD58 and WZD59 pedigrees). Of the novel 30 variants, as shown in Table 3, there were one polymorphism in the D-loop region, one variant in the 16S rRNA gene, the 12224C>T variant in the tRNASer(AGY) gene, 17 silent variants and 19 missense mutations in the protein encoding genes. These variants in RNAs and polypeptides were further evaluated by phylogenetic analysis of these variants and sequences from other organisms including mouse (Bibb et al. 1981), bovine (Gadaleta et al. 1989), and Xenopus laevis (Roe et al. 1985). The 7331C>A (F476L) variants in the CO1 gene and the 12224C>T variant in the tRNASer(AGY) gene showed evolutionary conservation in these species. The 12224C>T variant, as shown in Fig. 2, occurs adjacent to anticodon stem, corresponding to conventional position 32 of the tRNASer(AGY) (Florentz et al. 2003). In fact, an cytosine at this position is a highly conserved base in sequenced tRNASer(AGY) from bacteria to human mitochodria (Sprinzl et al. 1998). It is anticipated that the 12224C>T mutation creates a novel base-pairing (32U-38A) on the anticodon stem of this tRNASer(AGY), thus causing a failure in tRNA metabolism. Allele frequency analysis in 262 hearing normal Chinese control subjects showed that three subjects carried the 7331C>A (F476L) variants in the CO1 gene, while none of 262 Chinese controls harbored the 12224C>T variant in the tRNASer(AGY) gene.
Table 2.
mtDNA variants in 43 hearing-impaired Chinese pedigrees carrying the A1555G mutation
| Gene | Position | Replacement | CRSb | WZD13 | WZD14 |
|---|---|---|---|---|---|
| D-LOOP | 73 | A to G | A | G | G |
| 94 | G to A | G | |||
| 139 | T to C | T | |||
| 146 | T to C | T | |||
| 150 | C to T | C | |||
| 152 | T to C | T | C | C | |
| 193 | A to G | A | |||
| 194 | C to T | C | |||
| 195 | T to C | T | |||
| 199 | T to C | T | |||
| 200 | A to G | A | |||
| 207 | G to A | G | |||
| 215 | A to G | A | |||
| 227 | A to T | A | T | ||
| 235 | A to G | A | |||
| 240 | A to G | A | |||
| 248 | del A | A | |||
| 249 | del A | A | |||
| 263 | A to G | A | G | G | |
| 279 | T to C | T | |||
| 298 | C to T | C | |||
| 307 | C to T | C | |||
| 310 | T to A/CTC/CCTC/TC | T | CTC | CTC | |
| 311 | del CC | CC | |||
| 312 | C to TC | ||||
| 319 | T to G | ||||
| 456 | C to T | C | |||
| 482 | T to C | T | |||
| 489 | T to C | T | C | C | |
| 499 | G to A | G | |||
| 513 | G to A | G | |||
| 514 | del C | C | |||
| 515 | del A | A | |||
| 514-517DEL | 514-517del | ||||
| 521-524 | del ACAC | ||||
| 523 | del A | A | |||
| 524 | del C | C | |||
| 566? | C-CCCC | ||||
| 568 | C-CCCC | ||||
| 574 | A to G | A | |||
| 16038 | A to G | A | |||
| 16066 | A to G | A | |||
| 16086 | T to C | T | |||
| 16092 | T to C | T |
Table 3.
Novel mtDNA variants in 43 hearing-impaired Chinese pedigrees carrying the A1555G mutation
| Gene | Position | Replacement | Conservation (H/B/M/X)a | Pedigrees |
|---|---|---|---|---|
| D-loop | 139 | T to C | WZD29 | |
| 16S rRNA | 1677 | C to T | A/A/T/A | WZD30 |
| ND1 | 4084 | G to A (V260I) | V/M/M/L | WZD20 |
| ND2 | 5131 | T to C (L221S) | L/A/L/T | WZD28 |
| CO1 | 5985 | G to A | WZD15 | |
| 6020 | C to T | WZD40 | ||
| 6113 | A to G | WZD16, WZD47 | ||
| 6191 | C to T | WZD20 | ||
| 6228 | C to T (L109F) | L/F/F/F | WZD39 | |
| 6269 | A to C | WZD36, WZD13 | ||
| 6918 | C to T (L339P) | L/M/L/L | WZD18 | |
| 7181 | C to T | WZD36, WZD13 | ||
| 7331 | C to A (P476L) | F/F/F/F | WZD23 | |
| ATP8 | 8412 | T to C (M16T) | M/M/S/S | WZD50 |
| 8488 | C to T | WZD54 | ||
| ATP6 | 8586 | A to G | WZD37 | |
| 8883 | T to C | WZD21 | ||
| CO3 | 9980 | A to G | WZD37 | |
| ND3 | 10327 | C to T (S90L) | S/M/M/W | WZD42 |
| ND4 | 10776 | T to C (V6A) | V/I/L/L | WZD50 |
| 11471 | C to T | WZD35 | ||
| tRNASer(AGY) | 12224 | C to T | C/C/C/C | WZD50 |
| ND5 | 12489 | A to G | WZD59 | |
| 12781 | A to G (I149V) | I/I/I/V | WZD59 | |
| 13442 | C to T (T369I) | T/T/T/I | WZD59 | |
| 13677 | C to T | WZD48 | ||
| 13959 | C to T | WZD52 | ||
| 14042 | A to G (H589R) | H/A/L/Q | WZD39 | |
| Cytb | 15373 | A to G | WZD25 | |
| 15445 | T to C | WZD15 |
Conservation of amino acid for polypeptides or nucleotide for RNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X).
Figure 2. Sites of secondary mtDNA mutations in Chinese pedigrees.
(a) The 12224C>T mutation in the secondary structure of tRNASer(AGY); (b) The 15908T>C and 15927G>A mutations in the secondary structure of tRNAThr; (c) The 5821C>T and 5802A>G mutations in the secondary structure of tRNACys; (d) The 14693A>G mutation in the secondary structure of tRNAGlu; (e) The 10454T>C mutation in the secondary structure of tRNAArg. (f). The 7444G>A mutation in the CO1 and tRNASer(UCN); (g). A schema of mtDNA sequence at position 12338 and adjacent sequence of ND5 and tRNALeu(CUN) from wild-type (WT) and mutant (MT). Arrows indicate the positions of the mtDNA mutations.
Phylogenetic and haplogroup analysis of mtDNAs carrying the 1555A>G mutation
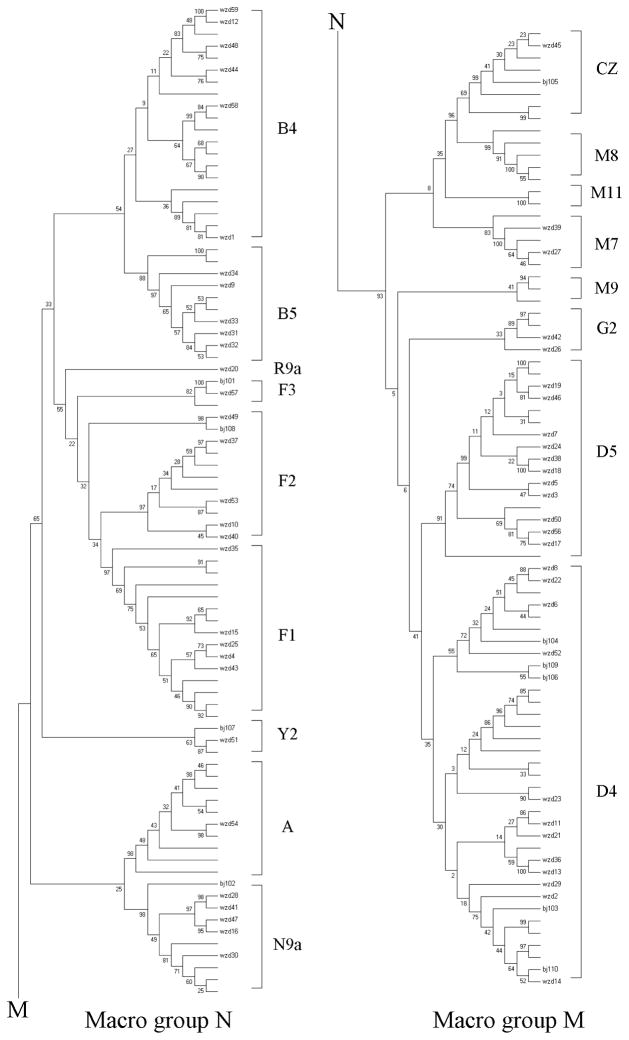
As shown in Table 1, the mtDNAs from 69 Chinese families carrying the 1555A>G mutation were distributed among ten different haplogroups (Tanaka et al. 2004; Kong et al. 2006). To determine if this distribution of deafness pedigrees corresponding to that of general Chinese population, 93 hearing normal Chinese control mtDNAs recruited from the same region were sequenced and assigned to certain Asian haplogroups. The phylogenetic tree including 69 Chinese pedigrees carrying the 1555A>G mutation and 93 Chinese controls is shown in Figure 3. Here, all mtDNA lineages fall into two macro-haplogroups M and N, of which Eastern Asian mtDNAs were subdivided (Tanaka et al. 2004; Kong et al. 2006). Indeed, the 1555A>G mutation is widely dispersed among ten common Eastern Asian subhaplogroups. As shown in Table 4, the frequencies of mtDNA haplogroups A, B, C, D, F, G, M, N, R and Y in 69 hearing-impaired families were 1.4%, 15.9%, 2.9%, 40.6%, 18.8%, 1.4%, 2.9%, 8.7%, 1.4% and 2.9%, respectively, while those of 93 Chinese controls were 9.7%, 20.4%, 4.3%, 25.8%, 17.2%, 2.2%, 14%, 4.3%, 0% and 1.1%, respectively. Strikingly, the haplogroup D accounted for 40.6% of the patient’s mtDNA samples but only 25.8% of the Chinese control mtDNA samples in this study and 22.2% of a large cohort of Eastern Asian population (Tanaka et al. 2004; Kong et al. 2006). Of other haplogroups, the frequency of haplogroup Y and N in the patients’ mtDNA samples was higher than those of control’s mtDNA samples, while the frequency of haplogroups A, B, C, G and M in hearing-impaired mtDNA samples were lower than those of control mtDNA samples.
Figure 3. Phylogenetic tree based on complete mtDNA sequences of 69 hearing-impaired Chinese subjects and 93 Chinese controls.
Hearing-impaired subject’s origins were given in Table 1. The capital letters A, M, N, B, C, D, G, F, R, and Y indicate haplogroups. The Neighbor-Joining phylogenetic tree was constructed using Software Version 4.0. (Tamura et al. 2007). Phylogenetic relationships among these samples were determined by bootstrapping tests (1000 times) (Marshall 1991). The numbers at interior branches referred to the bootstrapping values.
Table 4.
Haplogroup distribution of mtDNA from 69 Han Chinese hearing-impaired families carrying the A1555G mutation and from 93 Chinese control subjects
| Group | A | B | C | D | F | G | M | N | R | Y | Z | Average | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency of mtDNA haplogroup (%) | Controls (n=93) | 9.7 | 20.4 | 4.3 | 25.8 | 17.2 | 2.2 | 14 | 4.3 | … | 1.1 | 2.2 | |
| Deafness subjects (n=69) | 1.4 | 15.9 | 2.9 | 40.6 | 18.8 | 1.4 | 2.9 | 8.7 | 1.4 | 2.9 | … | ||
| Average penetrance (including the use of drugs) | 40 | 42.8 | 50 | 24.0 | 27.4 | 14.3 | 33.5 | 26 | 12.5 | 37 | … | 29.4 | |
| Average penetrance (excluding the use of drugs) | 16 | 34.5 | 35 | 15.1 | 17.7 | 0 | 0 | 18.8 | 0 | 25.9 | … | 17.6 | |
| Average age-at-onset (excluding the use of drugs) (years) | 18 | 14.2 | 14 | 14.2 | 14 | … | … | 16.8 | … | 13 | … | 14.5 | |
Penetrance (%) = affected matrilineal relatives/total matrilineal relatives
Statistical analysis
As shown in Table 4, the average penetrances of hearing loss among 69 Chinese pedigrees carrying the 1555A>G mutation were 29.4% and 17.6%, when aminoglycoside-induced deafness was included or excluded, respectively and the average age-at-onset of hearing loss among 69 Chinese pedigrees was 14.5 years old. To further evaluate the role of mitochondrial genetic modifiers in deafness expression, statistical analysis was carried out using the unpaired, two-tailed Student’s t-test. Here, the average penetrances of hearing loss among 21 Chinese pedigrees carrying one of additional potentially functional significant mtDNA variants such as 12338T>C variant [50% and 34%, when aminoglycoside-induced deafness was included or excluded] showed significantly higher than 47 pedigrees lacking significant mtDNA variants [19.7% and 9.9%, when aminoglycoside-induced deafness was included or excluded] (p<0.0001) and (p<0.0001), respectively. Similarly, the average age-at-onset (12.9 years old) of hearing loss among 21 Chinese pedigrees carrying one of the additional mtDNA variants showed significantly younger than those (15.7 years old) among 47 pedigrees lacking significant mtDNA variants (p<0.0148). Furthermore, the average penetrance of hearing loss among mtDNA haplogroups A, B, C, D, F, G, M, N, R and Y ranged from 12.5% to 50% or 0% to 35%, when aminoglycoside-induced deafness was included or excluded, respectively, while the age-at-onset of hearing loss among ten haplogroups varied from 13 to 18 years old. In particular, the average penetrances of hearing loss among 11 Chinese pedigrees carrying haplogroup B [42.8% and 34.5%, when aminoglycoside-induced deafness was included or excluded] showed significantly higher than those among 69 pedigrees carrying the 1555A>G mutation (p<0.0001) and (p<0.0001), respectively. On the other hand, there were no statically significant difference of age-at-onset of hearing loss between 11 Chinese pedigrees carrying haplogroup B and average value of 69 Chinese pedigrees. In addition, the difference of average penetrances and age-at-onset of hearing loss between one of other 9 haplogroups and average value of 69 Chinese pedigrees was not statically significant, although the penetrances of hearing loss among pedigrees carrying the haplogroup C, Y and F2 were higher than the average values among 69 families carrying the 1555A>G mutation.
Mutational analysis of TRMU gene
Our previous study showed that the TRMU 10A>S mutation modulated the phenotypic manifestation of the 1555A>G mutation in the Israeli/European pedigrees (Guan et al. 2006). To determine if TRMU modulates the phenotypic expression of the 1555A>G in these Chinese families, we performed the PCR-amplification and sequence analysis of DNA fragments spanning 11 exons and their flanking sequences of TRMU using DNA samples derived from 69 affected matrilineal relatives, who did not have a history of exposure to aminoglycosides, of 69 Chinese pedigrees carrying the 1555A>G mutation and 11 unrelated Chinese controls lacking the 1555A>G mutation. However, no sequence changes in this gene were identified among these Chinese individuals. The absence of TRMU mutation suggested that TRMU may not play an important role in the phenotypic expression of the 1555A>G mutation in these Chinese families.
DISCUSSION
In the present study, we investigated the contribution of mitochondrial genetic modifiers to the phenotypic expression of deafness-associated 12S rRNA mutation. The incidence of the 1555A>G mutation was 3.96% in a cohort of 1742 hearing-impaired Chinese pediatric subjects. Deafness as a sole clinical phenotype was only present in the maternal lineage of 69 Chinese pedigrees carrying the 1555A>G mutation. A wide range of penetrance and expressivities of hearing impairment was observed in the Chinese families. The penetrances of hearing loss (affected matrilineal relatives/total matrilineal relatives) in 69 Chinese pedigrees ranged from 3.2% to 67%, with the average of 29.5%, when aminoglycoside-induced deafness was included. When the effect of aminoglycosides was excluded, the penetrances of hearing loss in these pedigrees ranged from 0% to 47.8%, with the average of 17.6%. In particular, none of matrilineal relatives in these Chinese 23 pedigrees developed hearing loss in the absence of aminoglycosides. By contrast, the penetrances of hearing loss in a large Arab-Israeli family and 19 Spanish pedigrees without a record of aminoglycoside treatment were 65.4% and 54.1%, respectively (Estivill et al. 1998; Bykhovkaya et al. 1998). Furthermore, when aminoglycoside-induced deafness was excluded, the average age-of-onset for hearing loss ranged from 5 and 30 years old, with the average of 14.5 years old in these 69 Chinese families. On the contrary, the average age-of-onset for hearing loss ranged from 1 and 65 years old, with the average of 20 years old, in 19 Spanish families in the absence of aminoglycoside exposure (Estivill et al. 1998).
The genetic and environmental modifier factors are apparently responsible for the variable penetrance and expressivity of hearing loss among these pedigrees carrying the 1555A>G mutation. The nuclear modifier genes most likely contributed to the phenotypic variability of matrilineal relatives within and among these Chinese families as described other pedigrees (Guan et al. 1996; 1998; 2006). In particular, the TRMU 10A>S variant worsened mitochondrial dysfunctions caused by the 1555A>G mutation, thereby leading to a deafness phenotype in the Arab-Israeli and European families (Guan et al. 2006). The lack of TRMU mutation in 69 affected matrilineal relatives in these Chinese families suggested that this gene may not play an important role in the phenotypic expression of the 1555A>G mutation. However, other nuclear modifier genes may modulate the disease expression of the 1555A>G mutation in these Chinese pedigrees, as in the case of other pedigrees (Bykhovkaya et al. 1998; Guan et al. 2006). The fact that some matrilineal relatives of these 69 pedigrees exhibited severe or profound hearing loss after an administration with aminoglycosides demonstrated that these antibiotics play a significant role in enhancing the penetrance and severity of hearing loss in these Chinese families. Therefore, children carrying the ototoxic mtDNA 1555A>G mutation are susceptible to the exposure of aminoglycosides, thereby inducing or worsening hearing impairment (Fischel-Ghodsian 2005; Guan 2005; Qian and Guan 2009). To diagnose whether children carry these ototoxic mtDNA mutations will enable us to predict which individuals are at risk for ototoxicity, to improve the safety of aminoglycoside therapy, and eventually to decrease the incidence of deafness.
Mitochondrial haplogroups have been shown to modulate the phenotypic expression of mtDNA mutation(s) associated with some clinical abnormalities including deafness (Guan et al. 1998; Li et al. 2004c), blindness (Torroni et al. 1997; Hudson et al. 2007), ageing (Coskun et al. 2003), Parkinson’s disease (Vander Walt et al. 2003), male infertility (Ruiz-Pesini et al. 2000) and diabetes (Fuku et al. 2007). Here, distinct sets of polymorphisms in their mtDNA of 69 Chinese pedigrees carrying the 1555A>G mutation, as shown in Figure 3, belonged to the ten Eastern Asian haplogroups A, B, C, D, F, G, M, N, R and Y, respectively (Tanaka et al. 2004; Kong et al. 2006). However, mtDNAs of Spanish pedigrees carrying the 1555A>G mutation belonged to the European haplogroups H, I, J, K, T, U, V and L (Torroni et al. 1999; de Castillo et al. 2003). These suggested that the 1555A>G mutation occurred sporadically and multiplied through the evolution of the mtDNA. A strong excess of haplogroup D-mtDNA carrying the 1555A>G mutation was comparable with the fact that mtDNA among hearing-impaired Spanish families carrying the 1555A>G mutation belonged to predominate H-haplogroup (Torroni et al. 1999; Achilli et al. 2004). In contrast, the lower frequency of haplogroups A, C, G, R and Y in these Chinese families suggested that the 1555A>G mutation on these mitochondrial backgrounds would be the independent mutational events. To further evaluate the role of haplogroups in deafness expression, the average penetrances and age-at-onset of hearing loss were compared between each haplogroup and average values of 69 Chinese pedigrees by using the unpaired, two-tailed Student’s t-test. Strikingly, the average penetrances of hearing loss among 11 Chinese pedigrees carrying haplogroup B showed significantly higher than those among 69 pedigrees carrying the 1555A>G mutation. However, the difference of average penetrances and age-at-onset of hearing loss between one of other 9 haplogroups and average value of 69 Chinese pedigrees was not statically significant, although the penetrances of hearing loss among pedigrees carrying the haplogroup C, Y and F2 were higher than the average values among 69 families carrying the 1555A>G mutation. In addition, there were no statically significant differences of age-at-onset of hearing loss between average value of 11 Chinese pedigrees carrying haplogroup B and average value of 69 Chinese pedigrees. These data suggest that the haplogroup B increases the penetrance of hearing loss in Chinese subjects carrying the 1555A>G mutation.
Mitochondrial haplogroup specific variants were implicated to contribute to the phenotypic manifestation of pathogenic mtDNA mutations. In particular, the haplogroup J specific variants 4216T>C and 13708G>A may increase the penetrance of vision loss associated ND4 11778G>A mutation (Torroni et al. 1997) or hearing loss associated with tRNASer(UCN) 7445A>G mutation (Guan et al. 1998), while the haplogroup L1b specific variants ND1 3308T>C and tRNAAla 5655T>C likely caused higher penetrance of deafness in an African pedigree than Japanese and French families carrying the 7511T>C mutation (Li et al. 2004c). In our investigations, ten haplogroup specific variants: tRNAThr 15927G>A of haplogroup B5b, ND1 12338T>C of haplogroup F2, tRNASer(UCN)/CO1 7444G>A of haplogroup B4, tRNACys 5802T>C, tRNAArg 10454T>C, tRNASer(AGY) 12224C>T and ND4 11696G>A of D4 haplogroup, tRNACys 5821G>A of haplogroup C, tRNAGlu 14693A>G of haplogroups Y2 and F, and tRNAThr 15908T>C of haplogroup Y2, were implicated to enhance the penetrance and expressivity of hearing loss among these Chinese families carrying the 1555A>G mutation. These variants localized at highly conserved nucleotides of tRNAs or amino acid of polypeptides and may cause potential structural and functional alterations. In addition, these mtDNA variants were present in homoplasmy only on the maternal lineage in these Chinese pedigrees and the allelic frequencies of these variants were less than 1% in the Chinese controls (Zhao et al. 2005; Young et al. 2006; Chen et al. 2008; Wang et al. 2008). Of these, the 15927G>A mutation locates at the fourth base in the anticodon stem (conventional position 42) of the tRNAThr. A guanine (G42) at this position of tRNAThr is highly conserved from bacteria to human mitochondria (Sprinzl et al. 1998; Florentz et al. 2003). The abolished base-pairing (28C-42G) of this tRNAThr by the 15927G>A mutation likely altered this tRNA metabolism. Functional significance of this variant was supported by the fact that the lower levels and altered electrophoretic mobility of tRNAThr were observed in cells carrying 1555A>G and 15927G>A mutations or only 15927G>A mutation but not cells carrying only 1555A>G mutation (Wang et al. 2008). Furthermore, the 7444G>A mutation resulted in a read-through of the stop codon AGA of the CO1 message, thereby adding three amino acids (Lys-Gln-Lys) to the C-terminal of polypeptide (Pandya et al. 1999; Yuan et al. 2005). Alternatively, the 7444G>A mutation is adjacent to the site of 3′ prime end endonucleolytic processing of L-strand RNA precursor, spanning tRNASer(UCN) and ND6 mRNA (Guan et a.. 1998; Yuan et al. 2005). Thus, the 7444G>A mutation may lead to a defect in the processing of the L-strand RNA precursor. Moreover, the haplogroup F2 specific variant ND5 12338T>C mutation resulted in the replacement of the first amino acid, translation-initiating methionine with a threonine (Chen et al. 2008). Thus, the truncated ND5 protein was shortened by two amino acids. The ND5 12338T>C mutation also locates in two nucleotides adjacent to the 3′ end of the tRNALeu(CUN) (Chen et al. 2008). As a result, the ND5 12338T>C mutation, similar to ND1 3308T>C mutation (Li et al. 2004c), may cause a decrease in ND5 mRNA level as well as alter the processing of RNA precursors, thereby leading to a reduction in tRNALeu(CUN) level. In addition, the 14693A>G mutation occurs at the first base (conventional position 54) of the TψC-loop of tRNAGlu, while the T-to-C mutation at position 10454 is located at the second base (conventional position 55) of the TψC loop of tRNAArg (Florentz et al. 2003). Indeed, nucleotides at position of 54, and 55 of the TψC loop of tRNA are often modified and thereby contribute to the structural formation and stabilization of functional tRNAs (Björk et al. 1995). As shown in Figure 2, the 5802T>C, 5821G>A and 15908T>C variants disrupted highly conserved base-pairings of these tRNAs (Ruiz-Pesini and Wallace 2006), while the 12224C>U variant occurs adjacent to anticodon (conventional position 32) of tRNASer(AGY) (Sprinzl et al. 1998; Florentz et al. 2003). Thus, the alteration of the tertiary structure of the mitochondrial tRNA by these variants may lead to a failure in this tRNA metabolism. In addition, the ND4 11696G>A mutation was associated with LHON and conferred a mild biochemical defect (De vries et al. 1996; Zhou et al. 2006). To further assess if the differences in penetrance and age-at-onset of hearing loss without aminoglycosides differ based on the presence and absence of additional mtDNA variant, a statistical analysis was performed by the unpaired, two-tailed Student’s t-test. The penetrances and age-at-onset of hearing loss among 22 Chinese pedigrees carrying one of additional mtDNA variants showed significantly higher or younger than 47 pedigrees lacking significant mtDNA variants, when aminoglycoside-induced deafness was excluded. Therefore, these deafness-associated secondary mutations, similar to the LHON-associated secondary tRNAMet 4435A>G and tRNAThr 15951A>G variants (Li et al. 2006; Qu et al. 2006), may worsen mitochondrial dysfunctions caused by the 1555A>G mutation, thereby increasing the penetrance and expressivity of hearing loss in these Chinese pedigrees.
Acknowledgments
FUNDING
This work was supported by Public Health Service grants RO1DC05230 and RO1DC07696 from the National Institute on Deafness and Other Communication Disorders, and grants from National Basic Research Priorities Program of China 2004CCA02200, Ministry of Public Heath of Zhejiang Province 2006A100 and Ministry of Science and Technology of Zhejiang Province 2007G50G2090026 to M.X.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achilli A, Rengo C, Magri C, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. 21 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33:D611–613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR. Biosynthesis and function of modified nucleotides. In: Söll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis and Function. Washington, DC: ASM Press; 1995. pp. 165–206. [Google Scholar]
- Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, Cantor RM, Aouizerat B, Bu X, Rotter JI, Jaber L, Fischel-Ghodsian N. Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet. 1998;77:421–426. doi: 10.1002/(sici)1096-8628(19980605)77:5<421::aid-ajmg13>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Chen B, Sun D, Yang L, et al. Mitochondrial ND5 12338T>C, tRNACys 5802T>C, and tRNAThr 15927G>A variants may have a modifying role in the phenotypic manifestation of deafness-associated 12S rRNA 1555A>G mutation in three Han Chinese pedigrees. Am J Med Genet. 2008;146A:1248–1258. doi: 10.1002/ajmg.a.32285. 16 co-authors. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Ruiz-Pesini E, Wallace DC. Control region mtDNA variants: longevity, climatic adaptation, and a forensic conundrum. Proc Natl Acad Sci USA. 2003;100:2174–2176. doi: 10.1073/pnas.0630589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, et al. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40:632–636. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Liu X, Han D, et al. Extremely low penetrance of deafness associated with the mitochondrial 12S rRNA mutation in 16 Chinese families: Implication for early detection and prevention of deafness. Biochem Biophys Res Commun. 2006;340:194–199. doi: 10.1016/j.bbrc.2005.11.156. 20 co-authors. [DOI] [PubMed] [Google Scholar]
- De Vries DD, Went LN, Bruyn GW, Scholte HR, Hofstra RM, Bolhuis PA, van Oost BA. Genetic and biochemical impairment of mitochondrial complex I activity in a family with Leber hereditary optic neuropathy and hereditary spastic dystonia. Am J Hum Genet. 1996;58:703–711. [PMC free article] [PubMed] [Google Scholar]
- Estivill X, Govea N, Barcelo E, Badenas C, Romero E, Moral L, Scozzari R, D’Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N. Genetic factors in aminoglycoside toxicity. Pharmacogenomics. 2005;6:27–36. doi: 10.1517/14622416.6.1.27. [DOI] [PubMed] [Google Scholar]
- Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuku N, Park KS, Yamada Y, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007;80:407–415. doi: 10.1086/512202. 13 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Guan MX. Prevalence of mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. The Voltra Review. 2005;105:211–237. doi: 10.1016/j.mito.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–997. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- Guan MX, Enriquez JA, Fischel-Ghodsian N, Puranam R, Lin CP, Marion MA, Attardi G. The Deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- Guan MX, Yan Q, Li X, Bykhovskaya Y, et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302. doi: 10.1086/506389. 22 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Carelli V, Spruijt L, et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–233. doi: 10.1086/519394. 25 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, Hutchin TP, Kappi T, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13:26–33. doi: 10.1038/sj.ejhg.5201250. 16 co- authors. [DOI] [PubMed] [Google Scholar]
- Kong Q-P, Bandelt H-J, Sun C, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. 12 co-authors. [DOI] [PubMed] [Google Scholar]
- Li R, Greinwald JH, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with nonsyndromic hearing loss. J Med Genet. 2004a;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Xing G, Yan M, Cao X, Liu XZ, Bu X, Guan MX. Cosegregation of C-insertion at position 961 with A1555G mutation of mitochondrial 12S rRNA gene in a large Chinese family with maternally inherited hearing loss. Am J Med Genet. 2004b;124A:113–117. doi: 10.1002/ajmg.a.20305. [DOI] [PubMed] [Google Scholar]
- Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res. 2004c;32:867–877. doi: 10.1093/nar/gkh226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li R, Chen J, Liao Z, et al. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside induced and non-syndromic hearing loss. Hum Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Qu J, Zhou X, Tong Y, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The mitochondrial tRNAThr A15951G mutation may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene. 2006;376:79–86. doi: 10.1016/j.gene.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Liao Z, Zhao J, Zhu Y, et al. The ND4 11696G>A mutation may influence the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation in a four-generation Chinese family. Biochem Biophys Res Commun. 2007;362:670–676. doi: 10.1016/j.bbrc.2007.08.034. 16 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR. Statistical tests and bootstrapping: assessing the reliability of phylogenies based on distance data. Mol Biol Evol. 1991;8:386–391. doi: 10.1093/oxfordjournals.molbev.a040655. [DOI] [PubMed] [Google Scholar]
- Matthijs G, Claes S, Longo-Bbenza B, Cassiman J-J. Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet. 1996;4:46–51. doi: 10.1159/000472169. [DOI] [PubMed] [Google Scholar]
- Morton ME. Genetic epidemiology of hearing impairment. Ann NY Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- Nance E, Sweeney A. Symposium on sensorineural hearing loss in children: early detection and intervention: genetic factors in deafness of early life. Otolaryngol Clin North Am. 1975;8:19–48. [PubMed] [Google Scholar]
- Pandya A, Xia X, Radnaabazar J, Batsuuri J, Dangaansuren B, Fischel-Ghodsian N, Nance WE. Mutation in the mitochondrial 12S ribosomal-RNA gene in 2 families from Mongolia with matrilineal aminoglycoside ototoxicity. J Med Genet. 1997;34:169–172. doi: 10.1136/jmg.34.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. 12 co-authors. [DOI] [PubMed] [Google Scholar]
- Qian Y, Guan MX. Interaction of aminoglycosides with human mitochondrial 12S ribosomal RNA carrying the deafness-associated mutation. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.00965-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Li R, Tong Y, Zhou X, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Invest Ophth Vis Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–696. doi: 10.1086/303040. 11 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch-Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Cabrera VM, González AM, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. 28 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Yang L, Zhu Y, et al. Very low penetrance of hearing loss in seven Han Chinese pedigrees carrying the deafness-associated 12S rRNA A1555G mutation. Gene. 2007;293:11–19. doi: 10.1016/j.gene.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D’Urbano L, et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Cruciani F, Rengo C, et al. The 1555A>G mutation in the 12S rRNA gene of human mtDNA: recurrent origins and founder events in families affected by sensorineural deafness. Am J Hum Genet. 1999;65:1349–1358. doi: 10.1086/302642. 15 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Walt JM, Nicodemus KK, Martin ER, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. 27 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu J, Zhu Y, et al. Mitochondrial tRNAThr 15927G>A mutation may modulate the phenotypic manifestation of ototoxic 12S rRNA 1555A>G mutation in four Chinese families. Pharmacogenet Genom. 2008;18:1059–1070. doi: 10.1097/FPC.0b013e3283131661. 12 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WY, Zhao L, Qian Y, Wang Q, Li N, Greinwald JH, Jr, Guan MX. Extremely low penetrance of hearing loss in four Chinese families with the mitochondrial 12S rRNA A1555G mutation. Biochem Biophys Res Commun. 2005;328:1244–1251. doi: 10.1016/j.bbrc.2005.01.085. [DOI] [PubMed] [Google Scholar]
- Young WY, Zhao L, Qian Y, Li R, Chen J, Yuan H, Dai P, Zhai S, Han D, Guan MX. Variants in mitochondrial tRNAGlu, tRNAArg and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Chinese families with hearing loss. Am J Med Genet. 2006;140A:2188–2197. doi: 10.1002/ajmg.a.31434. [DOI] [PubMed] [Google Scholar]
- Yuan H, Qian Y, Xu Y, et al. Cosegregation of the 7444G>A mutation in the mitochondrial COI/tRNASer(UCN) genes with the 12S rRNA 1555A>G mutation in a Chinese family with aminoglycoside-induced and non-syndromic hearing loss. Am J Med Genet. 2005;138A:133–140. doi: 10.1002/ajmg.a.30952. 15 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wang Q, Qian Y, Li R, Cao J, Hart LC, Zhai S, Han D, Young WY, Guan MX. Clinical evaluation and mitochondrial genome sequence analysis of two Chinese families with aminoglycoside-induced and nonsyndromic hearing loss. Biochem Biophys Res Commun. 2005;336:967–973. doi: 10.1016/j.bbrc.2005.08.199. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wei Q, Yang L, et al. Leber’s hereditary optic neuropathy is associated with the mitochondrial ND4 11696G>A mutation in five Chinese families. Biochem Biophys Res Commun. 2006;340:69–75. doi: 10.1016/j.bbrc.2005.11.150. 11 co- co-authors. [DOI] [PubMed] [Google Scholar]