Abstract
The study presents structural models for the complex of the chemotaxis inhibitory protein of Staphylococcus aureus, CHIPS, and receptor for anaphylotoxin C5a, C5aR. The models are based on the recently found NMR structure of the complex between CHIPS fragment 31-121 and C5aR fragment 7-28, as well as on previous results of molecular modeling of C5aR. Simple and straightforward modeling procedure selected low-energy conformations of the C5aR fragment 8-41 that simultaneously fit the NMR structure of the C5aR 10-18 fragment and properly orient the NMR structure of CHIPS31-121 relative to C5aR. Extensive repacking of the side chains of CHIPS31-121 and C5aR8-41 predicted specific residue-residue interactions on the interface between CHIPS and C5aR. Many of these interactions were rationalized with experimental data obtained by site-directed mutagenesis of CHIPS and C5aR. The models correctly showed that CHIPS binds only to the first binding site of C5a to C5aR not competing with C5a fragment 59-74, which binds the second binding site of C5aR. The models also predict two elements of CHIPS, fragments 48-58 and 97-111, may be used as structural templates for potential inhibitors of C5a.
Keywords: GPCRs, C5a receptor, chemotaxis inhibitory protein, molecular modeling
Introduction
CHIPS, a 121-membered chemotaxis inhibitory protein of Staphylococcus aureus, is a potent inhibitor of neutrophil and monocyte chemotaxis involving C5a anaphylatoxin or formylated peptides [1]. CHIPS selectively binds both C5a receptor (C5aR) and formylated peptide receptor (FPR) with nanomolar affinities [2], which makes it a promising lead for development of new anti-inflammatory compounds. Mutational studies have previously demonstrated that different epitopes of CHIPS inhibit signaling through C5aR or FPR: the first six residues of CHIPS blocked FPR but not C5aR [3], whereas the truncated CHIPS fragments, 31-121 and 31-113, inhibited C5a binding to C5aR as effectively as the entire CHIPS, but lacked FPR antagonism [4, 5].
C5a binds and activates the C5aR utilizing two distinct binding sites, the first located in the N-terminus of C5aR and the second located in a helical crevice between the extracellular loops which accommodate the C-terminus of the C5a ligand (residues 65–74) [6–8]. Mutational studies of C5aR have mapped CHIPS binding to the N-terminal fragment of C5aR (residues 1–38) and demonstrated an essential role for the C5aR fragment 10-18 [9]. In addition, CHIPS does not affect activation of C5aR by a peptide mimic of the C5a 65-74 fragment [9]. These studies demonstrated also that CHIPS blocks C5a activation by inhibiting the binding of the intact ligand to the N-terminus of the C5a receptor and also implied that CHIPS does not interact appreciably with the extracellular loops of the C5aR.
Previously, NMR spectroscopy provided the three-dimensional solution structure of the isolated CHIPS 31-121 [4] and, very recently, in complex with the C5aR 7-28 fragment (Tyr11 and Tyr14 of C5aR were sulfated) [10]. The NMR structure of the C5aR7-28:CHIPS31-121 complex confirmed the important role of C5aR fragment 10-18 in binding CHIPS. The C-terminal portion of the C5aR7-28 peptide was highly flexible, which prevented elucidation of specific interactions between residues 22–28 of C5aR7-28 with CHIPS in the NMR structure [10]. Also, mutational studies using tethered N-terminal peptides that either introduced point mutations into this region or deleted it altogether did not impact binding of CHIPS, thereby supporting the notion that this region does not interact directly with CHIPS [9]. However, in the context of the intact C5aR, residues 22 – 28 may adopt a very different set of conformations, especially when one considers that this region is directly connected to the transmembrane helix of C5aR (TM1, residues 38–63 [11]).
Our previous studies developed 3D models of the C5aR:C5a complex involving various possibilities for flexible extracellular loops and the N-terminal fragment of C5aR [11]. The models were validated by comparison with the available data of site-directed mutagenesis of C5aR [11] and the results of a novel technique of disulfide trapping by random mutagenesis [12]. In the present study, we employed the results of the previous modeling of the C5aR and the NMR data on the C5aR7-28:CHIPS31-121 complex to generate new models for interaction between CHIPS and the C5aR. Our modeling procedure selected low-energy conformations of the C5aR fragment 8-41 that simultaneously fit the NMR structure of the C5aR 10-18 fragment and properly orient the NMR structure of CHIPS31-121 relative to C5aR. Extensive repacking of the side chains of CHIPS31-121 and C5aR8-41 predicted specific residue-residue interactions on the interface between CHIPS and C5aR. Many of these interactions are rationalized with experimental data obtained by site-directed mutagenesis of CHIPS and C5aR.
Materials and methods
All energy calculations were performed using the ECEPP/2 force field with rigid valence geometry [13, 14] and trans-conformations of Pro residues; residues Arg, Lys, Glu and Asp were regarded as charged species. The 3D structures of the C5aR8-41:CHIPS31-121 complex were obtained by energy minimization of the system consisting of two components, namely the selected conformation(s) of C5aR8-41 and the NMR-derived structure of CHIPS31-121. The modeling procedure was essentially the same as that applied earlier for determining the 3D structures of the transmembrane regions of GPCRs (see, e.g., [11] for details). The backbone structures of each component were regarded as rigid bodies (with the fixed values of the backbone dihedral angles), while the side chains of both C5aR8-41 and CHIPS31-121 were extensively repacked prior to energy minimization. For CHIPS mutants, the corresponding side chains were replaced in the NMR structure. The algorithm of side chain repacking [15, 16] involved a stepwise grid search (with the grid step of 30°) in the space of the dihedral angles of the side chains, χi. To ensure thorough sampling of the χi space, two options of the search were applied independently. The first option sampled the grid varying sequentially all χ1 angles of the side chains of CHIPS31-121 and C5aR8-41, then all χ2 angles, then all χ3 angles, etc. until all side chain angles of both components were rotated. The second option sampled all χi angles for each side chain in order of their number in protein sequence, i.e., from 31 to 121 for CHIPS31-121 and from 8 to 41 for C5aR8-41. In both cases, sampling was performed until convergence to energetically optimal packing of side chains was achieved. If necessary, specific side chains (as those on CHPS:C5aR interface) were repacked additionally. Finally, energy minimization in the space of possible rotations and translations of the two components as well as in the space of the dihedral angles of the side chains followed. Results of all options were pooled together to cover energetically plausible conformational possibilities for the side chains. To account to some extent for the absence of solvent and membrane environment, energy calculations were performed using two values of macroscopic dielectric constant, ε = 2 (the generic value for the ECEPP force field) and ε = 80, which is closer to the water dielectric constant. In most cases, distributions of the side chain rotamers obtained by calculations with both values were identical.
Results and discussion
Computational models of the CHIPS31-121:C5aR8-41 complex
Previous modeling of the isolated N-terminal fragment C5aR8-41 identified 185 low-energy backbone structures [11]; it was reasonable to assume that possible conformations of C5aR8-41 in complex with CHIPS should be selected from this pool of structures. 59 of 185 structures contained spatial positions of fragment C5aR 10-18 that fit to that observed in the C5aR7-28:CHIPS31-121 complex by NMR spectroscopy (according to the rms cut-off of 2 Å calculated for the Cα atoms of residues Asp10, Tyr11, Gly12, Tyr14, Asp15 and Asp18; NMR model #1 from the PDB entry 2K3U was used for fitting). In turn, only 17 of 59 structures showed no steric clashes with the NMR structure of CHIPS31-121 (i.e., no intermolecular Cα-Cα distance was less than 3 Å). When these 17 structures were fit to the helical stem of TM1 (residues 38–41) of our model of C5aR, only four structures did not contain steric clashes with either the TM region of C5aR or with all possible conformations of the extracellular loops that have been found previously [11]. Visual inspection showed that, in two of the four structures, CHIPS31-121 was significantly embedded into the membrane space, which is highly unlikely; those two structures were discarded from subsequent considerations.
The remaining two models of the C5aR8-41:CHIPS31-121 complex are depicted in Figs 1A and 1B. The interfaces between C5aR and CHIPS were established by subjecting both models to extensive side chain repacking and energy calculations (see Methods). The system of possible residue-residue contacts on the interface in both models is described in Table 1 (a contact was defined as situation when at least one distance between atoms belonging to the corresponding side chains was less than 5.5 Å). The two models feature significantly different potential strong salt bridges and/or hydrogen bonding at the interfaces. The strongest interactions in the first model were between Y48/K51 of CHIPS and Asp10 of C5aR (the corresponding residues are shown in Table 1 in bold; different notations are applied to distinguish between amino acid residues of C5aR and CHIPS). In the second model, the strongest interactions were K51 - Thr8, K54 – Ser30, K100 – Thr29 and Y108 – Thr29. Importantly, many of interactions on the interface listed in Table 1 should be regarded as possibilities, since some side chains possess more conformational freedom and therefore may also interact with residues not on the interface (see also below).
Fig. 1.
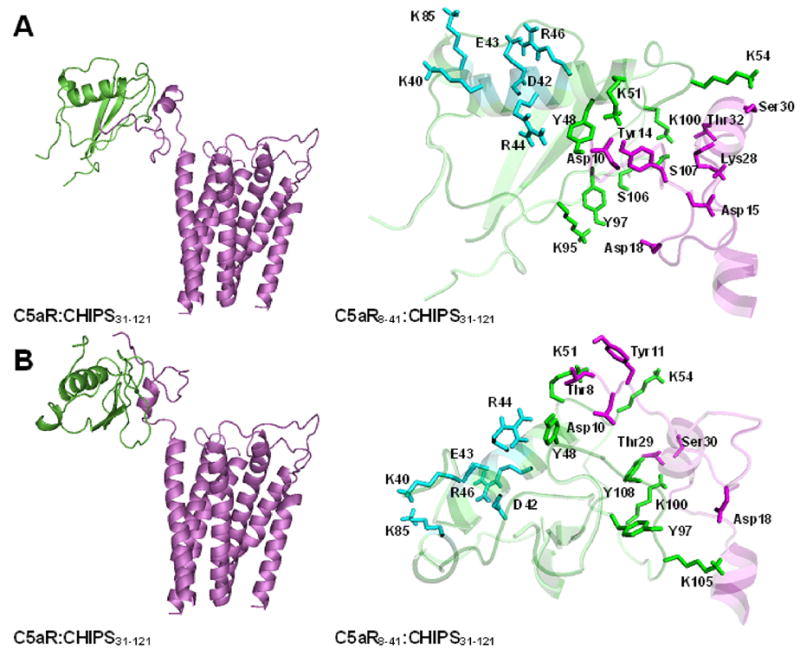
Right panels show cartoon sketches of complexes of CHIPS31-121 (green) and C5a (magenta). Left panels show more detailed sketches of complexes CHIPS31-121:C5aR8-41 focusing on possible strong residue-residue interactions. Side chains are shown as sticks and labeled by one letter notation (CHIPS residues) or by three-letter notation (C5aR residues). Residues K40 – R46 are shown in cyan, interface residues Y48 – Y108 are in green, Asp10 – Ser30 are in magenta. A and B relate to the first and second suggested models, respectively.
Table 1.
Contacts between side chains of CHIPS31-121 and C5aR8-41. Residues participating in strongest intermolecular interactions are shown in bold.
| Residues of CHIPS | Residues of C5aR8-41 |
|
|---|---|---|
| First model of complex | Second model of complex | |
| R44 | Asp10 | |
| Y48 | Pro9, Asp10, Tyr14 | Thr8, Pro9, Asp10 |
| K51 | Thr8, Pro9, Asp10, Tyr14 | Thr8, Tyr11 |
| T53 | Pro25, Thr29 | Tyr14, Thr29 |
| K54 | Val26, Thr29, Ser30 | Pro25, Val26, Ser30 |
| N55 | Thr29 | Thr29 |
| Q58 | Thr29, Thr32 | |
| K95 | Asp18 | |
| Y97 | Asp18 | |
| F98 | Tyr14 | |
| K100 | Thr32 | Pro25, Lys28, Thr29, Thr32, Leu33, Arg34 |
| E103 | Arg34 | |
| K105 | Asp18, Arg34 | |
| S107 | Lys28, Tyr14, Thr32 | Lys28, Arg34 |
| Y108 | Tyr14, Pro25 | Tyr14, Pro25, Thr29 |
| V109 | Tyr14 | Tyr14 |
| I110 | Tyr14 | Pro9, Asp10, Tyr14 |
| N111 | Asp10, Tyr14 | |
Rationalizing of site-directed mutagenesis data by computational models of the CHIPS31-121:C5aR8-41 complex
Interfaces between C5aR and CHIPS suggested by both models agree well with the two independent sets of data on NMR titration of C5aR1-37 in presence of 15N-CHIPS0-121 [17] and of the sulfated C5aR7-28 in presence of 15N-CHIPS31-121 [10]. Both studies demonstrated that two segments of CHIPS (ca. 45 – 61 and 98 – 111) directly interact with the C5aR fragments; these segments figure prominently in the binding interfaces of both computational models. Mutations of K95 (K95A-CHIPS1-121 [4], K95S-CHIPS31-113 [5]) or Y97 (Y97A-CHIPS31-121 [10], Y97K-CHIPS31-113 [5]) significantly lowered the ability of mutants to block activation of C5aR by C5a. K95 and Y97 likely are directly involved in interactions with the C5aR residues on C5aR:CHIPS interface, since CD spectroscopy showed that the above mutations do not disturb structural integrity of CHIPS (see Fig S3 in [10]). At the same time, interactions K95 - Asp18 and Y97 – Asp18 are available in the first computational model but not in the second one that features interaction K105 - Asp18 (see Table 1 and Fig. 1). Other experimental data on CHIPS mutants also support the first model. Specifically, mutations at Y48A-CHIPS31-121 [10] and K51A-CHIPS1-121 [4] led to significant loss of CHIPS ability to block C5aR activation by C5a by likely affecting the strong Y48 - Asp10 and K51 - Asp10 interactions present in the first model. Alternatively, the K51A mutation may interrupt the strong K51 – Thr8 interaction characteristic for the second model. Also, S107A-CHIPS31-121 [10] and S107N-CHIPS31-113 [5] only partially inhibit C5aR interaction with C5a, which may be due to loss of the possible interaction S107 - Lys28 present in both models (Lys28 may alternatively interact either with S107 or with Asp15, which is shown in Fig. 1A). In both models, S106 closely contacts the side chain of Y97 that may change orientation in the S106A-CHIPS31-121 mutant. This mutation significantly impairs CHIPS ability to block C5aR activation [10], which might reflect loss of interaction Y97 - Asp18 present in the first but not in the second model. At the same time, mutation K105A in CHIPS1-121 [4] also disrupts CHIPS interactions with C5aR, which supports the proposed contact K105 - Asp18 in the second model.
The mutational data obtained from mutational studies of the C5aR demonstrated essential roles for residues Asp10, Asp15 and Asp18 for interaction with CHIPS [9]. These observations also are more consistent with the first model, where Asp10 and Asp18 may be involved into direct interactions with Y48 and K51 as well as with K95 and Y97, respectively, and Asp15 may interact with Lys28 influencing possible interaction of the latter with S107. In the second model, on the contrary, only Asp18 may interact directly with the K105 residue of CHIPS.
The mutations in R44A-CHIPS1-121 and R46A-CHIPS1-121 also impair the ability of CHIPS to block C5aR activation by C5a [4], while not altering the secondary structure of CHIPS31-121 [10]. In both computational models, R44 and R46 are not directly involved in interactions with the C5aR residues. To gain some insights into how these mutations might impair C5aR interactions, we separately modeled the complexes between these two CHIPS mutants and C5aR8-41. Extensive side chain repacking for the mutant complexes C5aR8-41:R44A-CHIPS31-121 and C5aR8-41:R46A-CHIPS31-121 revealed more subtle influences of R44A or R46A mutations on possible interactions between C5aR and CHIPS. The side chains of both residues are involved in plethora of potential salt bridges/hydrogen bonds with the side chains of other residues of CHIPS (see Fig. 1). The negatively charged D42 and E43 residues may interact with the positively charged residues K40, R44 and R46 located in the same helical stretch of CHIPS (segment 38–51), forming the salt bridges K40 – E43, D42 – R46 and E43 – R46. They also may interact with K85 located in the loop connecting two β-sheets of CHIPS, 70–76 and 94–100. Also, the side chain of R44 may form strong hydrogen bond with the side chain of Y48, which participates in the direct interaction with Asp10 in the first model. Accordingly, removal of the R44 side chain in R44A-CHIPS1-121 may increase conformational flexibility of the Y48 side chain thus affecting direct interactions with C5aR. In R46A-CHIPS1-121, the salt bridges D42 – R46 and E43 – R46 are absent, and the salt bridges K40 – E43, D42 – K85 and E43 – K85 become dominant. Addition of the latter two strong residue-residue interactions between structural elements of CHIPS may influence overall conformational stability of the molecule, as was noted in the earlier experimental study [4]. On the other hand, conformational possibilities of the R44 side chain and, as a consequence, possibilities of the Y48 side chain still may be affected in R46A-CHIPS1-121.
Computational models of the CHIPS31-121:C5aR8-41 complex compared to NMR structure and to model of C5a:C5aR complex
Both computational models are not mutually exclusive. Both models are similar to that suggested by the NMR spectroscopy [10]. It is not surprising, since one important element of our models, namely spatial positions of Cα atoms in the C5aR 10-18 fragment, was required to be compatible to the NMR model. Nevertheless, many details of the C5aR:CHIPS interface suggested by our models and by the NMR model are different. The NMR study used the sulfated Tyr11 and Tyr14 in C5aR7-28 to ensure forming of strong complex with CHIPS31-121 (ca. 100 times stronger than that using the non-sulfated C5aR7-28 [10]), while our study considered both tyrosines as non-sulfated. (The role of the sulfated/non-sulfated Tyr11 and Tyr14 in C5aR in binding CHIPS is disputable, since mutations Y11F in C5aR1-37 and Y14F in the entire C5aR did not affect CHIPS binding [9]; this point recently was discussed further [10].) Accordingly, in the NMR model, the sulfate group of sTyr11 was coordinated by interactions with the side chains of R44, Y48 and K51 (the former, though, was not well-resolved in the NMR structure [10]), and the sulfate group of sTyr14 interacted with the backbone amide hydrogens of K54 and N55 as well as with the side chain of Q58. These interactions, obviously, are absent in our models; according to Table 1, we might rather expect interactions of K51 either with sTyr14 in the first model, or with sTyr11 in the second one.
Our models predict the specific orientations of CHIPS relative to C5aR (see Fig. 1) that exclude interactions between CHIPS and the transmembrane region of C5aR. This agrees with the experimental observation that CHIPS does not interfere with binding of C5a 59-74 to C5aR [9], which binds to a cavity in the transmembrane region of C5aR between TM3, TM4, TM5 and TM6 [11]. Interestingly, our first model features a backbone conformation of C5aR8-41 that was selected previously as capable to interact with C5a (conformation a, see Table 1 in [11]). However, the interface between C5aR8-41 and CHIPS31-121 in our first model includes Thr8, Pro9, Asp10, Tyr14, Asp18, Pro25, Val26, Lys28, Thr29, Ser30 and Thr32, while the interface between C5aR8-41 and C5a in conformation a includes different residues, namely Leu22, Asp27, Asn31, Val35, Pro36 and Asp37.
Concluding remarks
Our modeling study yielded two structural models of the C5aR:CHIPS31-121 complex based on conformational possibilities of the N-terminal fragment C5aR8-41, established earlier and on the NMR-derived structure of the C5aR7-28:CHIPS31-121 complex. The models elucidated the general orientation of CHIPS31-121 relative to C5aR and interface between C5aR8-41 and CHIPS31-121. The models provide plausible rationalization of the available data of site-directed mutagenesis of CHIPS and the N-terminal fragment of C5aR. In agreement with experimental data, the models showed that CHIPS binds only to the first binding site of C5a to C5aR, and, therefore, CHIPS does not compete with C5a 59-74, which binds the second binding site of C5aR. The models also predict that two elements of CHIPS, namely fragments 48-58 and 97-111 may be used as structural templates for potential inhibitors of C5a. The atomic coordinates of the C5aR7-28:CHIPS31-121 complexes are available from the authors by request.
Acknowledgments
The study was supported by the NIH grant GM 71634 (G.V.N. and T.B.) and GM 63720 (T.B.).
Abbreviations
- GPCR
G-protein-coupled receptor
- C5a
anaphylotoxin C5a
- C5aR
C5a receptor
- CHIPS
chemotaxis inhibitory protein of Staphylococcus aureus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Haas CJ, Veldkamp KE, Peschel A, Weerkamp F, Van Wamel WJ, Heezius EC, Poppelier MJ, Van Kessel KP, van Strijp JA. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. Journal of Experimental Medicine. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postma B, Poppelier MJ, van Galen JC, Prossnitz ER, van Strijp JA, de Haas CJ, van Kessel KP. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. Journal of Immunology. 2004;172:6994–7001. doi: 10.4049/jimmunol.172.11.6994. [DOI] [PubMed] [Google Scholar]
- 3.Haas PJ, de Haas CJ, Kleibeuker W, Poppelier MJ, van Kessel KP, Kruijtzer JA, Liskamp RM, van Strijp JA. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. Journal of Immunology. 2004;173:5704–5711. doi: 10.4049/jimmunol.173.9.5704. [DOI] [PubMed] [Google Scholar]
- 4.Haas PJ, de Haas CJ, Poppelier MJ, van Kessel KP, van Strijp JA, Dijkstra K, Scheek RM, Fan H, Kruijtzer JA, Liskamp RM, Kemmink J. The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules. Journal of Molecular Biology. 2005;353:859–872. doi: 10.1016/j.jmb.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Gustafsson E, Haas PJ, Walse B, Hijnen M, Furebring C, Ohlin M, van Strijp JA, van Kessel KP. Identification of conformational epitopes for human IgG on Chemotaxis inhibitory protein of Staphylococcus aureus. BMC Immunology. 2009;10:13. doi: 10.1186/1471-2172-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollison KW, Mandecki W, Zuiderweg ER, Fayer L, Fey TA, Krause RA, Conway RG, Miller L, Edalji RP, Shallcross MA. Identification of receptor-binding residues in the inflammatory complement protein C5a by site-directed mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:292–296. doi: 10.1073/pnas.86.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siciliano SJ, Rollins TE, DeMartino J, Konteatis Z, Malkowitz L, Van Riper G, Bondy S, Rosen H, Springer MS. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMartino JA, Van Riper G, Siciliano SJ, Molineaux CJ, Konteatis ZD, Rosen H, Springer MS. The amino terminus of the human C5a receptor is required for high affinity C5a binding and for receptor activation by C5a but not C5a analogs. Journal of Biological Chemistry. 1994;269:14446–14450. [PubMed] [Google Scholar]
- 9.Postma B, Kleibeuker W, Poppelier MJ, Boonstra M, Van Kessel KP, Van Strijp JA, de Haas CJ. Residues 10-18 within the C5a receptor N terminus compose a binding domain for chemotaxis inhibitory protein of Staphylococcus aureus. Journal of Biological Chemistry. 2005;280:2020–2027. doi: 10.1074/jbc.M412230200. [DOI] [PubMed] [Google Scholar]
- 10.Ippel JH, de Haas CLC, Bunschoten A, van Strijp JAG, Kruijtzer JAW, Liskamp RMJ, Kemmink J. Structure of the Tyrosine-sulfated C5a Receptor N Terminus in Complex with Chemotaxis Inhibitory Protein of Staphylococcus aureus. Journal of Biological Chemistry. 2009;284:12363–12372. doi: 10.1074/jbc.M808179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikiforovich GV, Marshall GR, Baranski TJ. Modeling Molecular Mechanisms of Binding of the Anaphylotoxin C5a to the C5a Receptor. Biochemistry. 2008;47:3117–3130. doi: 10.1021/bi702321a. [DOI] [PubMed] [Google Scholar]
- 12.Hagemann IS, Miller DL, Klco JM, Nikiforovich GV, Baranski TJ. Structure of the complement factor 5a receptor-ligand complex studied by disulfide trapping and molecular modeling. Journal of Biological Chemistry. 2008;283:7763–7775. doi: 10.1074/jbc.M709467200. [DOI] [PubMed] [Google Scholar]
- 13.Dunfield LG, Burgess AW, Scheraga HA. Energy Parameters in Polypeptides. 8. Empirical Potential Energy Algorithm for the Conformational Analysis of Large Molecules. J Phys Chem. 1978;82:2609–2616. [Google Scholar]
- 14.Nemethy G, Pottle MS, Scheraga HA. Energy Parameters in Polypeptides. 9. Updating of Geometrical Parameters, Nonbonded Interactions, and Hydrogen Bond Interactions for the Naturally Occurring Amino Acids. J Phys Chem. 1983;87:1883–1887. [Google Scholar]
- 15.Nikiforovich GV, Hruby VJ, Prakash O, Gehrig CA. Topographical Requirements for Delta-Selective Opioid Peptides. Biopolymers. 1991;31:941–955. doi: 10.1002/bip.360310804. [DOI] [PubMed] [Google Scholar]
- 16.Nikiforovich GV, Mihalik B, Catt KJ, Marshall GR. Molecular mechanisms of constitutive activity: mutations at position 111 of the angiotensin AT1 receptor. J Pept Res. 2005;66:236–248. doi: 10.1111/j.1399-3011.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- 17.Wright AJ, Higginbottom A, Philippe D, Upadhyay A, Bagby S, Read RC, Monk PN, Partridge LJ. Characterisation of receptor binding by the chemotaxis inhibitory protein of Staphylococcus aureus and the effects of the host immune response. Molecular Immunology. 2007;44:2507–2517. doi: 10.1016/j.molimm.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


