Abstract
Much evidence suggests that “developmental regulator” genes, like transcription factors and signaling molecules, are typically controlled by many modular, tissue-specific cis-regulatory elements that function during embryogenesis. These elements are often far from gene coding regions and promoters. Bone Morphogenetic Proteins (BMPs) drive many processes in development relating to organogenesis and differentiation. Four BMP family members, Bmp2, Bmp4, Bmp5, and Gdf6, are now known to be under control of distant cis-regulatory elements. BMP genes are thus firmly placed in the category of genes prone to this phenomenon. The analysis of distant BMP regulatory elements has provided insight into the many pleiotropic effects of BMP genes, and underscores the biological importance of noncoding genomic DNA elements.
Keywords: BMP, enhancer, cis-regulation, skeletal development
1. Introduction
To date, five members of the BMP family have been well documented to be controlled by long-range regulatory sequences: the fly dpp (decapentaplegic) gene, and the mouse Bmp2, Bmp4, Bmp5 and Gdf6 genes. The studies of the mouse genes are reviewed here, in the chronological order in which the cis-regulatory studies on long-range elements were initiated.
2. Bmp5
Bmp5 was one of the first mammalian genes for which direct evidence indicated long-range cis-regulatory elements. This discovery was facilitated in the pre-genomic era by short ear alleles that were structural rearrangements downstream of the gene, as well as transgenic analysis of well-mapped genomic fragments spanning the region. More recent analyses have demonstrated conservation of long-range Bmp5 enhancers and their roles in shaping skeletal morphology.
2.1. The short ear mouse
Bmp5 was identified as the gene mutated in short ear mice by David Kingsley and colleagues [1]. The short ear mouse mutant has impaired development of several seemingly-unrelated skeletal elements, such as the ear pinna, xiphisternum, and thyroid cartilage. Studies of Bmp5 mRNA showed that these phenotypes in the short ear mouse correlated with localized transcription of Bmp5 within the developing skeleton [2–5]. This suggested that the pleiotropic, anatomically-regional phenotypes caused by Bmp5 deficiency were largely explained by an elegant program of transcriptional regulation, whereby its effects are restricted to specific parts of the skeleton. Thus, identifying cis-acting sequences governing the transcription of Bmp5 became an important quest.
2.2. Transgenic and genetic analysis of Bmp5 cis-regulatory domains
Surprisingly, when 5 kilobases (kb) of DNA extending 5’ to the Bmp5 start site were tested for ability to activate a lacZ reporter gene in transgenic mouse embryos, no expression was detected [3]. For most genes studied in the pre-genomic era, this would likely have been the frustrating end of a cis-regulatory project. Fortunately, an astute observation prevented this from occurring. Specifically, 3 radiation-induced inversion short ear mutations were found to have breakpoints ranging from 6 to 105 kb downstream of the terminal Bmp5 exon. Moreover, the effects of the inversions on ear size were varied. The pinna was smaller in mice with the closest 3’ inversion (allele se30DThWb), similar to mice carrying deletions of the whole gene, while mice with the most distant 3’ inversion allele (se4CHLd) had larger (but not normal-sized) ears. This suggested the intervening region had regulatory effect(s) on Bmp5 expression despite being fully 3’ to the coding sequence.
Transgenic analysis of clones spanning this region was performed by linking DNA fragments to a minimal promoter/lacZ minigene, followed by X-gal staining of transgenic embryos [3]. This revealed several separate enhancer elements driving gene expression in the manubrium (anterior sternum), distal genital tubercle, thyroid cartilage, and lung mesenchyme. In each case, lacZ expression closely matched endogenous Bmp5 mRNA, indicating bona fide enhancer elements. Supporting these findings, Bmp5 mRNA was absent from genitalia, thyroid cartilage and lung mesenchyme of mice homozygous for the proximal short ear inversion breakpoint, while expression was normal or only partly reduced in these structures for mice carrying the more distal inversion. Moreover, each inversion reduced Bmp5 transcripts in embryonic pinna cartilage, while expression in thyroid gland and intestine was unaffected. This strongly supported a model in which the inversions act to separate pinna-specific cis-enhancers from the Bmp5 transcription unit and other enhancers (e.g., intestinal) found closer to the promoter. This also excluded a simple model in which the inversion alleles globally inhibited Bmp5 by a chromosomal position effect, as the effects of either inversion were highly tissue-specific. These studies demonstrated an elegant combined analysis of structural mutations, gene expression, and transgenic assays. They were also remarkable for showing that mammalian BMP genes, like the fly BMP paralog decapentaplegic (dpp) gene, could have a modular arrangement of distant regulatory elements that extended far beyond the exon structure of the gene [6].
Transgenesis with bacterial artificial chromosomes (BACs) allowed larger regions of the Bmp5 region to be tested for cis-regulatory function. Prior to the advent of bacterial recombination methods, modification of BACs with reporter genes was not practical. Therefore, DiLeone et al. used a co-injection transgenic assay whereby BAC clones from different regions of the Bmp5 locus were simply mixed with a minimal Hsp68 promoter/lacZ minigene and injected into mouse embryos [2]. Both constructs typically co-integrate at a single genomic location, potentially allowing regulatory elements in the BAC to cis-regulate the lacZ reporter. Results obtained using this strategy agreed with the previous studies and uncovered even more Bmp5 enhancers in the 3’ region and within the transcription unit itself. These included enhancers driving expression in the thyroid gland, ribs, and intestines. The ability of these enhancers to shape skeletal elements through Bmp5 expression was also shown in an elegant experiment, whereby a 3’ Bmp5 BAC clone was co-injected with a Bmp5 promoter/cDNA minigene, and the resulting co-integrated transgene array was crossed onto the short ear mutant background, resulting in partial rescue of ear length. This supported a model in which at least some pinna cartilage-specific enhancers are within the 3’ BAC. Moreover, it suggested that widely distributed Bmp5 enhancers promote ear length by driving its expression in the pinna cartilage primordium. The xiphisternum, a butterfly-shaped projection of the posterior sternum that is normally absent in short ear homozygotes, was also completely restored by the BAC/cDNA transgene. This, combined with BAC/lacZ reporter transgene analysis, indicated that a xiphisternum-specific enhancer was in the same BAC clone.
2.3. Modular Bmp5 enhancers allow fine-tuning of skeletal morphology
The previous studies suggested that Bmp5 enhancers are remarkably modular. Data uncovering even more layers of Bmp5 enhancer modularity was recently described by Guenther et al [7]. They showed that embryos carrying separate Bmp5 BAC transgenes expressed a reporter in distinct sub-patterns within the rib perichondrium. Thus, a BAC clone spanning most of the Bmp5 transcription unit drove reporter gene expression strongly in the lateral perichondrium of proximal ribs, while the same BAC drove expression strongly along the lateral and medial sides, and more weakly on the anterior/posterior sides of the distal ribs. Complementing this pattern, a distant, non-overlapping 3’ BAC clone drove gene expression in the anterior, medial, and posterior sides of the proximal ribs, and in the anterior and posterior sides only of the distal ribs. The combined action of both enhancers approaches the complete Bmp5 pattern. Thus, it appears that some of the rather localized Bmp5 mRNA expression domains are created from the combined action of separate enhancers that function in anatomically distinct sub-regions.
Many studies have shown that highly evolutionarily conserved regions (ECRs) often demarcate developmentally regulated enhancers [8]. Comparative sequence analysis of the Bmp5 gene revealed that numerous intronic noncoding elements are highly conserved in mammals [7]. By subcloning intronic Bmp5 ECRs and testing them individually in lacZ transgenic assays, a rib enhancer-containing fragment (Ex4r) was identified. Ex4r is about 1 kb in size, and is located just downstream of exon 4, approximately 99 kilobases from the Bmp5 promoter. In dorsal ribs, this enhancer drives expression in lateral perichondrium; in ventral ribs the expression becomes more diffuse and distributed around the rib. Interestingly, this pattern mirrors the developmental contribution of the posterior halves of each somite to the individual ribs, thus suggesting a connection between somite compartmentalization and Bmp5 regulation. Guenther et al. further showed that altering BMP signaling in the Ex4r-specific rib domain altered rib morphology. This was achieved by directing expression of either a constitutively active or dominant negative BMP receptor subunit under the control of the Ex4r enhancer.
Why is there such exquisite modularity of Bmp5 expression in rib perichondrium? The answer is tied to how ribs maintain their characteristic shape as they grow in size. As the ribs grow, the rate of osteogenesis (balanced by opposing bone resorption) is differentially controlled around the rib circumference and along the inside and outside edges of the developing bone collar. The Bmp5 Ex4r enhancer drives addition of bone to the lateral rib surface by regionally stimulating BMP signaling, while separate Bmp5 enhancers probably drive bone formation on the interior/medial rib surface facing the marrow cavity. Guenther et al. speculated that the discrete rib enhancers allow the patterned control of BMP activity that is needed for proper maintenance of curvature as the ribs grow. Furthermore, the modular nature of BMP rib enhancers probably facilitated evolution of rib morphology by fine-tuning rib curvature and diameter, which varies widely across species. A similar theme of modular Bmp5 enhancers was found to affect formation of bony turbinates, which are stemlike structures in the nasal cavity. Short ear mice lack anterior nasal turbinates and branchlike structures on other turbinates. Bmp5 expression in growing turbinates is controlled by at least two separate enhancers: one that acts in the proximal stemlike region of the turbinates, and one that is active in their branching tips [7].
3. Gdf6
In mammals, the Gdf5, Gdf6 and Gdf7 genes form a closely related subfamily within the BMP gene family and have been implicated in limb joint formation and chondrogenesis [9–13]. Strikingly, in embryonic mouse limb buds the Gdf genes are expressed in transverse stripes across the developing skeletal elements, corresponding to sites of imminent joint formation [9, 11, 14, 15]. These studies established the Gdf genes among the earliest known markers of limb joint formation, and are in general agreement with other studies that have demonstrated the chondrogenic activity of Gdf proteins [16–18]. Whereas Gdf5 is expressed in all developing limb joints [11, 15], Gdf6 and Gdf7 are restricted to specific joints. Gdf6, in particular, is expressed in wrist, ankle, elbow and knee joints; moreover, homozygous Gdf6 −/− mutant animals have characteristic fusions of wrist and ankle bones [14].
3.1. BAC transgenic analysis of Gdf6, modular joint control elements and extensive noncoding conservation
The precisely patterned expression and phenotypic effects of Gdf6 are reminiscent of Bmp5. Likewise, transgenic analysis of the Gdf6 locus has revealed that discrete enhancers control its expression in different subsets of joints. Since work on Bmp5 had revealed that distant regulatory elements were a theme, BAC constructs were used to dissect Gdf6 [19]. The compact nature of Gdf6 gene structure (two exons and one intron, spanning 18 kb) facilitated a “BAC scanning” approach, whereby BAC clones extending either far 5’ or far 3’ to the gene were modified by inserting a lacZ reporter into the Gdf6 coding region, and subsequently analyzed for ability to recapitulate Gdf6 expression in transgenic mouse embryos. The boundaries of the individual BAC clones could thus be used to map suspected Gdf6 enhancers in numerous anatomical domains (e.g. basisphenoid bone, mammary glands, tooth buds, whisker buds, dorsal retina, genitalia, and larynx). These enhancers were distributed across 5’ and 3’ regions extending approximately 50 kb on either side of the coding regions.
Remarkably, Gdf6 BACs that extended as far as 45 kb 5’, and 120 kilobases 3’ were unable to drive limb joint expression. Only BACs extending more than 45 kb 5’ were able to do so – specifically in proximal joints (e.g. elbow and knee but not ankle or wrist). By engineering deletions in these large 5’ BACs, a region of ~8 kb was found to be necessary for joint-specific activity. A fragment from this interval that contained the most highly conserved regions was subcloned, and transgenic tests of this subclone proved it could function as a joint enhancer, driving expression specifically in shoulder, elbow, hip, and knee joints only. Thus, Gdf6 expression in proximal vs. distal joints is apparently under the control of separate cis-elements. The 5’ joint enhancer function was refined to a 440 bp sequence termed the Proximal Joint Element (PJE) that is approximately 64 kb upstream of the mouse Gdf6 promoter [20]. The PJE is highly conserved across mammals and birds, and contains motifs for Hox/Pbx and TCF/Lef factors, suggesting this enhancer might integrate Hox-driven limb patterning cues with Wnt-mediated signals known to control joint differentiation [21].
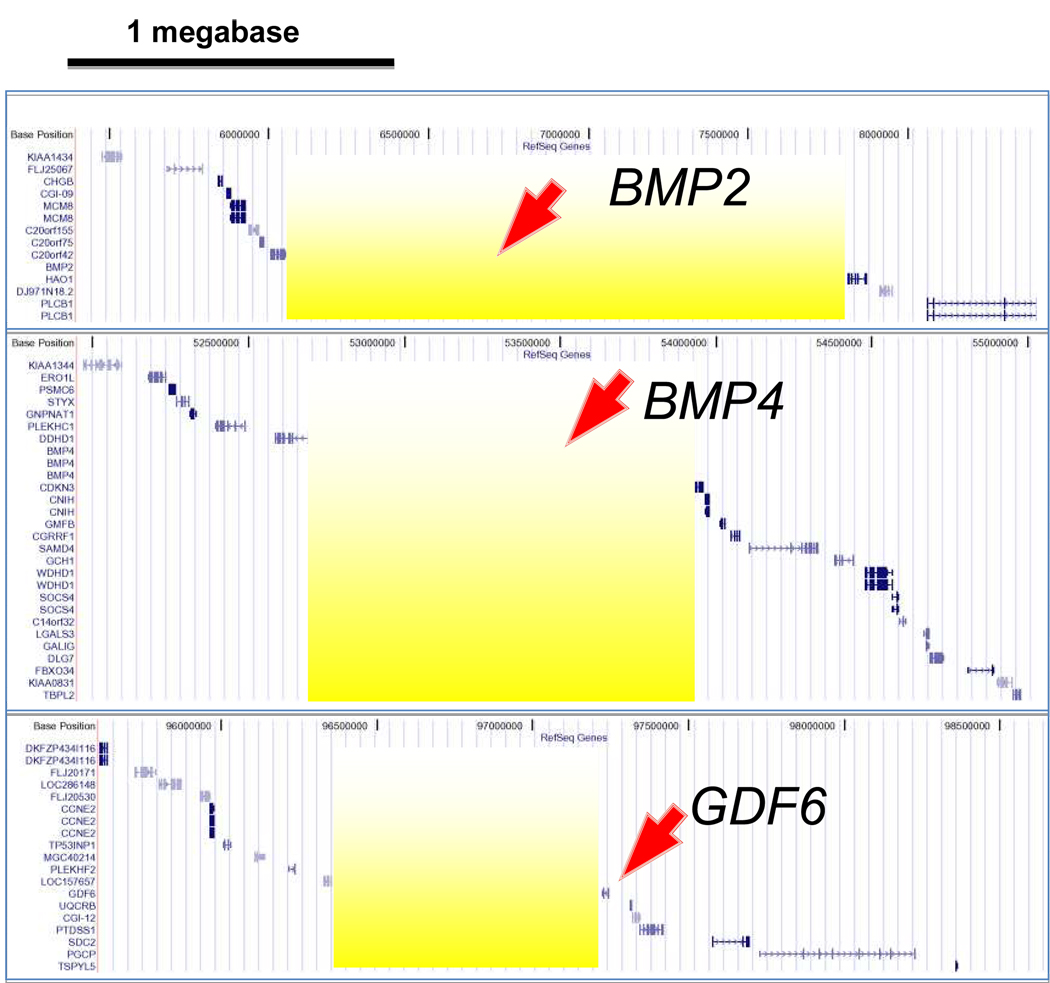
Although 270 kilobases of DNA were tested for Gdf6 cis-regulatory function, some enhancers were not uncovered. This suggested that some cis-elements were located even further from the gene. A syntenic analysis revealed that during rodent evolution, a chromosomal rearrangement broke the Gdf6 chromosome about 70 kb 5’ to Gdf6, just between the PJE and a cluster of unrelated genes [22]. Thus, it is unlikely that additional functionally conserved Gdf6 enhancers are more than 70 kb 5’ to the gene. In contrast, the 3’ flanking region of Gdf6 is a “gene desert” of approximately 900 kb (see Fig. 1). It has extensive cross-species synteny despite apparent lack of coding genes. This is a feature that is frequently associated with developmental control genes [23]. Portnoy et al. performed extensive multispecies sequence comparisons around Gdf6 using MultiPipmaker, WebMCS and ExactPlus software [20]. This revealed many candidate regulatory sequences flanking Gdf6, some of which are conserved in fish. An ongoing analysis confirms several mammal/fish noncoding ECRs are within the Gdf6 3’ gene desert, which is present next to the fish gdf6a/radar gene [N. Reed and D. Mortlock, manuscript in preparation].
Figure 1. Gene deserts around the human BMP2, BMP4 and GDF6 genes.
Shown are UCSC genome browser plots of roughly 3 megabases of DNA surrounding each gene and all annotated (RefSeq) genes. The yellow shading indicates “gene deserts” around or adjacent to each BMP gene. The gene deserts are similarly-sized in the mouse genome, and each gene desert has extensive regions of noncoding cross-species conservation (not shown).
3.2. Potential role of the Gdf6 “gene desert” in skeletal patterning
Tassabehji et al. described a chromosomal inversion in humans that breaks the GDF6 gene desert, causing a dominant subtype of Klippel-Feil Syndrome (KFS) [24]. Klippel-Feil Syndrome is characterized by congenital fusion of cervical vertebrae. Affected persons of pedigree KFS-02 also have fusions of wrist and ankle bones, impaired rotation and extension of proximal limb joints, vocal impairment due to larynx malformations, and hearing loss. The similar array of defects seen in the Gdf6 knockout mouse implies a heterozygous cis-regulatory effect on GDF6 is at least partly responsible. Remarkably, the human breakpoint is +697 kb 3’ to the GDF6 promoter and the homologous position in mouse is +793 kb. Missense mutations in GDF6 coding regions have been associated with congenital eye defects and KFS in humans, with varying penetrance and severity [25] although eye defects are not observed in the KFS-02 pedigree. Interestingly, retinal enhancers probably map close to the gene itself [19]. It is currently unclear whether the KFS-02 inversion physically separates Gdf6 enhancers from the locus. Alternatively, a position effect of transposed chromatin may intrude across the gene desert, reducing GDF6 expression in certain tissues. It is conceivable that the gene desert contains long-range enhancers that activate both Gdf6 and gene(s) on the other side of the desert that have related functions in skeletal development. The inversion might separate these genes from cis-enhancers on the Gdf6 side of the breakpoint. Further studies to map Gdf6 enhancers and examine expression of adjacent genes should help to shed light on these possibilities.
4. Bmp2
The founding member of the Bmp family, Bmp2, was discovered in the late 80’s as a potent bone-stimulating agent [26]. Together with Bmp4, it plays a critical role in skeletal development [27]. Early attempts to understand the cis-regulation of Bmp2 and Bmp4 focused on their respective promoter activities in osteoblasts. Interestingly, both genes have at least two major transcription start sites, resulting in the production of slightly different messenger RNAs [28–32]. For Bmp4, the distal promoter appears to be more active in osteoblasts [33], whereas the reverse appears to be true for Bmp2 [31, 34]. A construct containing 2.4 kb of sequence upstream from the distal promoter of Bmp4 can drive expression of a reporter gene in cultured osteoblasts [33], as can a 2.7 kb construct containing both the distal and proximal promoters of Bmp2 [35]. In contrast, neither construct has robust osteoblast activity when tested in vivo [36–38]. Moreover, there is no activity in several other tissues where endogenous Bmp2 and Bmp4 mRNA is observed. It is worth noting that the 2.7 kb Bmp2 construct was used as a tool to identify several compounds capable of stimulating bone formation in vivo, including statins [39–41]. Still, the inability of either the 2.7 kb Bmp2 or 2.4 kb Bmp4 constructs to recapitulate the complete expression pattern of these genes in vivo indicates that important cis-acting regulatory elements must be located outside of these regions.
4.1. Bmp2 and Bmp4 are embedded in conserved gene deserts
Similar to Gdf6, mammalian Bmp2 and Bmp4 are located within large gene deserts over a megabase across (Fig. 1). The large intergenic spaces on either side of both genes are observed across vertebrates, though they are smaller in fish species (e.g. pufferfish) that have significantly more compact genomes overall. The closely related Bmp2 and Bmp4 genes form a BMP subfamily homologous to the Drosophila gene decapentaplegic (dpp), which Dileone et al noted were regulated by several modular enhancers [3]. Many of these are located 3’ to the gene, separately controlling its expression within embryonic tissues and imaginal discs [6].
4.2. BAC transgenic analysis of Bmp2
The possibility that Bmp2 could be regulated by distant cis-acting elements was first addressed by R. Chandler et al [42]. In their report, they describe using BAC transgenesis to conduct a broad initial survey of the large gene desert in which the mouse Bmp2 gene is situated. Their approach was similar to that used for Gdf6; however, only 2 overlapping BAC clones were used. These contained the mouse Bmp2 coding sequence and approximately 200 kb of either 5’- or 3’-flanking sequence. As before, they were modified via homologous recombination in bacteria so that a lacZ reporter was inserted into the Bmp2 mature-peptide-coding sequence. Transgenic mice were generated with these constructs and were assayed at various embryonic stages for reporter expression.
When considered together, the two Bmp2 BAC lines displayed patterns of expression that recapitulated many known endogenous sites of Bmp2 expression in the embryo. Additionally, several novel sites of Bmp2 expression were revealed, and these were subsequently confirmed by in situ hybridization. When reporter activity did not appear where expected, it was usually at sites where Bmp2 is expressed at low levels, and the overall intensity of staining in the animals where this occurred was weaker. This, in turn, was due to lower quantities of transgene integration, rather than degradation of the transgene or positional effects. An important conclusion from this preliminary experiment was that many of the cis-acting regulatory elements needed to drive expression of Bmp2 in an appropriate spatiotemporal manner are located on the same chromosome as, and are within 200 kb of, the gene itself.
More significantly, the number of individual lacZ patterns unique to either BAC clone was greater than the number shared by both BACs, despite over 50 kb of overlapping DNA between the clones (including 2.7 kb of promoter sequence and 53.7 kb of downstream sequence, along with all exons and introns). This indicated that most of the enhancers located within 200 kb of Bmp2 are found at distances greater than 2.7 and 53.7 kb from the transcription start site in the 5’ and 3’ directions, respectively. Some of the expression domains unique to the BAC line containing mostly 5’ sequence (hereafter referred to as the 5’ BAC line) included the synovial joints, digit tips, skeletal muscle, gut epithelium, liver, thymus, adrenal glands, lung, gonads, vasculature, choroid plexus, and skin epithelium. Likewise, expression domains found only in the 3’ BAC line included the intervertebral discs, tooth bud enamel knot, kidney, pelage hair follicle placodes, mammary glands, midbrain, and interdigital mesenchyme. Tissues where both BAC constructs drove expression included the retinal pigmented epithelium, whisker hair shafts, ventral footpads, mesentery, pituitary gland, and tongue.
Reminiscent of Bmp5 expression in the ribs, the Bmp2 constructs drove expression in a complementary fashion in endochondral bones. Specifically, the 5’ BAC drove expression in the hypertrophic chondrocytes of the growth plate, whereas the 3’ BAC drove expression in the osteoblasts of the surrounding perichondrium. This, combined with the fact that the 3’ BAC also drove expression in osteoblasts of intramembranous bones strongly suggested that an osteoblast-specific enhancer was located more than 53.7 kb downstream from Bmp2. This is especially remarkable given that previous studies of Bmp2 cis-regulation in osteoblasts focused only on 2.7 kb of upstream sequence.
4.3. BAC deletion revealed modular Bmp2 regulatory domains and led to cloning of a conserved osteoblast-specific enhancer
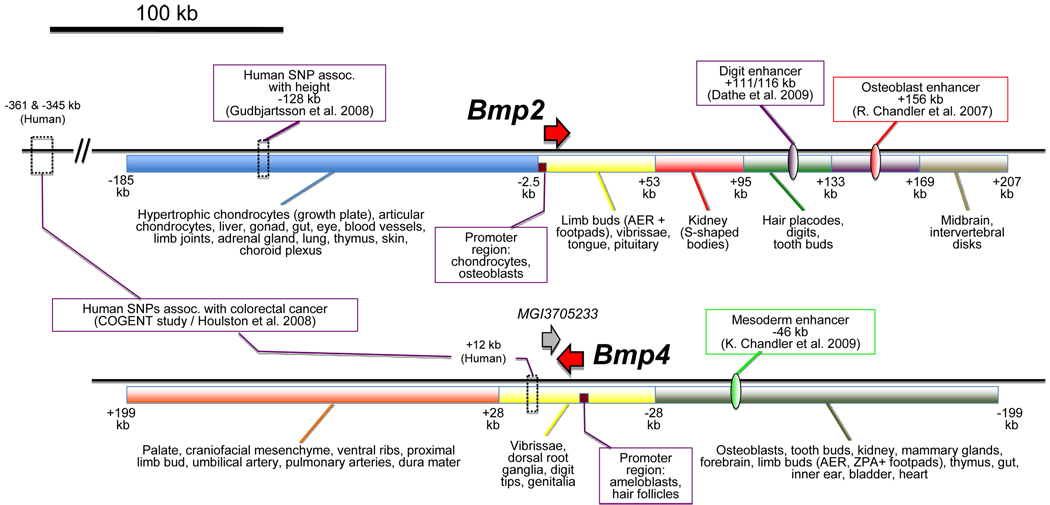
In order to parse out the cis-regulatory elements found more than 53.7 kb downstream from Bmp2, R. Chandler et al generated 4 separate deletion constructs based on the full-length 3’ BAC, each of which lacked approximately 40 kb of sequence. Together, the four deletions covered roughly 160 kb of sequence without any gaps or overlaps. Transgenic analysis of each deletion construct revealed a conspicuous lack of reporter expression in one or more tissues where the full-length construct was expressed. For example, a deletion construct targeting the region between 132.8 and 168.8 kb downstream from the Bmp2 transcription start site resulted in a loss of osteoblast expression. It was not lost from any other tissues; moreover, osteoblast expression was not abrogated by any of the other deletion constructs. Figure 2 summarizes the sites of expression for which each deleted region appears to be critical.
Figure 2. Schematic diagram of the cis-regulatory landscape surrounding the mouse Bmp2 and Bmp4 genes.
Shown is a summary of the locations of important cis-regulatory features that have been identified to date for Bmp2 and Bmp4. The colored horizontal bars represent large domains that were shown by BAC transgenesis to contain elements driving expression in the indicated tissues. Smaller domains that were tested for enhancer activity are shown as upright ovals. SNPs with significant correlations to human disease and development are highlighted by dashed boxes. Promoter regions that have been tested for ability to drive reporter gene expression in vivo are shown as purple boxes. Red arrows correspond to the coding sequences for Bmp2 and Bmp4.
Developmental enhancers typically have a high degree of evolutionary conservation [8]. Having placed the osteoblast enhancer between 132.8 and 168.8 kb downstream from Bmp2, R. Chandler et al searched within this region for sequences bearing a high degree of homology across several vertebrate species. Although many conserved sequences were present, 2 in particular stood out from the rest as possessing the highest degree of conservation. These sequences, termed ECR1 and ECR2, are conserved across mammals and are roughly 469 and 296 bp, respectively. ECR1 also contains a core sequence of 272 bp that is conserved in chicken. Neither ECR1 nor ECR2 is conserved in frog or teleost fish. To test these regions for functionality, they subcloned a 4.5 kb fragment containing ECR1 and ECR2 along with the intervening sequence into an Hsp68-lacZ minigene vector. Transgenic embryos with this construct showed remarkably consistent staining in endochondral osteoblasts, as well as mosaic staining in intramembranous osteoblasts of the mandible. Tests of each ECR separately showed that ECR1–but not ECR2–was sufficient to confer enhancer activity in skeletal tissues. This strongly suggests that an osteoblast-specific Bmp2 enhancer element is located within ECR1, which is 156.3 kb downstream from the promoter.
5. Bmp4
In a sister study to that of Bmp2 by K. Chandler et al, the role of distant cis-regulatory elements in Bmp4 expression was addressed with the same initial approach used for Bmp2 [43]. The 2 BAC clones used for this study collectively spanned about 400 kb, centered on the Bmp4 gene. Separately, they contained a reporter gene in place of the Bmp4 coding sequence, along with roughly 230 kb of flanking sequence. Many domains of endogenous Bmp4 expression were recapitulated in transgenic mouse embryos generated with the pair of BAC constructs; however, there was a conspicuous absence of reporter expression in the extra-embryonic ectoderm, eye, trachea, and anterior limb bud. Lack of expression in these tissues was not likely due to positional effects because it was consistent across multiple independent lines.
As with Bmp2, the BACs each drove reporter expression in a mostly distinct subset of tissues where Bmp4 is typically expressed. For example, the 5’ BAC line drove expression in the mesoderm, tooth bud dermal papilla, kidney, pelage hair follicle placode, mammary glands, forebrain, apical ectodermal ridge, thymus, gut, inner ear, zone of polarizing activity, bladder, ventral pawpads, and heart outflow tract (Fig. 2). In contrast, the 3’ BAC drove expression in the roof palate mesenchyme, ventral ribs, vertebral column, proximal limb mesenchyme, umbilical artery, dura mater, dorsal aorta, pulmonary arteries, and craniofacial mesenchyme. Interestingly, expression in osteoblasts was driven by the 5’, but not the 3’ BAC. This situation mirrors that which was seen for Bmp2. Because the two BACs shared a 60 kb region of overlap extending out to 30 kb on either side of the Bmp4 transcription start site, regulatory elements located within this region were present in both BACs. Accordingly, there were several sites where reporter expression was consistently observed in lines harboring both constructs. These included the whisker hair shafts, dorsal root ganglia, digit tips, and genital tubercle. An important conclusion from these observations was that while several cis-regulatory elements are close to Bmp4, most are located more than 30 kb away.
5.1 Bmp4 expression in mesoderm is controlled by an ancient 5’ enhancer
To pinpoint the location of enhancers within the large areas defined by BAC transgenes, K. Chandler et al searched for sequences with high evolutionary conservation. They improved the specificity of their search by focusing on sequences that aligned not only across mammalian species but also with fish. Three such sequences were discovered, termed Evolutionarily-Conserved Regions (ECRs) 1, 2, and 3. These were located 101 and 46-kb upstream, and 80 kb downstream of the Bmp4 promoter, respectively. In addition to their nucleotide sequences, the spatial orientation of these elements relative to Bmp4 was conserved across species as well. This is true in spite of the fact that genes flanking the Bmp4 gene desert have undergone rearrangement between mammals and fish. Thus, ECRs 1, 2 and 3 are ancient vertebrate features.
To test the hypothesis that ECRs 1, 2, and 3 could function as developmental enhancers for Bmp4, K. Chandler et al took two complementary approaches. First, they deleted each ECR separately from the full-length BAC constructs via homologous recombination in bacteria. Remarkably, removal of ECR2 from the 5’ BAC resulted in a loss of reporter expression in the outflow tract of the heart, as well as mesoderm of the lateral plate and extra-embryonic tissues. In contrast, removal of ECR1 and ECR3 from the 5’ and 3’ full-length BACs, respectively, had no detectable effect on reporter gene expression during mouse development. ECR1 and ECR3 may be functionally redundant, either with each other or with enhancer elements elsewhere in the genome. This could explain why removal of either one fails to abrogate reporter expression. Alternatively, they may act at later stages of development. Regardless, ECR2 appears to be necessary for Bmp4 expression in several tissues during mouse development.
K. Chandler et al also tested each ECR separately for ability to drive tissue-specific expression of a reporter during development. In agreement with their previous results, the ECR2-containing transgene drove reporter expression in the extra-embryonic mesoderm and lateral plate mesoderm. Likewise, the transgenes containing ECR1 and ECR3 could not drive reproducible expression in any tissue. These results indicate that ECR2 is a mesoderm-specific enhancer element. Accordingly, ECR2 contains putative bindings sites for several transcription factors that regulate embryonic mesoderm development, including Gata4, Cdx1, Nfe2l1(Tcf11), Zic3 and the Hand1/E47 dimer. Moreover, genetic studies suggest that Bmp4 is important for mesodermal development and function [44]. The conservation of ECR2 in fish further suggests this is an ancient feature of vertebrate development.
6. Relevance to human biology and perspectives
The wide array of cis-regulatory elements near BMP genes suggests that in humans, mutations in specific elements could cause various tissue-specific effects. Dathe et al. recently discovered mutations in the BMP2 3’ flanking region that cause brachydactyly type A2 [45]. These were microduplications of a 5.5 kb segment 110 kb downstream of BMP2. The authors showed that this segment contains highly conserved sequences (across mammals, chick and frog) and can function as an enhancer in transgenic mouse assays, recapitulating Bmp2 expression in the interdigital regions of developing limb buds. Altered BMP2 expression may interfere with GDF5-mediated signaling through the BMPRIB receptor, which is critical for phalangeal development [45].
Recently, a meta-analysis of genome-wide polymorphisms was performed to identify variants influencing human height [46]. Among the 30 or so most consistently associated variants was a single nucleotide polymorphism (rs967417) located 128 kb 5’ to BMP2, although the effect of this SNP on BMP2 expression is unknown. In another genome-wide association study, Houlston et al. identified SNPS at −345 and −361 kb upstream of BMP2 that define one of four loci associated with colorectal cancer susceptibility [47]. One of the other three loci was defined by a SNP only 12 kb 3’ to BMP4. As BMP signaling regulates the renewal of intestinal stem cells, variation in BMP2/4 cis-regulatory elements may affect this process by subtly altering BMP expression.
What is a gene? For the purposes of annotation, it is generally not easy to incorporate non-coding cis-regulatory sequences as part of an individual gene’s structure for a variety of reasons. However, the BMPs are examples of genes for which there is probably more noncoding than coding sequence devoted to their overall, myriad function(s). New genomic technologies, such as high-resolution chromatin immunoprecipitation and chromosome conformation assays, will undoubtedly be useful in helping define potential functions of distant regulatory elements for the BMP gene family.
Biographies

Steven Pregizer is a postdoctoral research fellow in the lab of Doug Mortlock at Vanderbilt University. He earned his PhD in molecular biology from the University of Southern California. His research interests include epigenomics, transcriptional regulation, and skeletal development.

Douglas Mortlock, Ph.D. is an Assistant Professor in the Center for Human Genetics Research and the Department of Molecular Physiology and Biophysics at Vanderbilt University School of Medicine, in Nashville, Tennessee. He completed his bachelor’s degree at Cornell and obtained his Ph.D. at the University of Michigan, where he studied mutations in the Hoxa13 gene that affect limb and urogenital patterning in mice and humans. From 1998–2002 he obtained postdoctoral training at Stanford the lab of David Kingsley where he initiated cis-regulatory studies of the Gdf6 gene. Since 2002 he has worked at Vanderbilt where his lab continues studies on Gdf6, Bmp2 and Bmp4 cis-regulation and function relating to skeletal development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kingsley DM, Bland AE, Grubber JM, et al. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 2.DiLeone RJ, Marcus GA, Johnson MD, Kingsley DM. Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci U S A. 2000;97:1612–1617. doi: 10.1073/pnas.97.4.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiLeone RJ, Russell LB, Kingsley DM. An extensive 3' regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King JA, Marker PC, Seung KJ, Kingsley DM. BMP5 and the molecular, skeletal, and soft-tissue alterations in short ear mice. Dev Biol. 1994;166:112–122. doi: 10.1006/dbio.1994.1300. [DOI] [PubMed] [Google Scholar]
- 5.King JA, Storm EE, Marker PC, Dileone RJ, Kingsley DM. The role of BMPs and GDFs in development of region-specific skeletal structures. Ann N Y Acad Sci. 1996;785:70–79. doi: 10.1111/j.1749-6632.1996.tb56245.x. [DOI] [PubMed] [Google Scholar]
- 6.Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM. An extensive 3' cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development. 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- 7.Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4:e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 9.Chang SC, Hoang B, Thomas JT, et al. Cartilage-derived morphogenetic proteins. New members of the transforming growth factor-beta superfamily predominantly expressed in long bones during human embryonic development. J Biol Chem. 1994;269:28227–28234. [PubMed] [Google Scholar]
- 10.Polinkovsky A, Robin NH, Thomas JT, et al. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat Genet. 1997;17:18–19. doi: 10.1038/ng0997-18. [DOI] [PubMed] [Google Scholar]
- 11.Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JT, Kilpatrick MW, Lin K, et al. Disruption of human limb morphogenesis by a dominant negative mutation in CDMP1. Nat Genet. 1997;17:58–64. doi: 10.1038/ng0997-58. [DOI] [PubMed] [Google Scholar]
- 13.Thomas JT, Lin K, Nandedkar M, Camargo M, Cervenka J, Luyten FP. A human chondrodysplasia due to a mutation in a TGF-beta superfamily member. Nat Genet. 1996;12:315–317. doi: 10.1038/ng0396-315. [DOI] [PubMed] [Google Scholar]
- 14.Settle SH, Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 15.Storm EE, Kingsley DM. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- 16.Hotten GC, Matsumoto T, Kimura M, et al. Recombinant human growth/differentiation factor 5 stimulates mesenchyme aggregation and chondrogenesis responsible for the skeletal development of limbs. Growth Factors. 1996;13:65–74. doi: 10.3109/08977199609034567. [DOI] [PubMed] [Google Scholar]
- 17.Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- 18.Tsumaki N, Tanaka K, Arikawa-Hirasawa E, et al. Role of CDMP-1 in skeletal morphogenesis: promotion of mesenchymal cell recruitment and chondrocyte differentiation. J Cell Biol. 1999;144:161–173. doi: 10.1083/jcb.144.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortlock DP, Guenther C, Kingsley DM. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 2003;13:2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portnoy ME, McDermott KJ, Antonellis A, et al. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics. 2005;86:295–305. doi: 10.1016/j.ygeno.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortlock DP, Portnoy ME, Chandler RL, Green ED. Comparative sequence analysis of the Gdf6 locus reveals a duplicon-mediated chromosomal rearrangement in rodents and rapidly diverging coding and regulatory sequences. Genomics. 2004;84:814–823. doi: 10.1016/j.ygeno.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tassabehji M, Fang ZM, Hilton EN, et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum Mutat. 2008;29:1017–1027. doi: 10.1002/humu.20741. [DOI] [PubMed] [Google Scholar]
- 25.Asai-Coakwell M, French CR, Ye M, et al. Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Hum Mol Genet. 2009;18:1110–1121. doi: 10.1093/hmg/ddp008. [DOI] [PubMed] [Google Scholar]
- 26.Wozney JM, Rosen V, Celeste AJ, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, Feng JQ, Feng M, Harris MA, Mundy GR, Harris SE. Cloning and sequence of bone morphogenetic protein 4 cDNA from fetal rat calvarial cell. Biochim Biophys Acta. 1993;1174:289–292. doi: 10.1016/0167-4781(93)90200-w. [DOI] [PubMed] [Google Scholar]
- 29.Feng JQ, Chen D, Esparza J, Harris MA, Mundy GR, Harris SE. Deer antler tissue contains two types of bone morphogenetic protein 4 mRNA transcripts. Biochim Biophys Acta. 1995;1263:163–168. doi: 10.1016/0167-4781(95)00106-q. [DOI] [PubMed] [Google Scholar]
- 20.Feng JQ, Harris MA, Ghosh-Choudhury N, Feng M, Mundy GR, Harris SE. Structure and sequence of mouse bone morphogenetic protein-2 gene (BMP-2): comparison of the structures and promoter regions of BMP-2 and BMP-4 genes. Biochim Biophys Acta. 1994;1218:221–224. doi: 10.1016/0167-4781(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh-Choudhury N, Choudhury GG, Harris MA, et al. Autoregulation of mouse BMP-2 gene transcription is directed by the proximal promoter element. Biochem Biophys Res Commun. 2001;286:101–108. doi: 10.1006/bbrc.2001.5351. [DOI] [PubMed] [Google Scholar]
- 32.Kurihara T, Kitamura K, Takaoka K, Nakazato H. Murine bone morphogenetic protein-4 gene: existence of multiple promoters and exons for the 5'-untranslated region. Biochem Biophys Res Commun. 1993;192:1049–1056. doi: 10.1006/bbrc.1993.1523. [DOI] [PubMed] [Google Scholar]
- 33.Feng JQ, Chen D, Cooney AJ, et al. The mouse bone morphogenetic protein-4 gene. Analysis of promoter utilization in fetal rat calvarial osteoblasts and regulation by COUP-TFI orphan receptor. J Biol Chem. 1995;270:28364–28373. doi: 10.1074/jbc.270.47.28364. [DOI] [PubMed] [Google Scholar]
- 34.Feng JQ, Chen D, Ghosh-Choudhury N, Esparza J, Mundy GR, Harris SE. Bone morphogenetic protein 2 transcripts in rapidly developing deer antler tissue contain an extended 5' non-coding region arising from a distal promoter. Biochim Biophys Acta. 1997;1350:47–52. doi: 10.1016/s0167-4781(96)00178-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhao M, Qiao M, Harris SE, Chen D, Oyajobi BO, Mundy GR. The zinc finger transcription factor Gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol Cell Biol. 2006;26:6197–6208. doi: 10.1128/MCB.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng JQ, Xing L, Zhang JH, et al. NF-kappaB specifically activates BMP-2 gene expression in growth plate chondrocytes in vivo and in a chondrocyte cell line in vitro. J Biol Chem. 2003;278:29130–29135. doi: 10.1074/jbc.M212296200. [DOI] [PubMed] [Google Scholar]
- 37.Feng JQ, Zhang J, Tan X, Lu Y, Guo D, Harris SE. Identification of cis-DNA regions controlling Bmp4 expression during tooth morphogenesis in vivo. J Dent Res. 2002;81:6–10. doi: 10.1177/002203450208100103. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Tan X, Contag CH, et al. Dissection of promoter control modules that direct Bmp4 expression in the epithelium-derived components of hair follicles. Biochem Biophys Res Commun. 2002;293:1412–1419. doi: 10.1016/S0006-291X(02)00416-3. [DOI] [PubMed] [Google Scholar]
- 39.Garrett IR, Chen D, Gutierrez G, et al. Selective inhibitors of the osteoblast proteasome stimulate bone formation in vivo and in vitro. J Clin Invest. 2003;111:1771–1782. doi: 10.1172/JCI16198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mundy G, Garrett R, Harris S, et al. Stimulation of bone formation in vitro and in rodents by statins. Science. 1999;286:1946–1949. doi: 10.1126/science.286.5446.1946. [DOI] [PubMed] [Google Scholar]
- 41.Zhao M, Ko SY, Liu JH, et al. Inhibition of microtubule assembly in osteoblasts stimulates bone morphogenetic protein 2 expression and bone formation through transcription factor Gli2. Mol Cell Biol. 2009;29:1291–1305. doi: 10.1128/MCB.01566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007;27:2934–2951. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandler KJ, Chandler RL, Mortlock DP. Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev Biol. 2009;327:590–602. doi: 10.1016/j.ydbio.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 45.Dathe K, Kjaer KW, Brehm A, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am J Hum Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 47.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]